A Comparison of Reproductive Performances in Young and Old Females: A Case Study on the Atlantic Bluefin Tuna in the Mediterranean Sea
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Experimental Design and Sampling Activities
2.2. H&E Histological Processing and Image Analysis
2.3. RNA Extraction and cDNA Synthesis
2.4. miRNA Identification
2.5. Reproductive Markers
2.6. Quantitative RT-PCR
2.7. Fluorescent In Situ Hybridization (FISH)
2.8. Statistical Analysis
3. Results
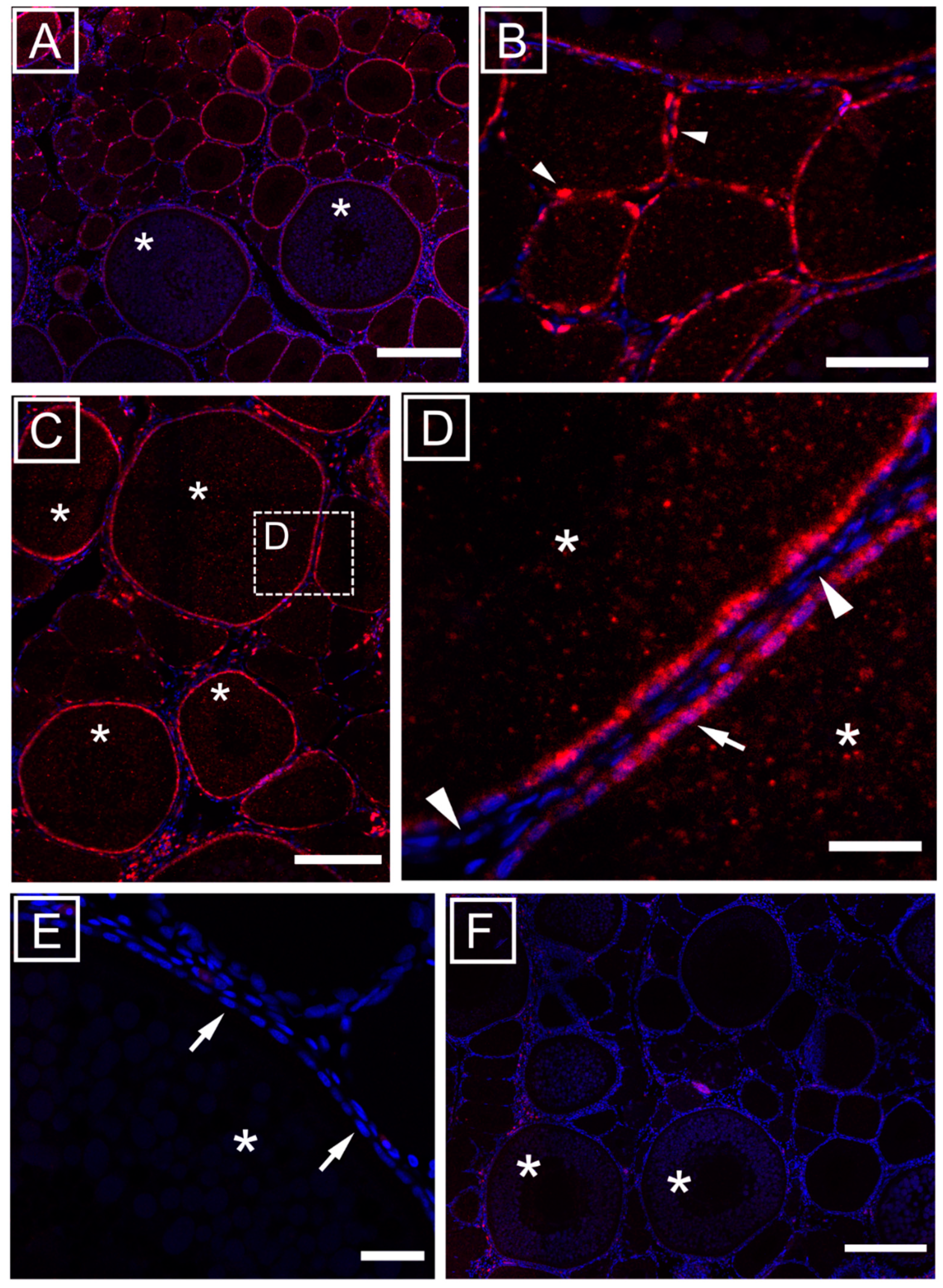
3.1. Ovarian Localization of mir-202-5p
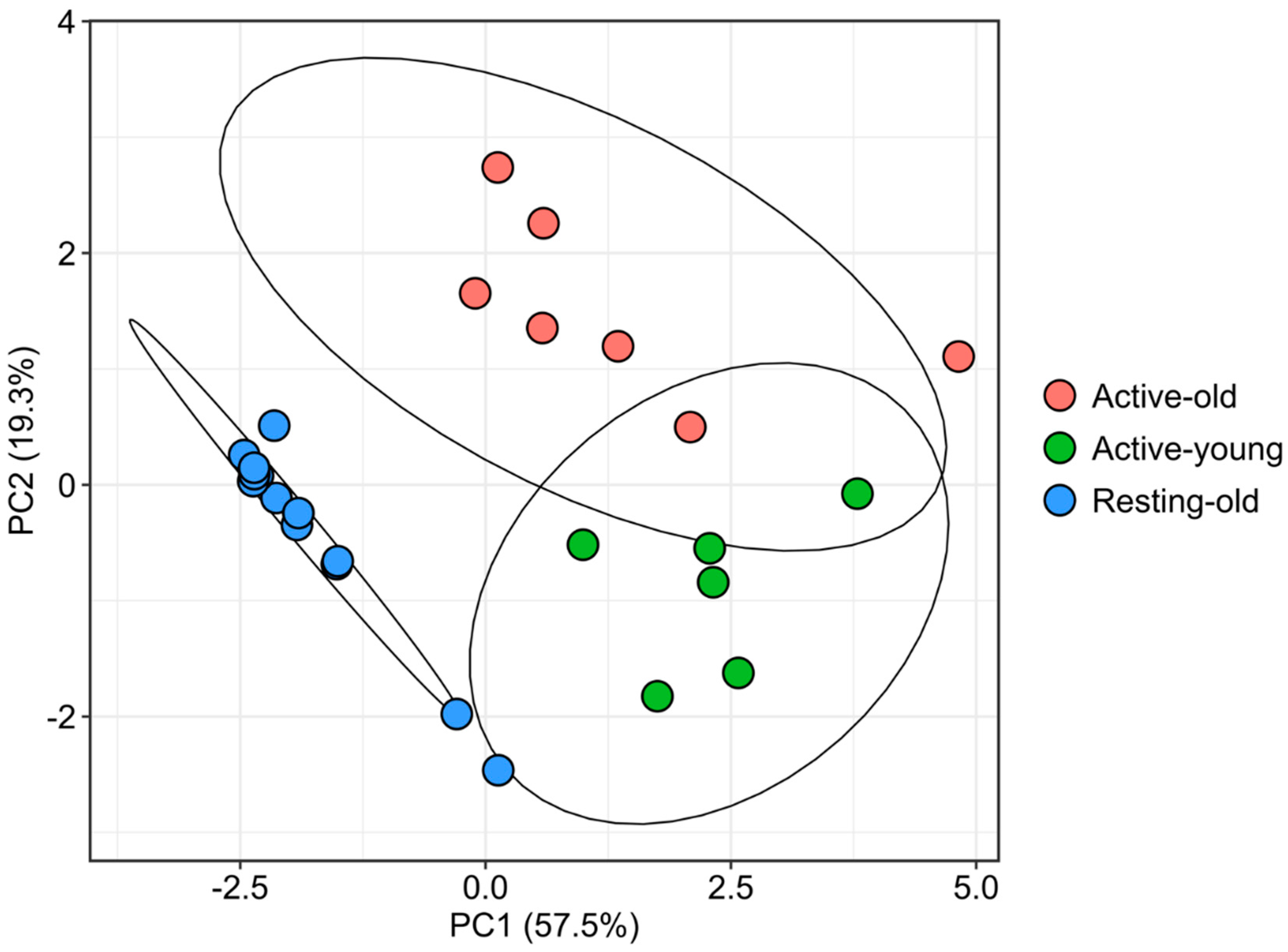
3.2. Multivariate Classification Reveals Two Subgroups during the Reproductive Period
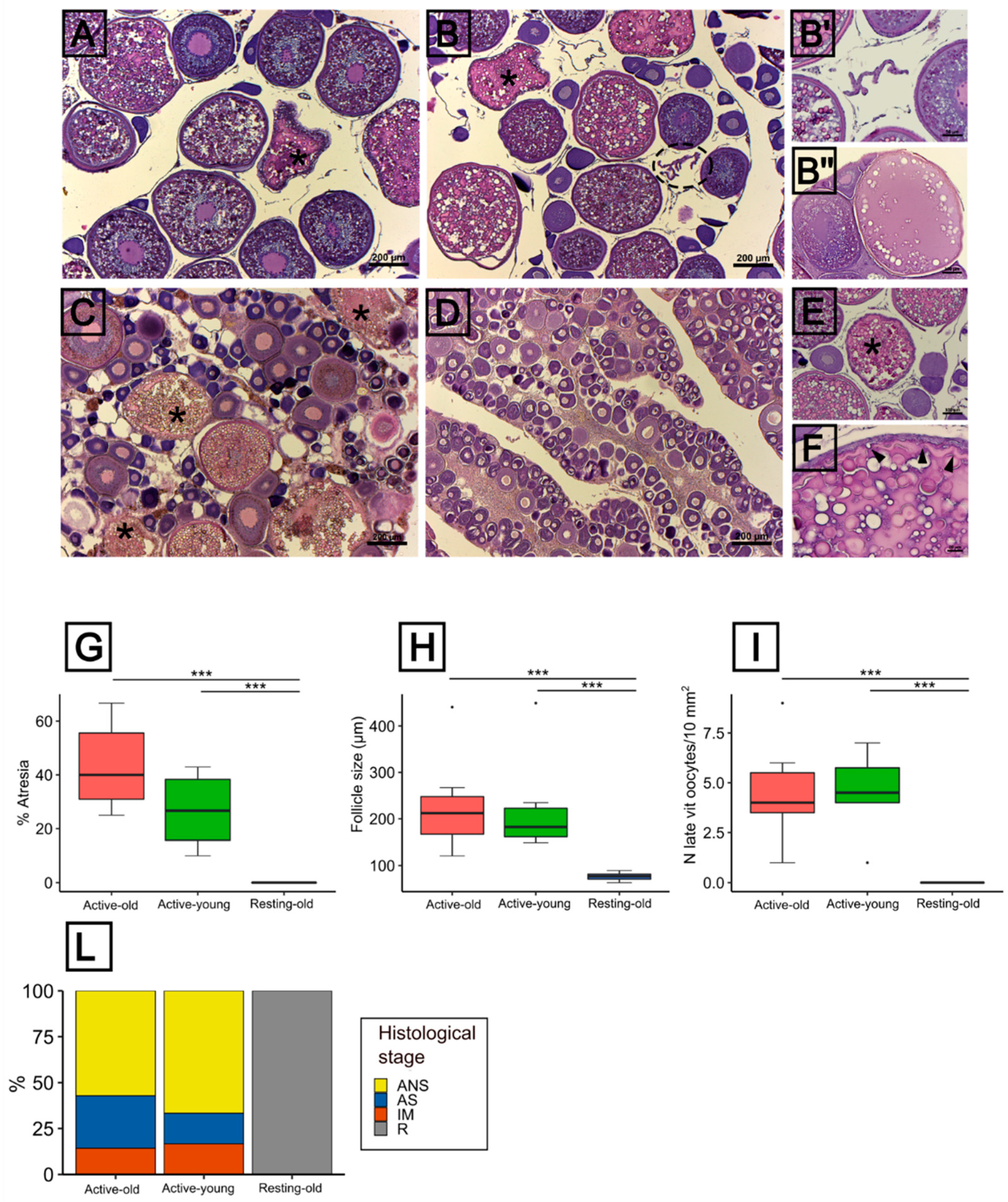
3.3. Histological Features of the Ovarian Condition
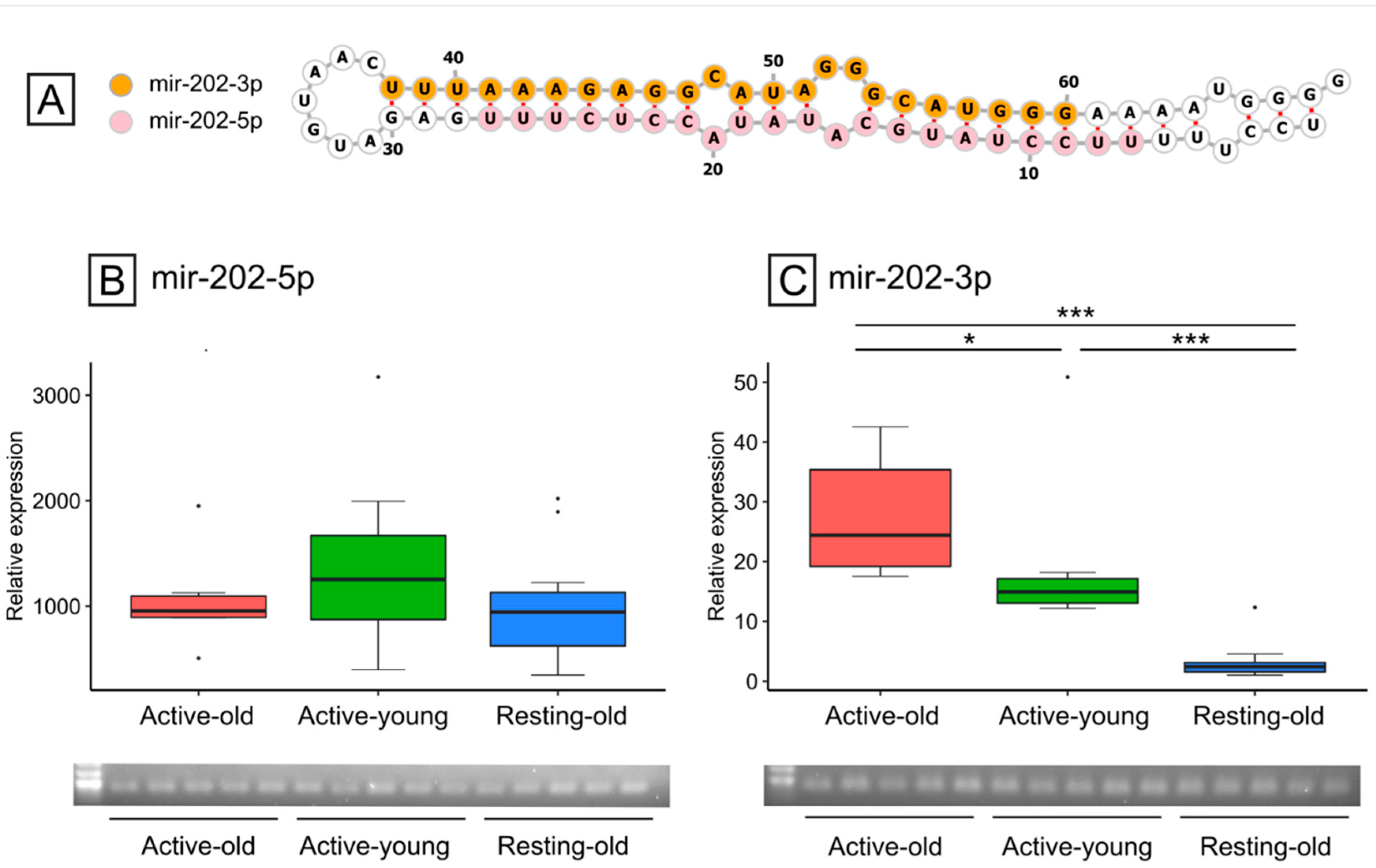
3.4. mir-202 Identification and Expression during Reproductive and Non-Reproductive Periods
3.5. Gene Expression Analysis of Reproductive Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jachowski, D.S.; Singh, N.J. Toward a mechanistic understanding of animal migration: Incorporating physiological measurements in the study of animal movement. Conserv. Physiol. 2015, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pankhurst, N.W. The endocrinology of stress in fish: An environmental perspective. Gen. Comp. Endocrinol. 2011, 170, 265–275. [Google Scholar] [CrossRef]
- Oliveira, R.F.; Hirschenhauser, K.; Carneiro, L.A.; Canario, A.V.M. Social modulation of androgen levels in male teleost fish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2002, 132, 203–215. [Google Scholar] [CrossRef]
- Lubzens, E.; Bobe, J.; Young, G.; Sullivan, C. V Maternal investment in fish oocytes and eggs: The molecular cargo and its contributions to fertility and early development. Aquaculture 2017, 472, 107–143. [Google Scholar] [CrossRef]
- Lubzens, E.; Young, G.; Bobe, J.; Cerdà, J. Oogenesis in teleosts: How fish eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef]
- Charlier, C.; Montfort, J.; Chabrol, O.; Brisard, D.; Nguyen, T.; Le Cam, A.; Richard-Parpaillon, L.; Moreews, F.; Pontarotti, P.; Uzbekova, S.; et al. Oocyte-somatic cells interactions, lessons from evolution. BMC Genomics 2012, 13, 560. [Google Scholar] [CrossRef] [PubMed]
- Houwing, S.; Kamminga, L.M.; Berezikov, E.; Cronembold, D.; Girard, A.; van den Elst, H.; Filippov, D.V.; Blaser, H.; Raz, E.; Moens, C.B.; et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell 2007, 129, 69–82. [Google Scholar] [CrossRef]
- Armisen, J.; Gilchrist, M.J.; Wilczynska, A.; Standart, N.; Miska, E.A. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009, 19, 1766–1775. [Google Scholar] [CrossRef][Green Version]
- Rauwerda, H.; Wackers, P.; Pagano, J.F.B.; De Jong, M.; Ensink, W.; Dekker, R.; Nehrdich, U.; Spaink, H.P.; Jonker, M.; Breit, T.M. Mother-specific signature in the maternal transcriptome composition of mature, unfertilized zebrafish eggs. PLoS ONE 2016, 11, e0147151. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef]
- Flynt, A.S.; Lai, E.C. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat. Rev. Genet. 2008, 9, 831–842. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Jin, Y.; Jia, P.; Jia, K.; Yi, M. MiR-202-5p is a novel germ plasm-specific microRNA in zebrafish. Sci. Rep. 2017, 7, 7055. [Google Scholar] [CrossRef]
- Juanchich, A.; Le Cam, A.; Montfort, J.; Guiguen, Y.; Bobe, J. Identification of Differentially Expressed miRNAs and Their Potential Targets During Fish Ovarian Development1. Biol. Reprod. 2013, 88. [Google Scholar] [CrossRef] [PubMed]
- Bouchareb, A.; Le Cam, A.; Montfort, J.; Gay, S.; Nguyen, T.; Bobe, J.; Thermes, V. Genome-wide identification of novel ovarian-predominant miRNAs: New insights from the medaka (Oryzias latipes). Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Farlora, R.; Valenzuela-Miranda, D.; Alarcón-Matus, P.; Gallardo-Escárate, C. Identification of microRNAs associated with sexual maturity in rainbow trout brain and testis through small RNA deep sequencing. Mol. Reprod. Dev. 2015, 82, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Ro, S.; Song, R.; Park, C.; Zheng, H.; Sanders, K.M.; Yan, W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA 2007, 13, 2366–2380. [Google Scholar] [CrossRef]
- Gay, S.; Bugeon, J.; Bouchareb, A.; Henry, L.; Delahaye, C.; Legeai, F.; Montfort, J.; Le Cam, A.; Siegel, A.; Bobe, J.; et al. MiR-202 controls female fecundity by regulating medaka oogenesis. PLOS Genet. 2018, 14, e1007593. [Google Scholar] [CrossRef]
- Jin, Y.; Liu, W.; Xiang, Y.; Zhang, W.; Zhang, H.; Jia, K.; Yi, M. Maternal miR-202-5p is required for zebrafish primordial germ cell migration by protecting small GTPase Cdc42. J. Mol. Cell Biol. 2019, 12, 530–542. [Google Scholar] [CrossRef]
- Rooker, J.R.; Alvarado Bremer, J.R.; Block, B.A.; Dewar, H.; de Metrio, G.; Corriero, A.; Kraus, R.T.; Prince, E.D.; Rodríguez-Marín, E.; Secor, D.H. Life History and Stock Structure of Atlantic Bluefin Tuna (Thunnus thynnus). Rev. Fish. Sci. 2007, 15, 265–310. [Google Scholar] [CrossRef]
- Block, B.A.; Dewar, H.; Blackwell, S.B.; Williams, T.D.; Prince, E.D.; Farwell, C.J.; Boustany, A.; Teo, S.L.H.; Seitz, A.; Walli, A.; et al. Migratory Movements, Depth Preferences, and Thermal Biology of Atlantic Bluefin Tuna. Science 2001, 293, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Cort, J.L.; Deguara, S.; Galaz, T.; Mèlich, B.; Artetxe, I.; Arregi, I.; Neilson, J.; Andrushchenko, I.; Hanke, A.; Neves dos Santos, M.; et al. Determination of L max for Atlantic Bluefin Tuna, Thunnus thynnus (L.), from Meta-Analysis of Published and Available Biometric Data. Rev. Fish. Sci. 2013, 21, 181–212. [Google Scholar] [CrossRef]
- Rooker, J.R.; Secor, D.H.; DeMetrio, G.; Kaufman, A.J.; Ríos, A.B.; Tičina, V. Evidence of trans-Atlantic movement and natal homing of bluefin tuna from stable isotopes in otoliths. Mar. Ecol. Prog. Ser. 2008, 368, 231–239. [Google Scholar] [CrossRef]
- Rooker, J.R.; Secor, D.H.; Zdanowicz, V.S.; De Metrio, G.; Relini, L.O. Identification of Atlantic bluefin tuna (Thunnus thynnus) stocks from putative nurseries using otolith chemistry. Fish. Oceanogr. 2003, 12, 75–84. [Google Scholar] [CrossRef]
- Brophy, D.; Rodríguez-Ezpeleta, N.; Fraile, I.; Arrizabalaga, H. Combining genetic markers with stable isotopes in otoliths reveals complexity in the stock structure of Atlantic bluefin tuna (Thunnus thynnus). Sci. Rep. 2020, 10, 14675. [Google Scholar] [CrossRef]
- Cort, J.L.; Abaunza, P. The Bluefin Tuna Fishery in the Bay of Biscay: Its Relationship with the Crisis of Catches of Large Specimens in the East Atlantic Fisheries from the 1960s; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- De Metrio, G.; Arnold, G.P.; Block, B.A.; Megalofonou, P.; Lutcavage, M.; Oray, I.; Deflorio, M. Movements of Bluefin Tuna (Thunnus Thynnus L.) Tagged in the Mediterranean Sea With Pop-Up Satellite Tags. ICCAT Collect. Vol. Sci. Pap. 2005, 58, 1337–1340. [Google Scholar]
- Cermeño, P.; Quílez-Badia, G.; Ospina-Alvarez, A.; Sainz-Trápaga, S.; Boustany, A.M.; Seitz, A.C.; Tudela, S.; Block, B.A. Electronic Tagging of Atlantic Bluefin Tuna (Thunnus thynnus, L.) Reveals Habitat Use and Behaviors in the Mediterranean Sea. PLoS ONE 2015, 10, e0116638. [Google Scholar] [CrossRef]
- Rey, J.C. El paso del atún rojo, Thunnus thynnus (Linnaeus, 1758), a través del Estrecho de Gibraltar y su relación. Esquemas de migración. Boletín Inst. Español Oceanogr. 1983, 1, 85–94. [Google Scholar]
- Rouyer, T.; Brisset, B.; Bonhommeau, S.; Fromentin, J.M. Update of the abundance index for juvenile fish derived from aerial surveys of bluefin tuna in the western Mediterranean sea. Collect. Vol. Sci. Pap. ICCAT 2018, 74, 2887–2902. [Google Scholar]
- Piccinetti, C. The bluefin seine fishery in the Adriatic. Collect. Vol. Sci. Pap. ICCAT 1980, 11, 124–128. [Google Scholar]
- Orsi Relini, L.; Cima, C.; Palandri, G. Preliminary observations on bluefin tuna catches in the Ligurian Sea. Catches in 1992. Size/Age for young. Length/Weight relationship. Collect. Vol. Sci. Pap. ICCAT 1995, 4, 355–357. [Google Scholar]
- Rooker, J.; Arrizabalaga, H.; Fraile, I.; Secor, D.; Dettman, D.; Abid, N.; Addis, P.; Deguara, S.; Karakulak, F.; Kimoto, A.; et al. Crossing the line: Migratory and homing behaviors of Atlantic bluefin tuna. Mar. Ecol. Prog. Ser. 2014, 504, 265–276. [Google Scholar] [CrossRef]
- Furnestin, J.; Dardignac, J. Le thon rouge du Maroc Atlantique (Thunnus thynnus Linné). Rev. Trav. l’Institut Pêches Marit. 1962, 26, 382–398. [Google Scholar]
- Aloncle, H. Marquage de thons dans le Golfe de Gascogne. Collect. Vol. Sci. Pap. ICCAT 1973, 1, 445–458. [Google Scholar]
- Arregui, I.; Galuardi, B.; Goñi, N.; Lam, C.H.; Fraile, I.; Santiago, J.; Lutcavage, M.; Arrizabalaga, H. Movements and geographic distribution of juvenile bluefin tuna in the Northeast Atlantic, described through internal and satellite archival tags. ICES J. Mar. Sci. 2018, 75, 1560–1572. [Google Scholar] [CrossRef]
- Mather III, F.J.; Bartlett, M.R.; Beckett, J.S. Transatlantic Migrations of Young Bluefin Tuna. J. Fish. Res. Board Canada 1967, 24, 1991–1997. [Google Scholar] [CrossRef]
- Chapman, E.W.; Jørgensen, C.; Lutcavage, M.E.; Hilborn, R. Atlantic bluefin tuna (Thunnus thynnus): A state-dependent energy allocation model for growth, maturation, and reproductive investment. Can. J. Fish. Aquat. Sci. 2011, 68, 1934–1951. [Google Scholar] [CrossRef]
- Medina, A. Reproduction of Atlantic bluefin tuna. Fish Fish. 2020, 21, 1109–1119. [Google Scholar] [CrossRef]
- Barneche, D.R.; Robertson, D.R.; White, C.R.; Marshall, D.J. Fish reproductive-energy output increases disproportionately with body size. Science 2018, 360, 642–645. [Google Scholar] [CrossRef]
- Addis, P.; Secci, M.; Locci, I.; Cau, A.; Sabatini, A. Analysis of Atlantic bluefin tuna catches from the last Tonnara in the Mediterranean Sea: 1993–2010. Fish. Res. 2012, 127–128, 133–141. [Google Scholar] [CrossRef]
- Cort, J.L.; Arregui, I.; Estruch, V.D.; Deguara, S. Validation of the Growth Equation Applicable to the Eastern Atlantic Bluefin Tuna, Thunnus thynnus (L.), Using Lmax, Tag-Recapture, and First Dorsal Spine Analysis. Rev. Fish. Sci. Aquac. 2014, 22, 239–255. [Google Scholar] [CrossRef]
- Schaefer, K.M. Reproductive biology of yellowfin tuna (Thunnus albacares) in the eastern Pacific Ocean. Inter-American Trop. Tuna Comm. 1998, 21, 201–272. [Google Scholar]
- Pousis, C.; De Giorgi, C.; Mylonas, C.C.; Bridges, C.R.; Zupa, R.; Vassallo-Agius, R.; de la Gándara, F.; Dileo, C.; De Metrio, G.; Corriero, A. Comparative study of liver vitellogenin gene expression and oocyte yolk accumulation in wild and captive Atlantic bluefin tuna (Thunnus thynnus L.). Anim. Reprod. Sci. 2011, 123, 98–105. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yang, Q.; Zhao, W.J.; Du, Q.Y.; Chang, Z.J. Selection of suitable candidate genes for miRNA expression normalization in Yellow River Carp (Cyprinus carpio. var). Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Y.L.; Chen, D.X.; Wu, P.; Yi, T.; Chen, T.; Zhang, J.S.; Chu, W.Y. Selection of reference genes for MicroRNA quantitative expression analysis in Chinese Perch, Siniperca chuatsi. Int. J. Mol. Sci. 2015, 16, 8310–8323. [Google Scholar] [CrossRef]
- Paul, S.C.; Sharma, A.; Mehta, R.; Paul, S. Genome Wide Computational Identification of Tuna (Thunnus orientalis) MicroRNAs and Their Targets. Ocean Sci. J. 2018, 53, 727–734. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pousis, C.; Santamaria, N.; Zupa, R.; De Giorgi, C.; Mylonas, C.C.; Bridges, C.R.; de la Gándara, F.; Vassallo-Agius, R.; Bello, G.; Corriero, A. Expression of vitellogenin receptor gene in the ovary of wild and captive Atlantic bluefin tuna (Thunnus thynnus). Anim. Reprod. Sci. 2012, 132, 101–110. [Google Scholar] [CrossRef]
- Marisaldi, L.; Basili, D.; Gioacchini, G.; Canapa, A.; Carnevali, O. De novo transcriptome assembly, functional annotation and characterization of the Atlantic bluefin tuna (Thunnus thynnus) larval stage. Mar. Genomics 2021, 58, 100834. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis 2009; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S-PLUS; Springer: New York, NY, USA, 2002; ISBN 0-387-95457-0. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Medina, A.; Abascal, F.J.; Megina, C.; Garcia, A. Stereological assessment of the reproductive status of female atlantic northern bluefin tuna during migration to mediterranean spawning grounds through the Strait of Gibraltar. J. Fish Biol. 2002, 60, 203–217. [Google Scholar] [CrossRef]
- Corriero, A.; Desantis, S.; Deflorio, M.; Acone, F.; Bridges, C.R.; De la Serna, J.M.; Megalofonou, P.; De Metrio, G. Histological investigation on the ovarian cycle of the bluefin tuna in the western and central Mediterranean. J. Fish Biol. 2003, 63, 108–119. [Google Scholar] [CrossRef]
- Addis, P.; Secci, M.; Biancacci, C.; Loddo, D.; Cuccu, D.; Palmas, F.; Sabatini, A. Reproductive status of Atlantic bluefin tuna, Thunnus thynnus, during migration off the coast of Sardinia (western Mediterranean). Fish. Res. 2016, 181, 137–147. [Google Scholar] [CrossRef]
- Idrissi, M.; Noureddine, R. Preliminary analysis of the size data of bluefin tuna (Thunnus thynnus) caught by the Moroccan Atlantic traps during 2009. ICCAT Collect. Vol. Sci. Pap. 2010, 65, 968–974. [Google Scholar]
- Nishida, T.; Tsuji, S.; Segawa, K. Spatial data analyses of Atlantic bluefin tuna larval surveys in the 1994 ICCAT BYP. ICCAT Collect. Vol. Sci. Pap. 1998, 48, 107–110. [Google Scholar]
- Suzuki, Z.; Kai, M. Movement of Atlantic Bluefin Tuna Toward the Strait of Gibraltar Inferred From Japanese Longline Data. ICCAT Collect. Vol. Sci. Pap. 2012, 67, 322–330. [Google Scholar]
- Aranda, G.; Abascal, F.J.; Varela, J.L.; Medina, A. Spawning Behaviour and Post-Spawning Migration Patterns of Atlantic Bluefin Tuna (Thunnus thynnus) Ascertained from Satellite Archival Tags. PLoS ONE 2013, 8, e76445. [Google Scholar] [CrossRef]
- Stokesbury, M.J.W.; Cosgrove, R.; Boustany, A.; Browne, D.; Teo, S.L.H.; O’Dor, R.K.; Block, B.A. Results of satellite tagging of Atlantic bluefin tuna, Thunnus thynnus, off the coast of Ireland. Hydrobiologia 2007, 582, 91–97. [Google Scholar] [CrossRef]
- Horton, T.W.; Block, B.A.; Drumm, A.; Hawkes, L.A.; O’Cuaig, M.; Maoiléidigh, N.Ó.; O’Neill, R.; Schallert, R.J.; Stokesbury, M.J.W.; Witt, M.J. Tracking Atlantic bluefin tuna from foraging grounds off the west coast of Ireland. ICES J. Mar. Sci. 2020, 77, 2066–2077. [Google Scholar] [CrossRef]
- Walli, A.; Teo, S.L.H.; Boustany, A.; Farwell, C.J.; Williams, T.; Dewar, H.; Prince, E.; Block, B.A. Seasonal Movements, Aggregations and Diving Behavior of Atlantic Bluefin Tuna (Thunnus thynnus) Revealed with Archival Tags. PLoS ONE 2009, 4, e6151. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.J.; Medina, A.; De La Serna, J.M.; Godoy, D.; Aranda, G. Tracking bluefin tuna reproductive migration into the Mediterranean Sea with electronic pop-up satellite archival tags using two tagging procedures. Fish. Oceanogr. 2016, 25, 54–66. [Google Scholar] [CrossRef]
- Fromentin, J.-M.; Lopuszanski, D. Migration, residency, and homing of bluefin tuna in the western Mediterranean Sea. ICES J. Mar. Sci. 2014, 71, 510–518. [Google Scholar] [CrossRef]
- Tudela, S.; Trápaga, S.S.; Cermeño, P.; Hidas, E.; Graupera, E.; Quilez-Badia, G. Bluefin tuna migratory behavior in the western and central Mediterranean Sea revealed by electronic tags. ICCAT Collect. Vol. Sci. Pap. 2011, 66, 1157–1169. [Google Scholar]
- Heinisch, G.; Corriero, A.; Medina, A.; Abascal, F.J.; de la Serna, J.-M.; Vassallo-Agius, R.; Ríos, A.B.; García, A.; de la Gándara, F.; Fauvel, C.; et al. Spatial–temporal pattern of bluefin tuna (Thunnus thynnus L. 1758) gonad maturation across the Mediterranean Sea. Mar. Biol. 2008, 154, 623–630. [Google Scholar] [CrossRef]
- Medina, A.; Aranda, G.; Gherardi, S.; Santos, A.; Mèlich, B.; Lara, M. Assessment of spawning of Atlantic bluefin tuna farmed in the western Mediterranean Sea. Aquac. Environ. Interact. 2016, 8, 89–98. [Google Scholar] [CrossRef]
- Gordoa, A.; Carreras, G. Determination of Temporal Spawning Patterns and Hatching Time in Response to Temperature of Atlantic Bluefin Tuna (Thunnus thynnus) in the Western Mediterranean. PLoS ONE 2014, 9, e90691. [Google Scholar] [CrossRef]
- Gordoa, A.; Olivar, M.P.; Arevalo, R.; Viñas, J.; Molí, B.; Illas, X. Determination of Atlantic bluefin tuna (Thunnus thynnus) spawning time within a transport cage in the western Mediterranean. ICES J. Mar. Sci. 2009, 66, 2205–2210. [Google Scholar] [CrossRef]
- Carnevali, O.; Maradonna, F.; Sagrati, A.; Candelma, M.; Lombardo, F.; Pignalosa, P.; Bonfanti, E.; Nocillado, J.; Palma, P.; Gioacchini, G.; et al. Insights on the seasonal variations of reproductive features in the Eastern Atlantic Bluefin Tuna. Gen. Comp. Endocrinol. 2019, 282, 113216. [Google Scholar] [CrossRef]
- Morais, R.D.V.S.; Thomé, R.G.; Lemos, F.S.; Bazzoli, N.; Rizzo, E. Autophagy and apoptosis interplay during follicular atresia in fish ovary: A morphological and immunocytochemical study. Cell Tissue Res. 2012, 347, 467–478. [Google Scholar] [CrossRef]
- Sales, C.F.; Melo, R.M.C.; Pinheiro, A.P.B.; Luz, R.K.; Bazzoli, N.; Rizzo, E. Autophagy and Cathepsin D mediated apoptosis contributing to ovarian follicular atresia in the Nile tilapia. Mol. Reprod. Dev. 2019, 86, 1592–1602. [Google Scholar] [CrossRef]
- Gohin, M.; Bobe, J.; Chesnel, F. Comparative transcriptomic analysis of follicle-enclosed oocyte maturational and developmental competence acquisition in two non-mammalian vertebrates. BMC Genomics 2010, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Senthilkumaran, B.; Yoshikuni, M.; Nagahama, Y. A shift in steroidogenesis occurring in ovarian follicles prior to oocyte maturation. Mol. Cell. Endocrinol. 2004, 215, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, Y.; Yamashita, M. Regulation of oocyte maturation in fish. Dev. Growth Differ. 2008, 50, S195–S219. [Google Scholar] [CrossRef]
- Luong, X.G.; Daldello, E.M.; Rajkovic, G.; Yang, C.-R.; Conti, M. Genome-wide analysis reveals a switch in the translational program upon oocyte meiotic resumption. Nucleic Acids Res. 2020, 48, 3257–3276. [Google Scholar] [CrossRef]
- Stebbins-Boaz, B.; Hake, L.E.; Richter, J.D. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996, 15, 2582–2592. [Google Scholar] [CrossRef] [PubMed]
- Farley, J.H.; Davis, T.L.O.; Bravington, M.V.; Andamari, R.; Davies, C.R. Spawning Dynamics and Size Related Trends in Reproductive Parameters of Southern Bluefin Tuna, Thunnus maccoyii. PLoS ONE 2015, 10, e0125744. [Google Scholar] [CrossRef]
- Farley, J.H.; Williams, A.J.; Hoyle, S.D.; Davies, C.R.; Nicol, S.J. Reproductive Dynamics and Potential Annual Fecundity of South Pacific Albacore Tuna (Thunnus alalunga). PLoS ONE 2013, 8, e60577. [Google Scholar] [CrossRef]
- Aranda, G.; Medina, A.; Santos, A.; Abascal, F.J.; Galaz, T. Evaluation of Atlantic bluefin tuna reproductive potential in the western Mediterranean Sea. J. Sea Res. 2013, 76, 154–160. [Google Scholar] [CrossRef]
- Hixon, M.A.; Johnson, D.W.; Sogard, S.M. BOFFFFs: On the importance of conserving old-growth age structure in fishery populations. ICES J. Mar. Sci. 2014, 71, 2171–2185. [Google Scholar] [CrossRef]
- Wang, X.; Song, X.; Bhandari, R.K. Distinct expression patterns of seven crucial microRNAs during early embryonic development in medaka (Oryzias latipes). Gene Expr. Patterns 2020, 37, 119133. [Google Scholar] [CrossRef] [PubMed]
- Zayed, Y.; Qi, X.; Peng, C. Identification of Novel MicroRNAs and Characterization of MicroRNA Expression Profiles in Zebrafish Ovarian Follicular Cells. Front. Endocrinol. (Lausanne) 2019, 10, 518. [Google Scholar] [CrossRef]
- Wong, Q.W.L.; Sun, M.A.; Lau, S.W.; Parsania, C.; Zhou, S.; Zhong, S.; Ge, W. Identification and characterization of a specific 13-miRNA expression signature during follicle activation in the zebrafish ovary. Biol. Reprod. 2018, 98, 42–53. [Google Scholar] [CrossRef]
- Presslauer, C.; Bizuayehu, T.T.; Kopp, M.; Fernandes, J.M.O.; Babiak, I. Dynamics of miRNA transcriptome during gonadal development of zebrafish. Sci. Rep. 2017, 7, 43850. [Google Scholar] [CrossRef] [PubMed]

| Group | SFL (cm) | Total Weight (kg) | Age |
|---|---|---|---|
| Active-old (n = 7) | 194.6 ± 33.9 a | 147.1 ± 71.9 a | 11.3 ± 2.7 a |
| Active-young (n = 6) | 139.3 ± 18.8 b | 51.4 ± 28.7 b | 8.4 ± 1.1 b |
| Resting-old (n = 13) | 210.8 ± 27.4 a | 204.8 ± 63.4 a | 12.7 ± 2.3 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marisaldi, L.; Iorillo, O.; Basili, D.; Gioacchini, G.; Bobe, J.; Thermes, V.; Maradonna, F.; Carnevali, O. A Comparison of Reproductive Performances in Young and Old Females: A Case Study on the Atlantic Bluefin Tuna in the Mediterranean Sea. Animals 2021, 11, 3340. https://doi.org/10.3390/ani11123340
Marisaldi L, Iorillo O, Basili D, Gioacchini G, Bobe J, Thermes V, Maradonna F, Carnevali O. A Comparison of Reproductive Performances in Young and Old Females: A Case Study on the Atlantic Bluefin Tuna in the Mediterranean Sea. Animals. 2021; 11(12):3340. https://doi.org/10.3390/ani11123340
Chicago/Turabian StyleMarisaldi, Luca, Orsola Iorillo, Danilo Basili, Giorgia Gioacchini, Julien Bobe, Violette Thermes, Francesca Maradonna, and Oliana Carnevali. 2021. "A Comparison of Reproductive Performances in Young and Old Females: A Case Study on the Atlantic Bluefin Tuna in the Mediterranean Sea" Animals 11, no. 12: 3340. https://doi.org/10.3390/ani11123340
APA StyleMarisaldi, L., Iorillo, O., Basili, D., Gioacchini, G., Bobe, J., Thermes, V., Maradonna, F., & Carnevali, O. (2021). A Comparison of Reproductive Performances in Young and Old Females: A Case Study on the Atlantic Bluefin Tuna in the Mediterranean Sea. Animals, 11(12), 3340. https://doi.org/10.3390/ani11123340







