Small RNA-Seq Analysis Reveals miRNA Expression of Short Distance Transportation Stress in Beef Cattle Blood
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Model of Short-Distance Transport Stress
2.2. RNA Extraction, sRNA Library Construction and Sequencing
2.3. Primer Design and qPCR
2.4. Small RNA-Seq Analysis and Statistics
3. Results
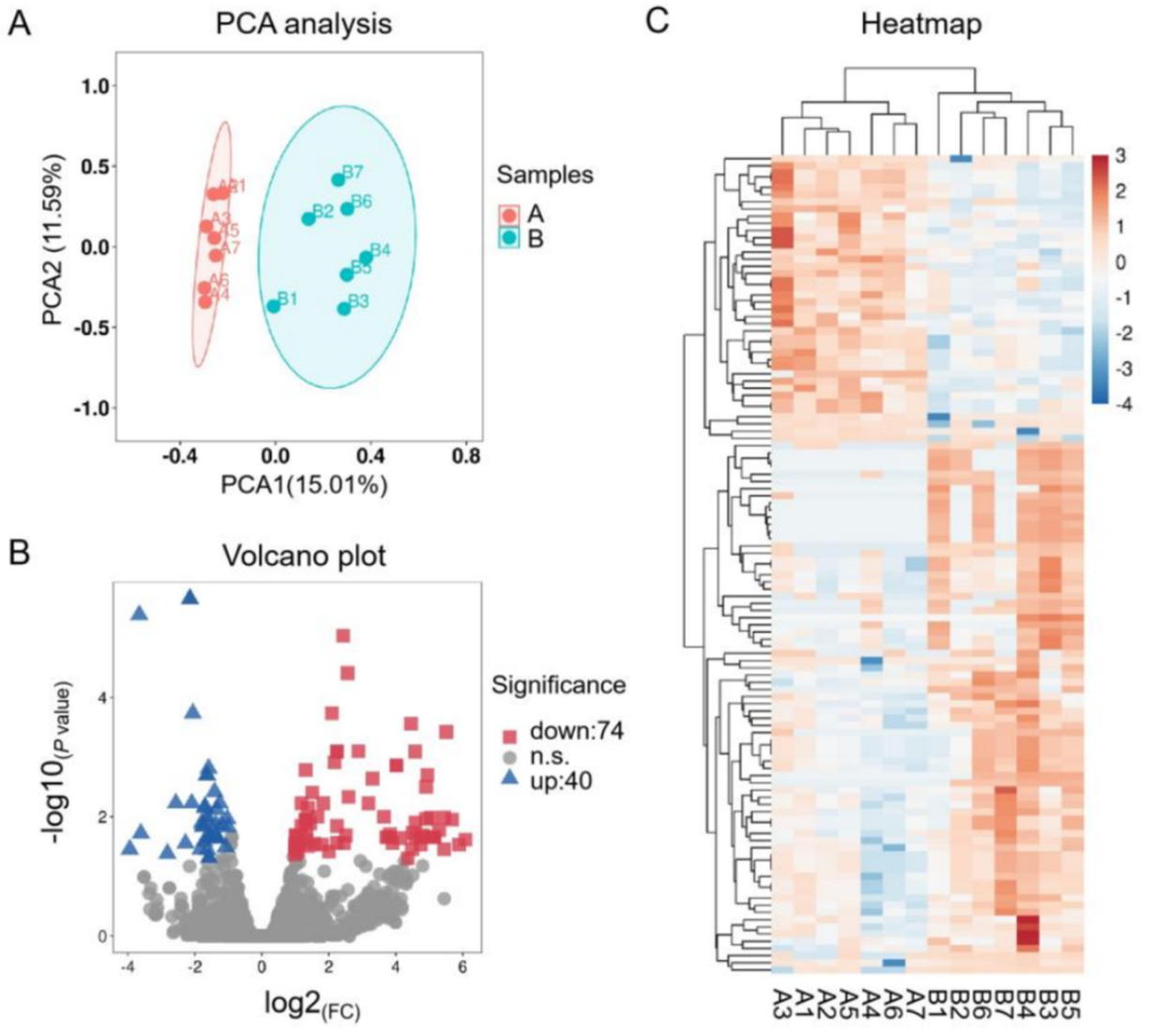
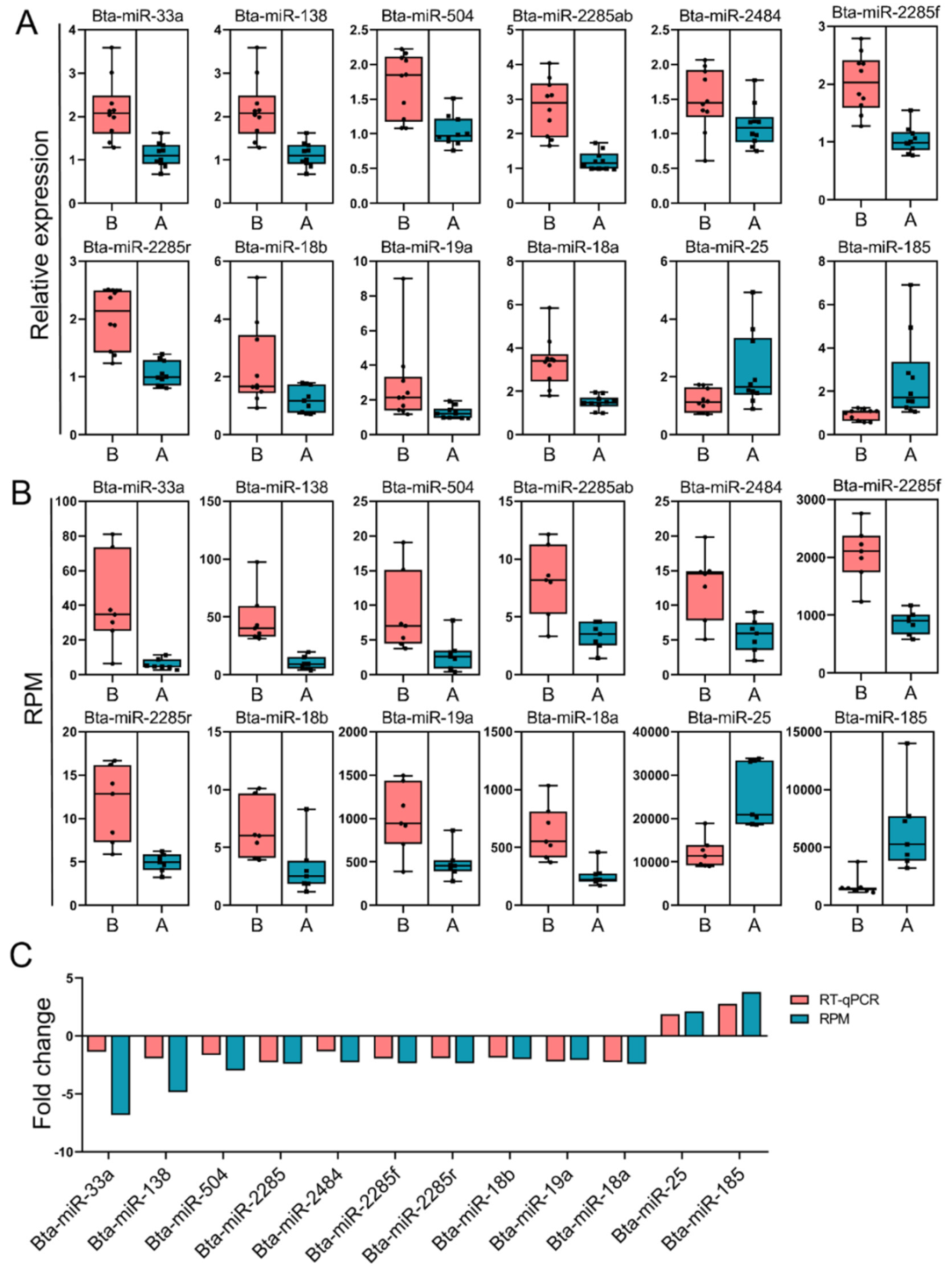
3.1. Preliminary Analysis of RNA-Seq Data and Verification by qPCR
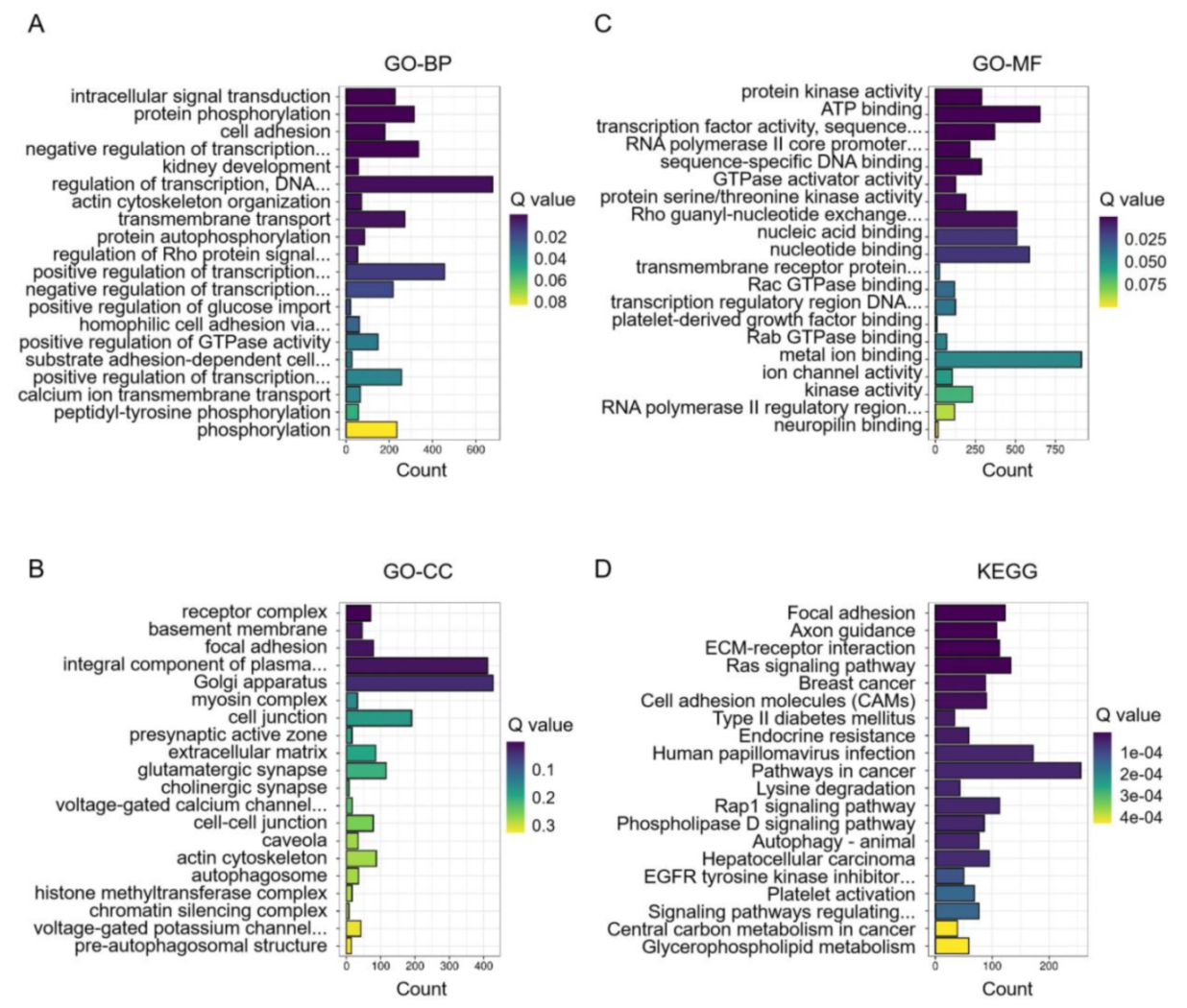
3.2. Enrichment of DEMs between before and after Transportation
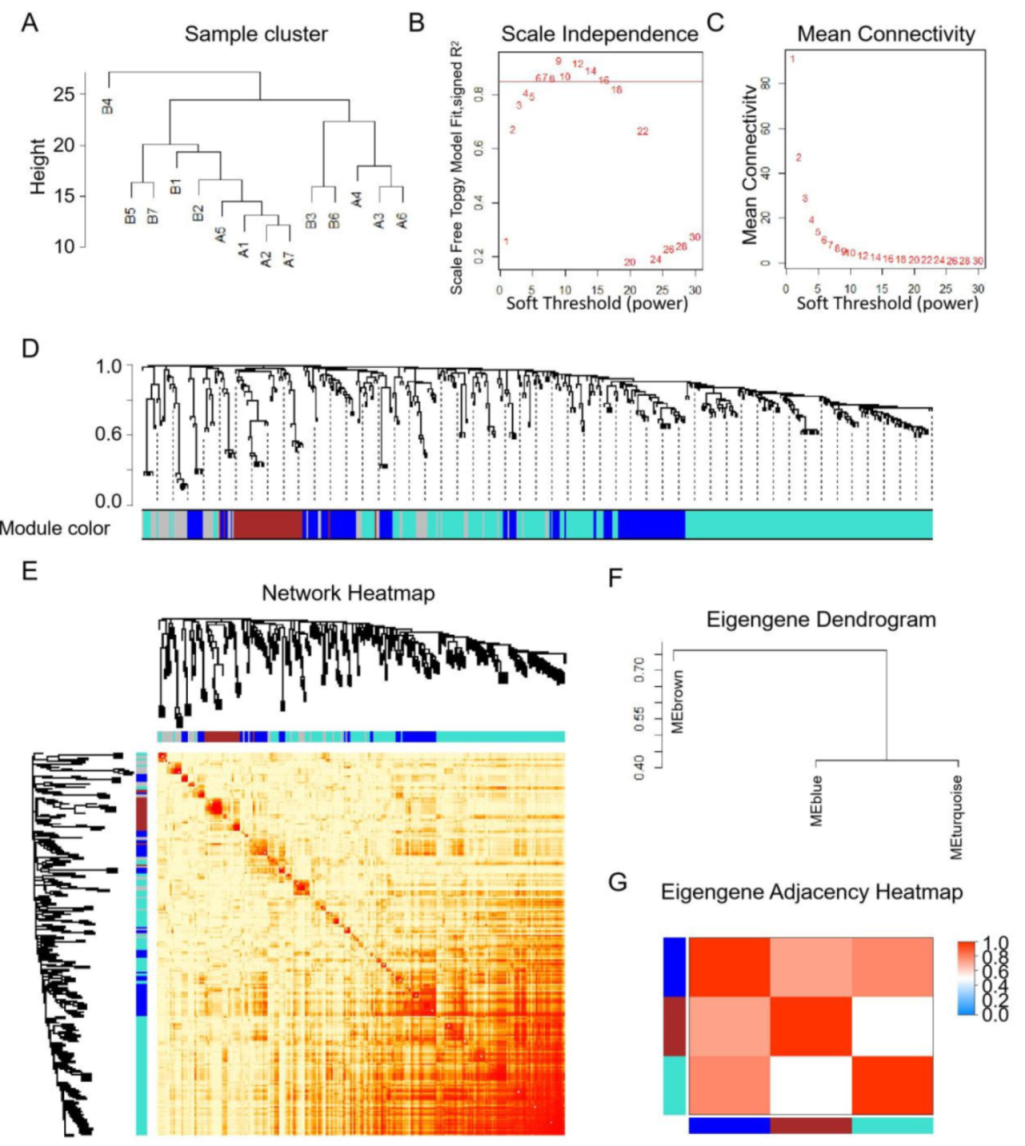
3.3. Co-Expression Analysis and Their Relationship with Hub Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, L.; He, C.; Zhou, Y.; Xu, L.; Xiong, H. Ground transport stress affects bacteria in the rumen of beef cattle: A real-time PCR analysis. Anim. Sci. J. 2017, 88, 790–797. [Google Scholar] [CrossRef]
- Van Engen, N.K.; Coetzee, J.F. Effects of transportation on cattle health and production: A review. Anim. Health Res. Rev. 2018, 19, 142–154. [Google Scholar] [CrossRef]
- Deters, E.L.; Hansen, S.L. Pre-transit vitamin C injection improves post-transit performance of beef steers. Animal 2020, 14, 2083–2090. [Google Scholar] [CrossRef]
- Li, F.; Shah, A.M.; Wang, Z.; Peng, Q.; Hu, R.; Zou, H.; Tan, C.; Zhang, X.; Liao, Y.; Wang, X.; et al. Effects of land transport stress on variations in ruminal microbe diversity and immune functions in different breeds of cattle. Animals 2019, 9, 599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alfaro, G.F.; Novak, T.E.; Rodning, S.P.; Moisá, S.J. Preconditioning beef cattle for long-duration transportation stress with rumen-protected methionine supplementation: A nutrigenetics study. PLoS ONE 2020, 15, e0235481. [Google Scholar] [CrossRef] [PubMed]
- Fike, K.; Spire, M.F. Transportation of cattle. Vet. Clin. Food Anim. Pract. 2006, 22, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Odore, R.; Badino, P.; Re, G.; Barbero, R.; Cuniberti, B.; D’Angelo, A.; Girardi, C.; Fraccaro, E.; Tarantola, M. Effects of housing and short-term transportation on hormone and lymphocyte receptor concentrations in beef cattle. Res. Vet. Sci. 2011, 90, 341–345. [Google Scholar] [CrossRef]
- Takemoto, S.; Tomonaga, S.; Funaba, M.; Matsui, T. Effect of long-distance transportation on serum metabolic profiles of steer calves. Anim. Sci. J. 2017, 88, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, J.B.; Cooke, R.F.; Brandão, A.P.; Schubach, K.M.; Colombo, E.A.; Daigle, C.L.; Duff, G.C.; Gouvêa, V.N. Impacts of commingling cattle from different sources on their physiological, health, and performance responses during feedlot receiving. Transl. Anim. Sci. 2020, 4, txaa204. [Google Scholar] [CrossRef]
- Pratelli, A.; Cirone, F.; Capozza, P.; Trotta, A.; Corrente, M.; Balestrieri, A.; Buonavoglia, C. Bovine respiratory disease in beef calves supported long transport stress: An epidemiological study and strategies for control and prevention. Res. Vet. Sci. 2021, 135, 450–455. [Google Scholar] [CrossRef]
- Zhao, H.; Tang, X.; Wu, M.; Li, Q.; Yi, X.; Liu, S.; Jiang, J.; Wang, S.; Sun, X. Transcriptome Characterization of Short Distance Transport Stress in Beef Cattle Blood. Front. Genet. 2021, 12, 124. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 11, R106. [Google Scholar]
- Jiao, X.; Sherman, B.T.; Huang, D.W.; Stephens, R.M.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID-WS: A stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012, 28, 1805–1806. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Delić, N.; Aleksić, S.; Petrović, M.M.; Pantelić, V.; Ostojić Andrić, D.; Petričević, M.; Nikšić, D. Methods for determining stress syndrome in beef cattle and its relevance to quality of meat. Biotechnol. Anim. Husb. 2014, 30, 37–44. [Google Scholar] [CrossRef]
- Marti, S.; Wilde, R.E.; Moya, D.; Heuston, C.E.M.; Brown, F.; Schwartzkopf-Genswein, K.S. Effect of rest stop duration during long-distance transport on welfare indicators in recently weaned beef calves. J. Anim. Sci. 2017, 95, 636–644. [Google Scholar] [CrossRef]
- Padalino, B.; Raidal, S.; Carter, N.; Celi, P.; Muscatello, G.; Jeffcott, L.; de Silva, K. Immunological, clinical, haematological and oxidative responses to long distance transportation in horses. Res. Vet. Sci. 2017, 115, 78–87. [Google Scholar] [CrossRef]
- Miranda-de la Lama, G.C.; Rodríguez-Palomares, M.; Cruz-Monterrosa, R.G.; Rayas-Amor, A.A.; Pinheiro, R.S.B.; Galindo, F.M.; Villarroel, M. Long-distance transport of hair lambs: Effect of location in pot-belly trailers on thermo-physiology, welfare and meat quality. Trop. Anim. Health Prod. 2018, 50, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Liao, Y.; Sun, B.; Guo, Y.; Deng, M.; Li, Y.; Liu, D. Effects of chronic heat stress on mRNA and miRNA expressions in dairy cows. Gene 2020, 742, 144550. [Google Scholar] [CrossRef]
- Lee, J.; Lee, S.; Son, J.; Lim, H.; Kim, E.; Kim, D.; Ha, S.; Hur, T.; Lee, S.; Choi, I. Analysis of circulating-microRNA expression in lactating Holstein cows under summer heat stress. PLoS ONE 2020, 15, e0231125. [Google Scholar] [CrossRef]
- Clariget, J.; Banchero, G.; Luzardo, S.; Fernández, E.; Pérez, E.; La Manna, A.; Saravia, A.; del Campo, M.; Ferrés, A.; Canozzi, M.E.A. Effect of pre-slaughter fasting duration on physiology, carcass and meat quality in beef cattle finished on pastures or feedlot. Res. Vet. Sci. 2021, 136, 158–165. [Google Scholar] [CrossRef]
- Meléndez, D.M.; Marti, S.; Haley, D.B.; Schwinghamer, T.D.; Schwartzkopf-Genswein, K.S. Effects of conditioning, source, and rest on indicators of stress in beef cattle transported by road. PLoS ONE 2021, 16, e0244854. [Google Scholar] [CrossRef]
- Ak, P.; Levine, A.J. p53 and NF-κB: Different strategies for responding to stress lead to a functional antagonism. FASEB J. 2010, 24, 3643–3652. [Google Scholar] [CrossRef]
- Scagnolari, C.; Zingariello, P.; Vecchiet, J.; Selvaggi, C.; Racciatti, D.; Taliani, G.; Riva, E.; Pizzigallo, E.; Antonelli, G. Differential expression of interferon-induced microRNAs in patients with chronic hepatitis C virus infection treated with pegylated interferon alpha. Virol. J. 2010, 7, 311. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Sharp, P.A. MicroRNA functions in stress responses. Mol. Cell. 2010, 40, 205–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehry, M.R. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J. Immunol. 1996, 156, 2345–2348. [Google Scholar] [PubMed]
- Vincent-Fabert, C.; Saintamand, A.; David, A.; Alizadeh, M.; Boyer, F.; Arnaud, N.; Zimber-Strobl, U.; Feuillard, J.; Faumont, N. Reproducing indolent B-cell lymphoma transformation with T-cell immunosuppression in LMP1/CD40-expressing mice. Cell. Mol. Immunol. 2019, 16, 412–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schurmans, S.; Pouillon, V.; Maréchal, Y. Regulation of B cell survival, development and function by inositol 1, 4, 5-trisphosphate 3-kinase B (Itpkb). Adv. Enzyme Regul. 2011, 51, 66. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.T.; Sandberg, M.; Huang, Y.; Young, M.; Sutton, S.; Sauer, K.; Cooke, M.P. Production of Ins (1, 3, 4, 5) P 4 mediated by the kinase Itpkb inhibits store-operated calcium channels and regulates B cell selection and activation. Nat. Immunol. 2007, 8, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Salunkhe, S.; Vaidya, T. CD40-miRNA axis controls prospective cell fate determinants during B cell differentiation. Mol. Immunol. 2020, 126, 46–55. [Google Scholar] [CrossRef] [PubMed]


| Gene Symbol | Sequence (5′-3′) | Accession |
|---|---|---|
| bta-miR-33a-F | CGGTGCATTGTAGTTGCATTGCA | NR_031206.1 |
| bta-miR-138-F | AGCTGGTGTTGTGAATCAGGC | NR_030863.1 |
| bta-miR-504-F | AGACCCTGGTCTGCACTC | NR_037357.1 |
| bta-miR-2285ab-F | CGCGAAAACCTGAATGAACTTCTTGG | NR_107841.1 |
| bta-miR-2484-F | CGGAGCTATGATGACTTTGATTGCAT | NR_031273.1 |
| bta-miR-2285f-F | CGCGAAAACCTGAATGAACTTTTTGG | NR_107730.1 |
| bta-miR-2285r-F | CGCAGAAACCTGGATGAACTTTTTGG | NR_107819.1 |
| bta-miR-18b-F | CGCTAAGGTGCATCTAGTGCAGTTA | NR_031266.1 |
| bta-miR-19a-F | CGCTGTGCAAATCTATGCAAAACTGA | NR_030794.1 |
| bta-miR-18a-F | CGCTAAGGTGCATCTAGTGCAGATA | NR_030892.1 |
| bta-miR-25-F | CATTGCACTTGTCTCGGTCTGA | NR_030941.1 |
| bta-miR-185-F | TGGAGAGAAAGGCAGTTCCTGA | NR_031178.1 |
| Samples | Total Reads | Clean Reads (%) | Average Length | Q20 | Q30 | GC Percentage |
|---|---|---|---|---|---|---|
| B1 | 14,094,654 | 11,203,765 (79.49) | 23.58 | 97.98% | 96.82% | 48.15% |
| B2 | 13,007,575 | 11,220,138 (86.26) | 23.38 | 98.06% | 97.07% | 46.54% |
| B3 | 17,654,231 | 14,020,104 (79.41) | 24.30 | 98.11% | 97.13% | 46.59% |
| B4 | 18,920,754 | 15,296,531 (80.85) | 24.14 | 98.13% | 97.16% | 45.83% |
| B5 | 12,983,637 | 10,692,413 (82.35) | 24.29 | 98.00% | 96.92% | 46.88% |
| B6 | 15,186,129 | 12,341,422 (81.27) | 23.67 | 98.11% | 97.15% | 45.81% |
| B7 | 16,294,446 | 14,284,501 (87.66) | 23.38 | 98.17% | 97.21% | 44.80% |
| A1 | 16,721,024 | 14,557,475 (87.06) | 23.08 | 98.14% | 97.16% | 45.85% |
| A2 | 15,938,206 | 14,048,794 (88.15) | 23.01 | 98.12% | 97.15% | 45.79% |
| A3 | 18,044,894 | 16,096,861 (89.20) | 22.63 | 98.02% | 97.03% | 45.50% |
| A4 | 11,003,312 | 9,269,728 (84.24) | 23.22 | 98.03% | 97.04% | 47.14% |
| A5 | 13,898,637 | 12,190,422 (87.71) | 23.24 | 98.09% | 97.07% | 45.90% |
| A6 | 11,140,389 | 9,773,101 (87.73) | 22.50 | 98.07% | 97.10% | 45.66% |
| A7 | 11,160,148 | 9,815,072 (87.95) | 23.36 | 98.08% | 97.08% | 46.24% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Tang, X.; Raza, S.H.A.; Zhao, H.; Li, Q.; Yi, X.; Althobaiti, F.; Shukry, M.; Sun, X. Small RNA-Seq Analysis Reveals miRNA Expression of Short Distance Transportation Stress in Beef Cattle Blood. Animals 2021, 11, 2850. https://doi.org/10.3390/ani11102850
Wu M, Tang X, Raza SHA, Zhao H, Li Q, Yi X, Althobaiti F, Shukry M, Sun X. Small RNA-Seq Analysis Reveals miRNA Expression of Short Distance Transportation Stress in Beef Cattle Blood. Animals. 2021; 11(10):2850. https://doi.org/10.3390/ani11102850
Chicago/Turabian StyleWu, Mingli, Xiaoqin Tang, Sayed Haidar Abbas Raza, Haidong Zhao, Qi Li, Xiaohua Yi, Fayez Althobaiti, Mustafa Shukry, and Xiuzhu Sun. 2021. "Small RNA-Seq Analysis Reveals miRNA Expression of Short Distance Transportation Stress in Beef Cattle Blood" Animals 11, no. 10: 2850. https://doi.org/10.3390/ani11102850
APA StyleWu, M., Tang, X., Raza, S. H. A., Zhao, H., Li, Q., Yi, X., Althobaiti, F., Shukry, M., & Sun, X. (2021). Small RNA-Seq Analysis Reveals miRNA Expression of Short Distance Transportation Stress in Beef Cattle Blood. Animals, 11(10), 2850. https://doi.org/10.3390/ani11102850








