A Preliminary Study on the Relationship between Parasitaemia and Cytokine Expression of Peripheral Blood Cells in Trypanosoma vivax-Experimentally Infected Cattle
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Infection and Sample Collection
2.2. qPCR (Quantitative Real-Time PCR) for Trypanosoma vivax Detection and Quantification
2.3. RNA Extraction and cDNA Transcription
2.4. Cytokine Assessment by RT-qPCR
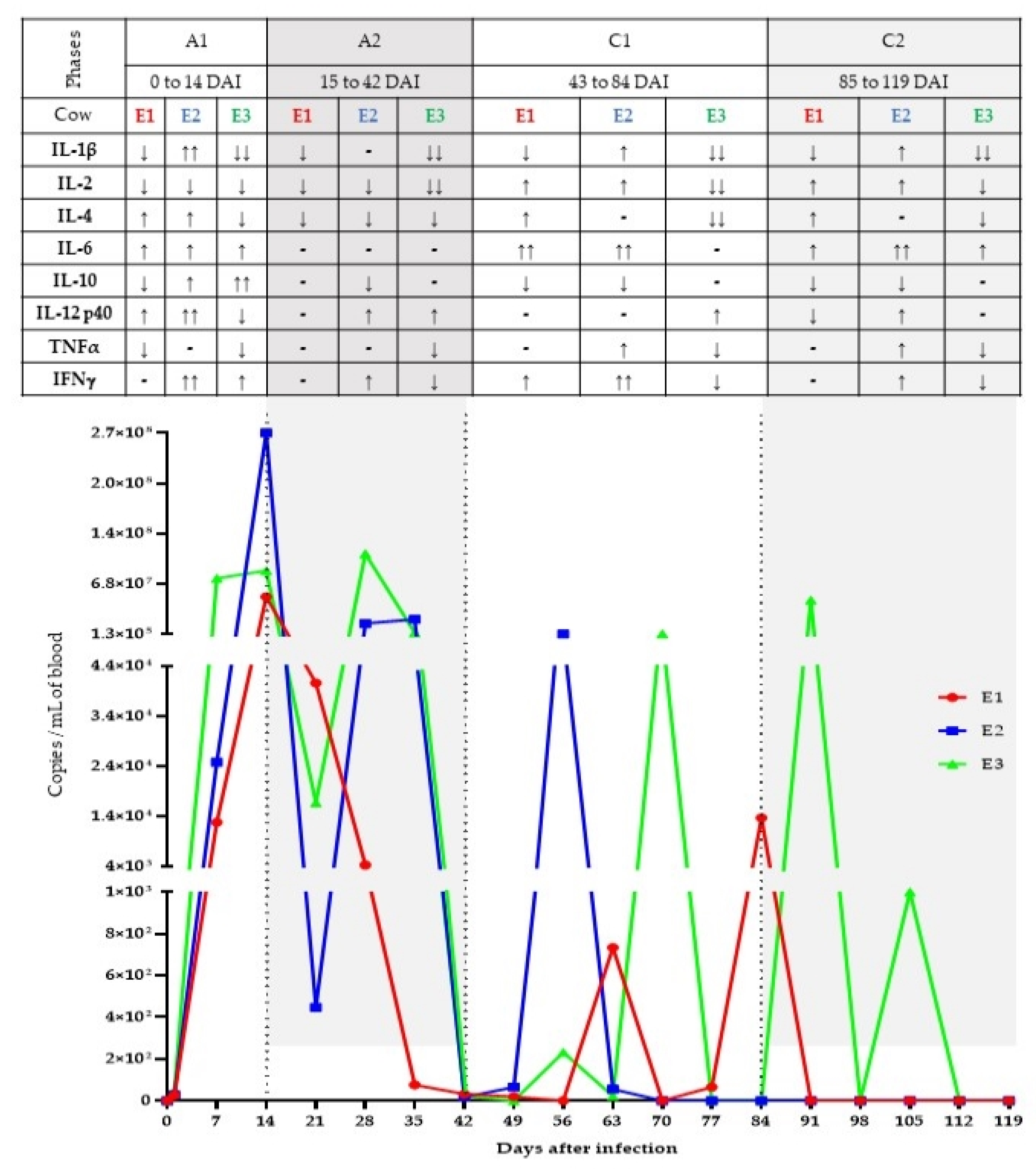
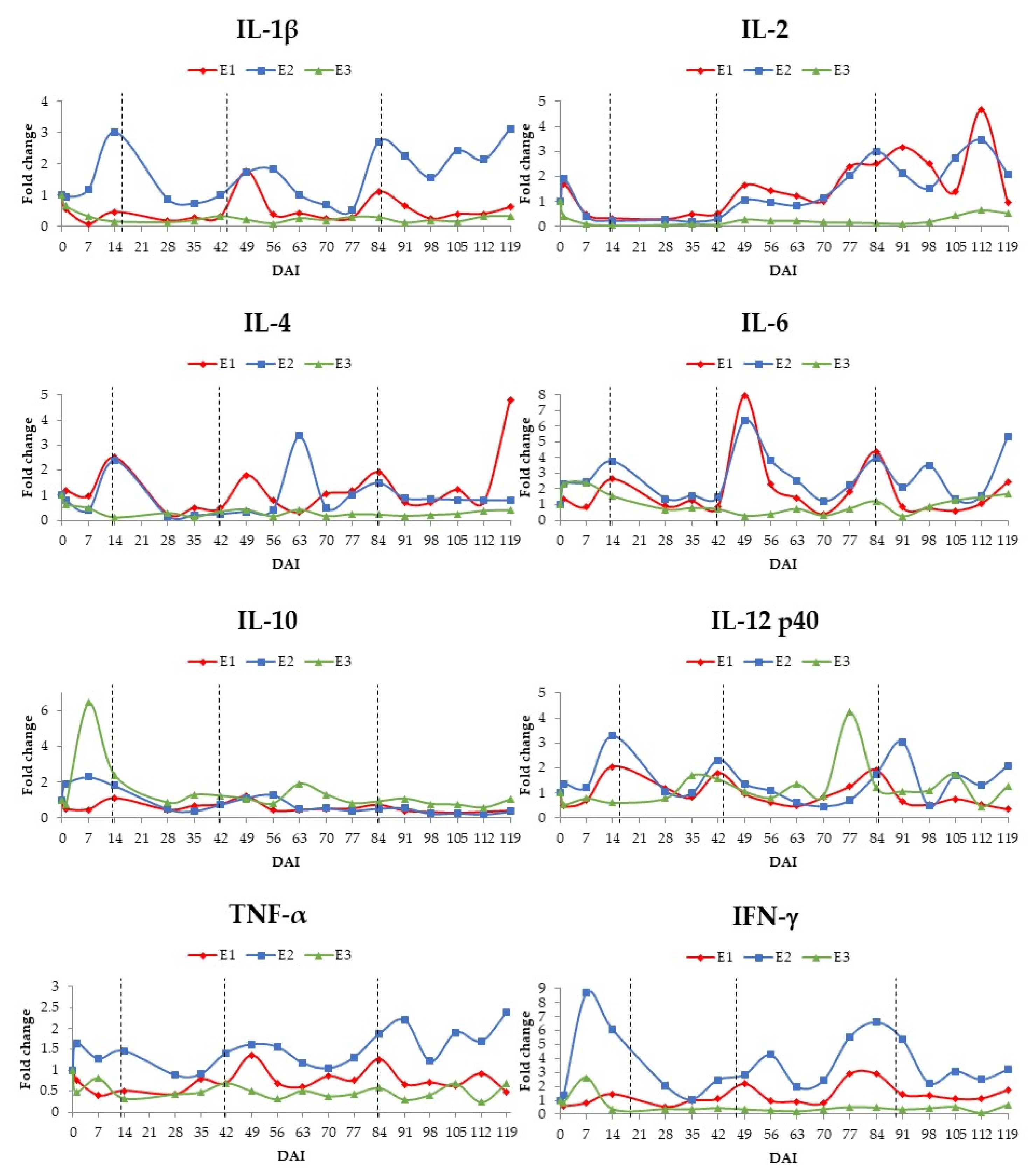
3. Results
3.1. qPCR for Trypanosoma vivax
3.2. Cytokine Assessment by RT-qPCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shaw, A.P.M.; Cecchi, G.; Wint, G.R.W.; Mattioli, R.C.; Robinson, T.P. Mapping the economic benefits to livestock keepers from intervening against bovine trypanosomosis in Eastern Africa. Prev. Vet. Med. 2014, 113, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; Koning, H.P.; Barrett, M.P. The animal trypanosomiases and their chemotherapy: A review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef]
- Dávila, A.M.R.; Silva, R.A.M.S. Animal trypanosomiasis in South America. Current status, partnership, and information technology. Ann. N. Y. Acad. Sci. 2000, 916, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Osório, A.L.A.R.; Madruga, C.R.; Desquesnes, M.; Soares, C.O.; Ribeiro, L.R.R.; Costa, S.C.G. Trypanosoma (Duttonella) vivax: Its biology, epidemiology, pathogenesis, and introduction in the New World—A review. Mem. Inst. Oswaldo Cruz. 2008, 103, 1–13. [Google Scholar] [CrossRef]
- Morrison, L.J.; Vezza, L.; Rowan, T.; Hope, J.C. Animal African Trypanosomiasis: Time to increase focus on clinically relevant parasite and host species. Trends Parasitol. 2016, 32, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.B.; Hernandéz-Gamboa, J.; Jiménez-Alfaro, C.; Zeledón, R.; Blandón, M.; Urbina, A. First report of Trypanosoma vivax infection in dairy cattle from Costa Rica. Vet. Parasitol. 2009, 163, 136–139. [Google Scholar] [CrossRef]
- Cadioli, F.A.; Barnabé, P.A.; Machado, R.Z.; Teixeira, M.C.A.; André, M.R.; Sampaio, P.H.; Fidelis Junior, O.L.; Teixeira, M.M.G.; Marques, L.C. First report of Trypanosoma vivax outbreak in dairy cattle in São Paulo state, Brazil. Bras. J. Vet. Parasitol. 2012, 21, 118–124. [Google Scholar] [CrossRef]
- Paguem, A.; Abanda, B.; Ndjonka, D.; Weber, J.S.; Ngomtcho, S.C.H.; Manchang, K.T.; Adoulmoumini, M.; Eisenbarth, A.; Renz, A.; Kelm, S.; et al. Widespread co-endemicity of Trypanosoma species infecting cattle in the Sudano-Sahelian and Guinea Savannah zones of Cameroon. BMC Vet. Res. 2019, 15, 344. [Google Scholar] [CrossRef]
- Desquesnes, M. Livestock Trypanosomoses and Their Vectors in Latin America, 1st ed.; OIE and CIRAD: Paris, France, 2004; p. 190. [Google Scholar]
- Cadioli, F.A.; Fidelis Junior, O.L.; Sampaio, P.H.; Santos, G.N.; André, M.R.; Castilho Neto, K.J.G.A.; Machado, R.Z. Detection of Trypanosoma vivax using PCR and LAMP during aparasitemic periods. Vet. Parasitol. 2015, 214, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Fidelis Junior, O.L.; Sampaio, P.H.; Gonçalves, L.R.; André, M.R.; Machado, R.Z.; Wijffels, G.; Cadioli, F.A. Comparison of conventional and molecular techniques for Trypanosoma vivax diagnosis in experimentally infected cattle. Bras. J. Vet. Parasitol. 2019, 28, 203–209. [Google Scholar] [CrossRef]
- Vincendeau, P.; Bouteille, B. Immunology and immunopathology of African trypanosomiasis. An. Acad. Bras. Cienc. 2006, 78, 645–665. [Google Scholar] [CrossRef] [PubMed]
- Stijlemans, B.; Vankrunkelsven, A.; Caljon, G.; Bockstal, V.; Guilliams, M.; Bosschaerts, T.; Beschin, A.; Raes, G.; Magez, S.; Baetselier, P. The central role of macrophages in trypanosomiasis-associated anemia: Rationale for therapeutical approaches. Endocr. Metab. Immune Disord. Drug Targets 2010, 10, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta 2014, 1843, 2563–2582. [Google Scholar] [CrossRef]
- Abrahamsohn, I.A. Cytokines in innate and acquired immunity to Trypanosoma cruzi infection. Braz. J. Med. Biol. Res. 1998, 31, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.A.; Mertens, B. Immune response of cattle infected with African trypanosomes. Mem. Inst. Oswaldo Cruz 1999, 94, 239–244. [Google Scholar] [CrossRef][Green Version]
- Musaya, J.; Matovu, E.; Nyirenda, M.; Chisi, J. Role of cytokines in Trypanosoma brucei-induced anaemia: A review of the literature. Malawi Med. J. 2015, 27, 45–50. [Google Scholar] [CrossRef]
- Fidelis Junior, O.L.; Sampaio, P.H.; Machado, R.Z.; André, M.R.; Marques, L.C.; Cadioli, F.A. Evaluation of clinical signs, parasitemia, hematologic and biochemical changes in cattle experimentally infected with Trypanosoma vivax. Braz. J. Vet. Parasitol. 2016, 25, 69–81. [Google Scholar] [CrossRef]
- Garcia, H.A.; Rodrigues, A.C.; Rodrigues, C.M.; Bengaly, Z.; Minervino, A.H.; Riet-Correa, F.; Machado, R.Z.; Paiva, F.; Batista, J.S.; Neves, L.; et al. Microsatellite analysis supports clonal propagation and reduced divergence of Trypanosoma vivax from asymptomatic to fatally infected livestock in South America compared to West Africa. Parasit. Vectors 2014, 7, 13. [Google Scholar] [CrossRef]
- Silbermayr, K.; Li, F.; Soudré, A.; Müller, S.; Sölkner, J. A novel qPCR assay for the detection of African animal trypanosomosis in trypanotolerant and trypanosusceptible cattle breeds. PLoS Negl. Trop. Dis. 2013, 7, 1–9. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Konnai, S.; Usui, T.; Ohashi, K.; Onuma, M. The rapid quantitative analysis of bovine cytokine genes by real-time RT-PCR. Vet. Microbiol. 2003, 94, 283–294. [Google Scholar] [CrossRef]
- Puech, C.; Dedieu, L.; Chantal, I.; Rodrigues, V. Design and evaluation of a unique SYBR Green real-time RT-PCR assay for quantification of five major cytokines in cattle, sheep and goats. BMC Vet. Res. 2015, 11, 14. [Google Scholar] [CrossRef]
- Livack, K.J.; Schimittgen, T.D. Analysis of relative gene expression data using Real-Time quantitative PCR and the 2-ΔΔCq method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Moldoveanu, B.; Otmishi, P.; Jani, P.; Walker, J.; Sarmiento, X.; Guardiola, J.; Saad, M.; Yu, J. Inflammatory mechanisms in the lung. J. Inflamm. Res. 2009, 2, 1–11. [Google Scholar]
- Kato, C.D.; Matovu, E.; Mugasa, C.M.; Nanteza, A.; Alibu, V.P. The role of cytokines in the pathogenesis and staging of Trypanosoma brucei rhodesiense sleeping sickness. Allergy Asthma Clin. Immunol. 2016, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Magez, S.; Benoit, S.; Radwanska, M.; Pays, E.; Ferguson, M.A.J.; Baetselier, P. The glycosyl-inositol-phosphate and dimyristoylglycerol moieties of the glycosylphosphatidylinositol anchor of the trypanosome variant-specific surface glycoprotein are distinct macrophage-activating factors. J. Immunol. 1998, 160, 1949–1956. [Google Scholar] [PubMed]
- Naessens, J.; Kitani, H.; Nakamura, Y.; Yagi, Y.; Sekikawa, K.; Iraqi, F. TNFα mediates the development of anaemia in a murine Trypanosoma brucei rhodesiense infection, but not the anaemia associated with a murine Trypanosoma congolense infection. J. Clin. Exp. Immunol. 2005, 139, 405–410. [Google Scholar] [CrossRef]
- Darji, A.; Beschin, A.; Sileghem, M.; Heremans, H.; Brys, L.; Baetselier, P. In vitro simulation of immunosuppression caused by Trypanosoma brucei: Active involvement of gamma interferon and tumor necrosis factor in the pathway of suppression. Infect. Immun. 1996, 64, 1937–1943. [Google Scholar] [CrossRef]
- Naessens, J.; Kitani, H.; Momotani, E.; Sekikawa, K.; Nthale, J.M.; Iraqi, F. Susceptibility of TNF-alpha-deficient mice to Trypanosoma congolense is not due to a defective antibody response. Acta Tropica 2004, 92, 193–203. [Google Scholar] [CrossRef]
- Vignali, D.A.A.; Kuchroo, V. IL-12 family cytokines: Immunological playmakers. Nat. Immunol. 2012, 13, 722–728. [Google Scholar] [CrossRef]
- Abbas, A.; Lichtman, A.; Pillai, S. Cellular and Molecular Immunology, 9th ed.; Elsevier: Philadelphia, PA, USA, 2018; p. 608. [Google Scholar]
- Mosmann, T.R.; Subash, S. The expanding universe of T-cell subsets: Th1, Th2 and more. Immunol. Today 1996, 17, 138–146. [Google Scholar] [CrossRef]
- Namangala, B.; Noe, W.; Baetselier, P.; Brys, L.; Beschin, A. Relative contribution of Interferon-γ and Interleukin-10 to resistance to murine African trypanosomosis. J. Infect. Dis. 2001, 183, 1794–1800. [Google Scholar] [CrossRef]
- Hertz, C.J.; Filutowicz, H.; Mansfield, J.M. Resistance to the African trypanosomes is IFN-γ dependent. J. Immunol. 1998, 161, 6775–6783. [Google Scholar] [PubMed]
- Rodrigues, A.A.; Saosa, J.S.S.; Silva, G.K.; Martins, F.A.; Silva, A.A.; Souza Neto, C.P.S.; Horta, C.; Zamboni, D.S.; Silva, J.S.; Ferro, E.A.V.; et al. IFN-γ plays a unique role in protection against low virulent Trypanosoma cruzi strain. PLoS Negl. Trop. Dis. 2012, 6, e1598. [Google Scholar] [CrossRef]
- Kashimoto, T. IL-6: From its discovery to clinical applications. Int. Immunol. 2010, 22, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Kato, C.D.; Alibu, V.P.; Nanteza, A.; Mugasa, C.M.; Matovu, E. Interleukin (IL)-6 and IL-10 are up regulated in late stage Trypanosoma brucei rhodesiense sleeping sickness. PLoS Negl. Trop. Dis. 2015, 9, e0003835. [Google Scholar] [CrossRef] [PubMed]
- Tizard, I.R. Veterinary Immunology, 10th ed.; Elsevier: St. Louis, MO, USA, 2018; p. 552. [Google Scholar]
- Sternberg, J.M.; Rodgers, J.; Bradley, B.; Maclean, L.; Murray, M.; Kennedy, P.G.E. Meningoencephalitic African trypanosomiasis: Brain IL-10 and IL-6 are associated with protection from neuro-inflammatory pathology. J. Neuroimmunol. 2005, 167, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bakhiet, M.; Jansson, L.; Büscher, P.; Holmdahl, R.; Kristensson, K.; Olsson, T. Control of parasitemia and survival during Trypanosoma brucei brucei infection is related to strain-dependent ability to produce IL-4. J. Immunol. 1996, 157, 3518–3526. [Google Scholar] [PubMed]
- Liao, W.; Lin, J.; Wang, L.; LI, P.; Leonard, W.J. Cytokine receptor modulation by interleukin-2 broadly regulates T helper cell lineage differentiation. Nat. Immunol. 2011, 12, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Sileghem, M.; Hamers, R.; Baetselier, P. Active suppression of interleukin 2 secretion in mice infected with Trypanosoma brucei AnTat 1.1.E. Parasite Immunol. 1986, 8, 641–649. [Google Scholar] [CrossRef]
- Mitchell, L.A.; Pearson, T.W.; Gauldie, J. Interleukin-1 and interleukin-2 production in resistant and susceptible inbred mice infected with Trypanosoma congolense. Immunology 1986, 57, 291–296. [Google Scholar] [PubMed]
| Cytokine | Primers (5′-3′) | GenBank Access Number |
|---|---|---|
| IL-1β | F- CAAGGAGAGGAAAGAGACA R- TGAGAAGTGCTGATGTACCA | M37211 |
| IL-2 | F- TTTTACGTGCCCAAGGTTAA R- CGTTTACTGTTGCATCATCA | M12791 |
| IL-4 | F- CAAAGAACACAACTGAGAAG R- AGGTCTTTCAGCGTACTTGT | M77120 |
| IL-6 | F- TCCAGAACGAGTATGAGG R- CATCCGAATAGCTCTCAG | X57317 |
| IL-10 | F- TGCTGGATGACTTTAAGGG R- AGGGCAGAAAGCGATGACA | U00799 |
| IL-12 p40 | F- AACCTGCAACTGAGACCATT R- ATCCTTGTGGCATGTGACTT | U11815 |
| IFNγ | F- ATAACCAGGTCATTCAAAGG R- ATTCTGACTTCTCTTCCGCT | M29867 |
| TNFα | F- CCAGAGGGAAGAGCAGTCC R- GGCTACAACGTGGGCTACC | NM_173966 |
| gapdh | F- TCGGAGTGAACGGATTCG R- ATCTCGCTCCTGGAAGATG | NM_001034034 |
| β-actin | F- CGCACCACTGGCATTGTCAT R- TCCAAGGCGACGTAGCAGAG | K00622 |
| H3F3A | F- GAGGTCTCTATACCATGGCTC R- GTACCAGGCCTGTAACGATG | NM_00101489 |
| YWHAZ | F- GAAAGGGATTGTGGACCAG R- GGCTTCATCAAATGCTGTCT | NM_174814 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fidelis Junior, O.L.; Sampaio, P.H.; Gonçalves, L.R.; Machado, R.Z.; André, M.R.; Wijffels, G.; Cadioli, F.A. A Preliminary Study on the Relationship between Parasitaemia and Cytokine Expression of Peripheral Blood Cells in Trypanosoma vivax-Experimentally Infected Cattle. Animals 2021, 11, 3191. https://doi.org/10.3390/ani11113191
Fidelis Junior OL, Sampaio PH, Gonçalves LR, Machado RZ, André MR, Wijffels G, Cadioli FA. A Preliminary Study on the Relationship between Parasitaemia and Cytokine Expression of Peripheral Blood Cells in Trypanosoma vivax-Experimentally Infected Cattle. Animals. 2021; 11(11):3191. https://doi.org/10.3390/ani11113191
Chicago/Turabian StyleFidelis Junior, Otavio Luiz, Paulo Henrique Sampaio, Luiz Ricardo Gonçalves, Rosangela Zacarias Machado, Marcos Rogério André, Gene Wijffels, and Fabiano Antonio Cadioli. 2021. "A Preliminary Study on the Relationship between Parasitaemia and Cytokine Expression of Peripheral Blood Cells in Trypanosoma vivax-Experimentally Infected Cattle" Animals 11, no. 11: 3191. https://doi.org/10.3390/ani11113191
APA StyleFidelis Junior, O. L., Sampaio, P. H., Gonçalves, L. R., Machado, R. Z., André, M. R., Wijffels, G., & Cadioli, F. A. (2021). A Preliminary Study on the Relationship between Parasitaemia and Cytokine Expression of Peripheral Blood Cells in Trypanosoma vivax-Experimentally Infected Cattle. Animals, 11(11), 3191. https://doi.org/10.3390/ani11113191







