Natural Bovine Coronavirus Infection in a Calf Persistently Infected with Bovine Viral Diarrhea Virus: Viral Shedding, Immunological Features and S Gene Variations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Case
2.2. Clinical Score and Treatment Procedures
2.3. RT-qPCR for Virological and Bacteriological Analysis
2.4. Virus Detection
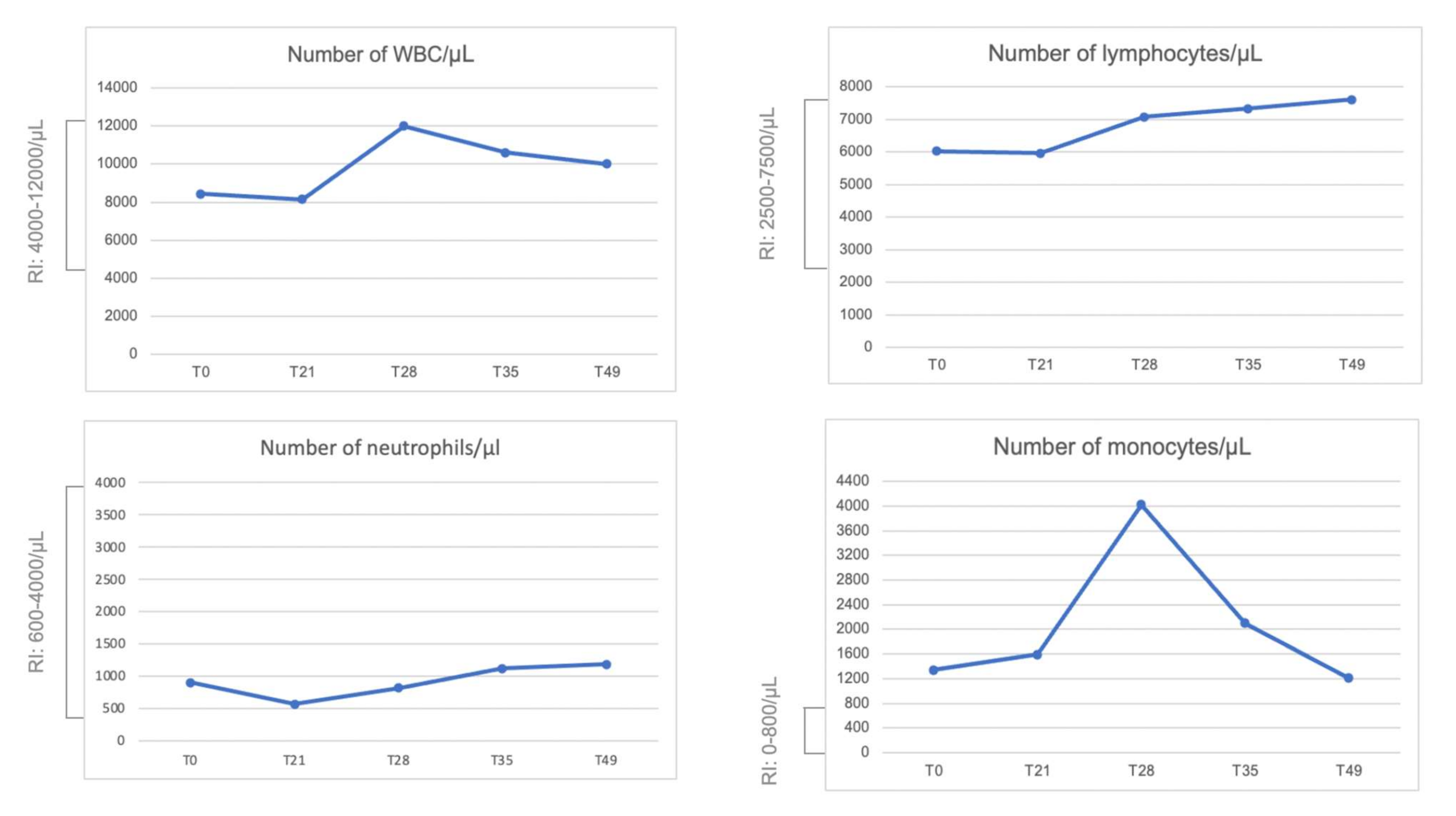
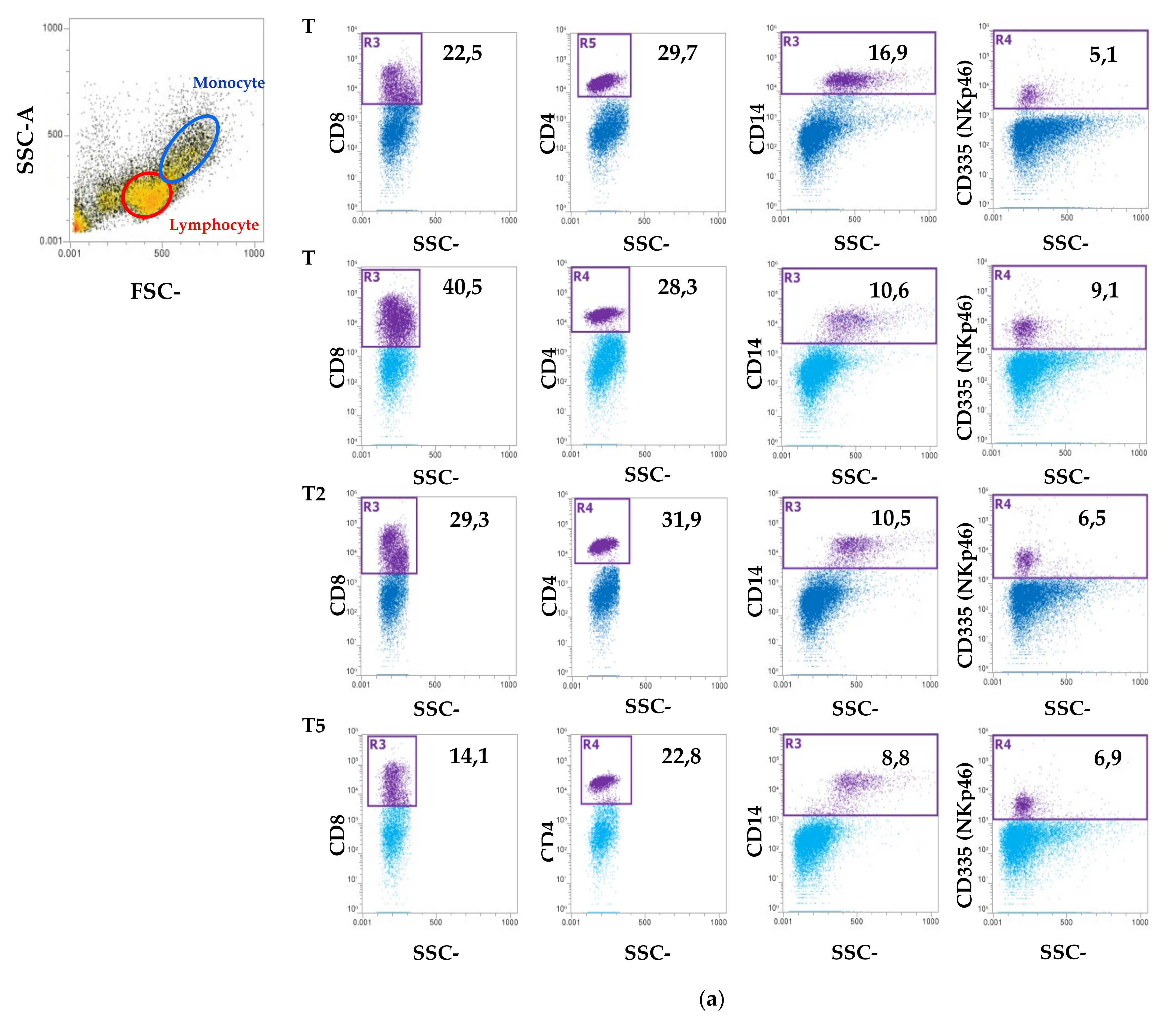
2.5. Blood Count and Peripheral Blood Mononuclear Cell (PBMC) Isolation Characterization Using Flow Cytometry
2.6. Serological Tests
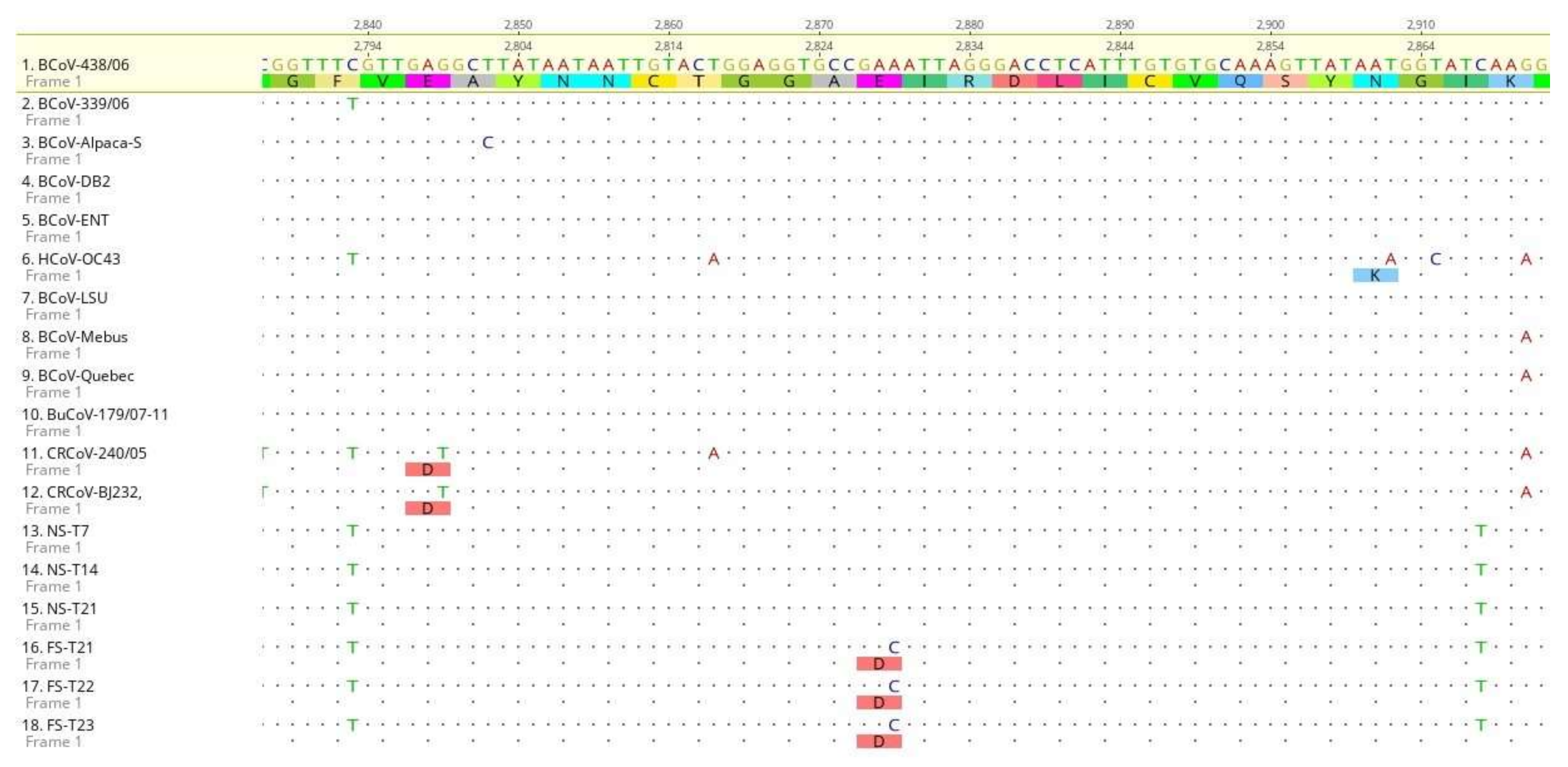
2.7. Sequence Analysis of BCoV S Protein
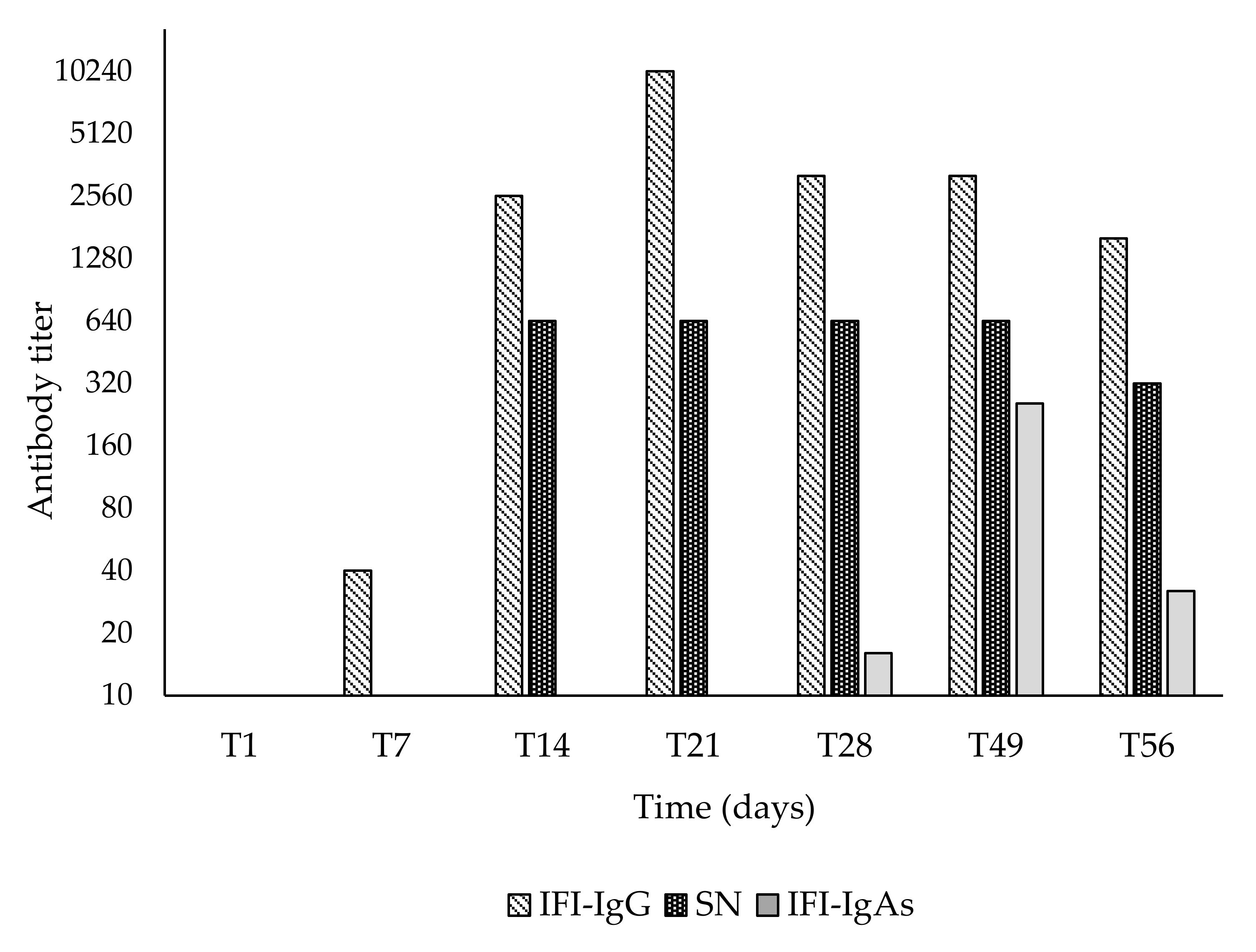
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pratelli, A. The evolutionary processes of canine coronaviruses. Adv. Virol. 2011, 2001, 562831. [Google Scholar] [CrossRef] [Green Version]
- Pratelli, A.; Martella, V.; Decaro, N.; Tinelli, A.; Camero, M.; Cirone, F.; Elia, G.; Cavalli, A.; Corrente, M.; Greco, G.; et al. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods 2003, 110, 9–17. [Google Scholar] [CrossRef]
- Terada, Y.; Matsui, N.; Noguchi, K.; Kuwata, R.; Shimoda, H.; Soma, T.; Mochizuki, M.; Maeda, K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE 2014, 9, e106534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimkin, V.; Beer, M.; Blome, S.; Hanke, D.; Hoper, D.; Jenckel, M.; Pohlmann, A. New chimeric porcine coronavirus in Swine Feces, Germany, 2012. Emerg. Infect. Dis. 2016, 22, 1314–1315. [Google Scholar] [CrossRef] [Green Version]
- Boniotti, M.B.; Papetti, A.; Lavazza, A.; Alborali, G.; Sozzi, E.; Chiapponi, C.; Faccini, S.; Bonilauri, P.; Cordioli, P.; Marthaler, D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg. Infect. Dis. 2016, 22, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Mandelik, R.; Sarvas, M.; Jackova, A.; Salamunova, S.; Novotny, J.; Vilcek, S. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia—lessons to learn. Acta Vet. Hung. 2018, 66, 488–492. [Google Scholar] [CrossRef]
- Cirone, F.; Padalino, B.; Tullio, D.; Capozza, P.; Lo Surdo, M.; Lanave, G.; Pratelli, A. Prevalence of Pathogens Related to Bovine Respiratory Disease Before and After Transportation in Beef Steers: Preliminary Results. Animals 2019, 9, 1093. [Google Scholar] [CrossRef] [Green Version]
- Pratelli, A.; Cirone, F.; Capozza, P.; Trotta, A.; Corrente, M.; Balestrieri, A.; Buonavoglia, C. Bovine respiratory disease in beef calves supported long transport stress: An epidemiological study and strategies for control and prevention. Res. Vet. Sci. 2021, 135, 450–455. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Saif, L.J. Bovine Coronavirus and the Associated Diseases. Front. Vet. Sci. 2021, 31, 643220. [Google Scholar] [CrossRef]
- Padalino, B.; Cirone, F.; Zappaterra, M.; Tullio, D.; Ficco, G.; Giustino, A.; Ndiana, L.A.; Pratelli, A. Factors affecting the development of Bovine Respiratory Disease: A cross-sectional study in beef steers shipped from France to Italy. Front. Vet. Sci. 2021, in press. [Google Scholar] [CrossRef]
- Fulton, R.W.; Herd, H.R.; Sorensen, N.J.; Confer, A.W.; Ritchey, J.W.; Ridpath, J.F.; Burge, L.J. Enteric disease in postweaned beef calves associated with Bovine coronavirus clade 2. J. Vet. Diagn. Investig. 2015, 27, 97–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domingo, E.; Martinez-Salas, E.; Sobrino, F.; de la Torre, J.C.; Portela, A.; Ortín, J.; López-Galindez, C.; Pérez-Brena, P.; Villanueva, N.; Nájera, R.; et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: Biological relevance—A review. Gene 1985, 40, 1–8. [Google Scholar] [CrossRef]
- Tsunemitsu, H.; Smith, D.R.; Saif, L.J. Experimental inoculation of adult dairy cows with bovine coronavirus and detection of coronavirus in feces by RT-PCR. Arch. Virol. 1999, 144, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Oma, V.S.; Tråvén, M.; Alenius, S.; Myrmel, M.; Stokstad, M. Bovine coronavirus in naturally and experimentally exposed calves; viral shedding and the potential for transmission. Virol. J. 2016, 13, 100. [Google Scholar] [CrossRef] [Green Version]
- Kanno, T.; Ishihara, R.; Hatama, S.; Uchida, I. A long-term animal experiment indicating persistent infection of bovine coronavirus in cattle. J. Vet. Med. Sci. 2018, 80, 1134–1137. [Google Scholar] [CrossRef] [Green Version]
- Workman, A.M.; Kuehn, L.A.; McDaneld, T.G.; Clawson, M.L.; Loy, J.D. Longitudinal study of humoral immunity to bovine coronavirus, virus shedding, and treatment for bovine respiratory disease in pre-weaned beef calves. BMC Vet. Res. 2019, 15, 161. [Google Scholar] [CrossRef] [PubMed]
- Peterhans, E.; Jungi, T.W.; Schweizer, M. BVDV and innate immunity. Biologicals 2003, 31, 107–111. [Google Scholar] [CrossRef]
- Brewoo, J.N.; Haase, C.J.; Sharp, P.; Schultz, R.D. Leukocyte profile of cattle persistently infected with bovine viral diarrhea virus. Vet. Immunol. Immunopathol. 2007, 115, 369–374. [Google Scholar] [CrossRef]
- Piccinini, R.; Luzzago, C.; Frigerio, M.; Frigerio, V.; Liandris, E.; Zecconi, A. Comparison of blood non-specific immune parameters in bovine virus diarrhoea virus (BVDV) persistently infected and in immune heifers. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Chase, C.C.L. The impact of BVDV infection on adaptive immunity. Biologicals 2013, 41, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, L.; Guida, M.; De Summa, S.; Di Fonte, R.; De Risi, I.; Garofali, M.; Caputo, M.; Negri, A.; Strippoli, S.; Serrati, S.; et al. uPAR+ extracellular vesicles: A robust biomarker of resistance to checkpoint inhibitor immunotherapy in metastatic melanoma patients. J. Immunother. Cancer 2021, 9, e002372. [Google Scholar] [CrossRef] [PubMed]
- Saif, L.J. Bovine Respiratory Coronavirus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Hodnik, J.J.; Ježek, J.; Starič, J. Coronaviruses in cattle. Trop. Anim. Health Prod. 2020, 52, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.-O.; Hasoksuz, M.; Nielsen, P.R.; Chang, K.-O.; Lathrop, S.; Saif, L.J. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch. Virol. 2001, 146, 2401–2419. [Google Scholar] [CrossRef] [PubMed]
- Traven, M.; Naslund, K.; Linde, N.; Linde, B.; Silvan, A.; Fossum, C.; Hedlund, K.O.; Larsson, B. Experimental reproduction of winter dysentery in lactating cows using BCV—comparison with BCV infection in milk-fed calves. Vet. Microbiol. 2001, 81, 127–151. [Google Scholar] [CrossRef]
- Thomas, C.J.; Hoet, A.E.; Sreevatsan, S.; Wittum, T.E.; Briggs, R.E.; Duff, G.C.; Saif, L.J. Transmission of bovine coronavirus and serologic responses in feedlot calves under field conditions. Am. J. Vet. Res. 2006, 67, 1412–1420. [Google Scholar] [CrossRef]
- Saif, L.J.; Redman, D.R.; Moorhead, P.D.; Theil, K.W. Experimentally induced coronavirus infections in calves: Viral replication in the respiratory and intestinal tracts. Am. J. Vet. Res. 1986, 47, 1426–1432. [Google Scholar]
- Traven, M.; Alenius, S.; Fossum, C.; Larsson, B. Primary bovine viral diarrhoea virus infection in calves following direct contact with a persistently viraemic calf. J. Vet. Med. B Infect. Dis. Vet. Public Health 1991, 38, 453–462. [Google Scholar] [CrossRef]
- Bidokhti, M.R.M.; Tråvén, M.; Krishna, N.K.; Munir, M.; Belák, S.; Alenius, S.; Cortey, M. Evolutionary dynamics of bovine coronaviruses: Natural selection pattern of the spike gene implies adaptive evolution of the strains. J. Gen. Virol. 2013, 94, 2036–2049. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Kim, G.Y.; Choy, H.E.; Hong, Y.J.; Saif, L.J.; Jeong, J.H.; Park, S.I.; Kim, H.H.; Kim, S.K.; Shin, S.S.; et al. Dual enteric and respiratory tropisms of winter dysentery bovine coronavirus in calves. Arch. Virol. 2007, 152, 1885–1900. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Score * | Body Temperature ° | Sensory | Diarrhea | Nasal Discharge | Cough |
|---|---|---|---|---|---|
| 0 | <38.5 | Bright, alert | Absent | Absent | Absent |
| 1 | 38.6–39.5 | Mildly depressed | Pasty | Serous/mucous | Sporadic |
| 2 | 39.6–40.0 | Moderately depressed | Runny | Mucopurulent | More than one cough every 10 min |
| 3 | >40.1 | Severely depressed | Watery | Purulent | Severe |
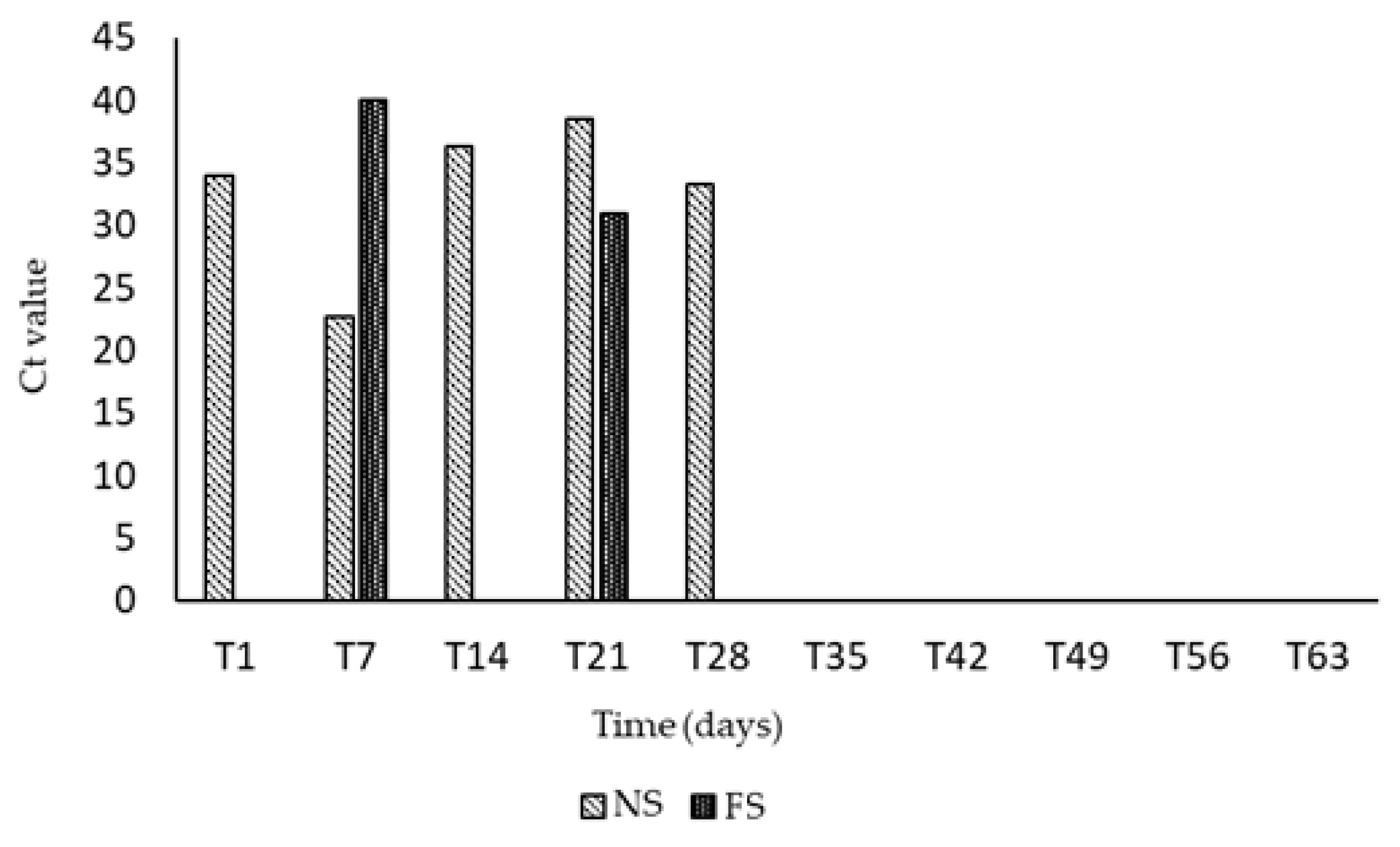
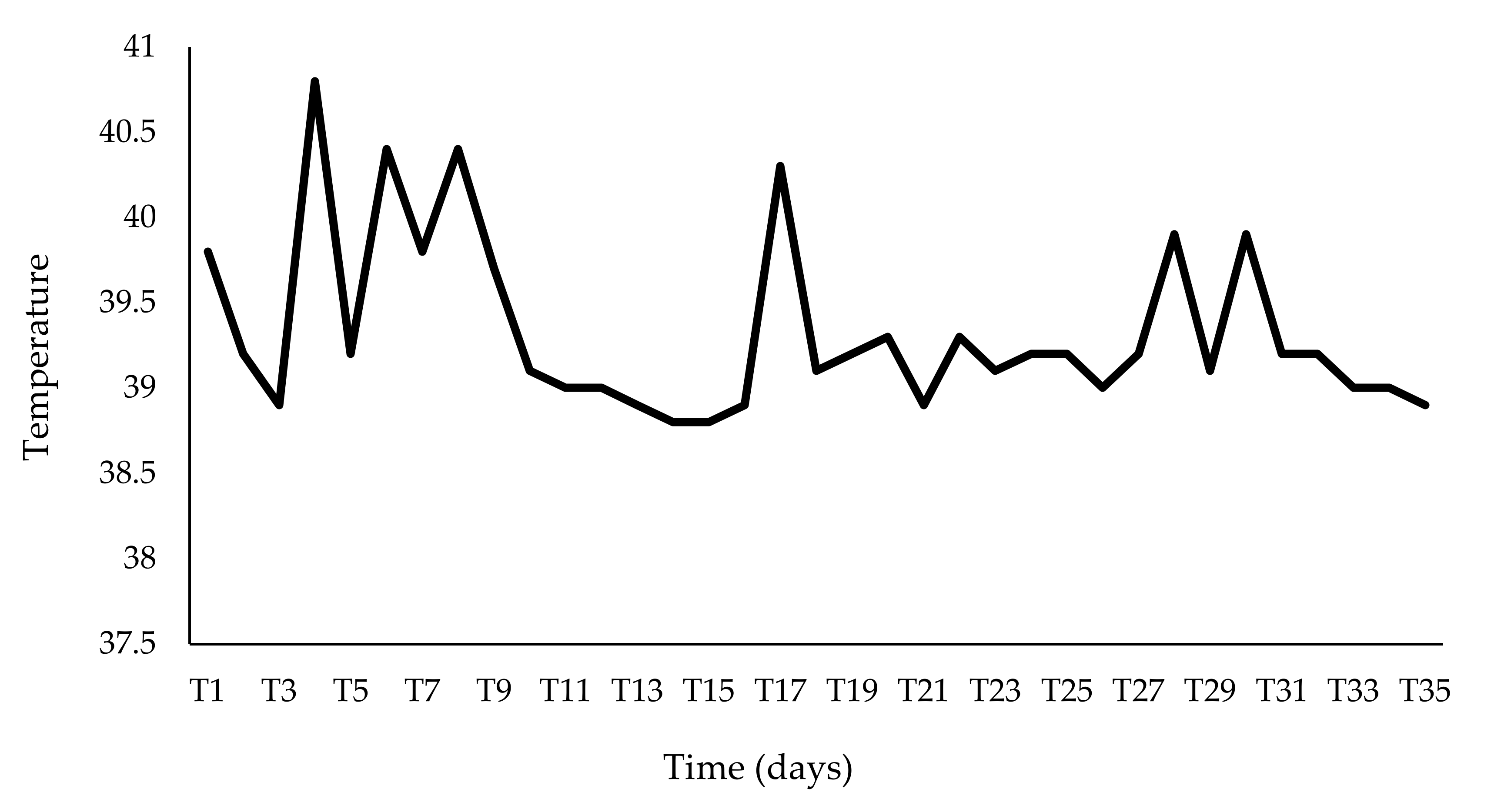
| dpi | Fever | Depression | Anorexia | Nasal Discharge | Cough | Diarrhea | Score ° | Ct |
|---|---|---|---|---|---|---|---|---|
| T1 | 2 | 1 | 0 | 0 | 1 | 0 | 4 | 34.0 |
| T7 | 2 | 2 | 1 | 0 | 1 | 0 | 6 | 22.69 |
| T14 | 1 | 1 | 1 | 0 | 0 | 0 | 3 | 36.28 |
| T21 | 0 | 1 | 0 | 0 | 1 | 1 | 3 | 38.48 |
| T28 | 2 | 2 | 0 | 0 | 1 | 0 | 5 | 33.35 |
| T35 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | neg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pratelli, A.; Lucente, M.S.; Cordisco, M.; Ciccarelli, S.; Di Fonte, R.; Sposato, A.; Mari, V.; Capozza, P.; Pellegrini, F.; Carelli, G.; et al. Natural Bovine Coronavirus Infection in a Calf Persistently Infected with Bovine Viral Diarrhea Virus: Viral Shedding, Immunological Features and S Gene Variations. Animals 2021, 11, 3350. https://doi.org/10.3390/ani11123350
Pratelli A, Lucente MS, Cordisco M, Ciccarelli S, Di Fonte R, Sposato A, Mari V, Capozza P, Pellegrini F, Carelli G, et al. Natural Bovine Coronavirus Infection in a Calf Persistently Infected with Bovine Viral Diarrhea Virus: Viral Shedding, Immunological Features and S Gene Variations. Animals. 2021; 11(12):3350. https://doi.org/10.3390/ani11123350
Chicago/Turabian StylePratelli, Annamaria, Maria Stella Lucente, Marco Cordisco, Stefano Ciccarelli, Roberta Di Fonte, Alessio Sposato, Viviana Mari, Paolo Capozza, Francesco Pellegrini, Grazia Carelli, and et al. 2021. "Natural Bovine Coronavirus Infection in a Calf Persistently Infected with Bovine Viral Diarrhea Virus: Viral Shedding, Immunological Features and S Gene Variations" Animals 11, no. 12: 3350. https://doi.org/10.3390/ani11123350
APA StylePratelli, A., Lucente, M. S., Cordisco, M., Ciccarelli, S., Di Fonte, R., Sposato, A., Mari, V., Capozza, P., Pellegrini, F., Carelli, G., Azzariti, A., & Buonavoglia, C. (2021). Natural Bovine Coronavirus Infection in a Calf Persistently Infected with Bovine Viral Diarrhea Virus: Viral Shedding, Immunological Features and S Gene Variations. Animals, 11(12), 3350. https://doi.org/10.3390/ani11123350








