De Novo Transcriptome Analysis Reveals Potential Thermal Adaptation Mechanisms in the Cicada Hyalessa fuscata
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Thermal Experiments
2.2. TruSeq mRNA Library Construction and Illumina Sequencing
2.3. De novo Transcriptome Assembly
2.4. Identification of Differentially Expressed Genes
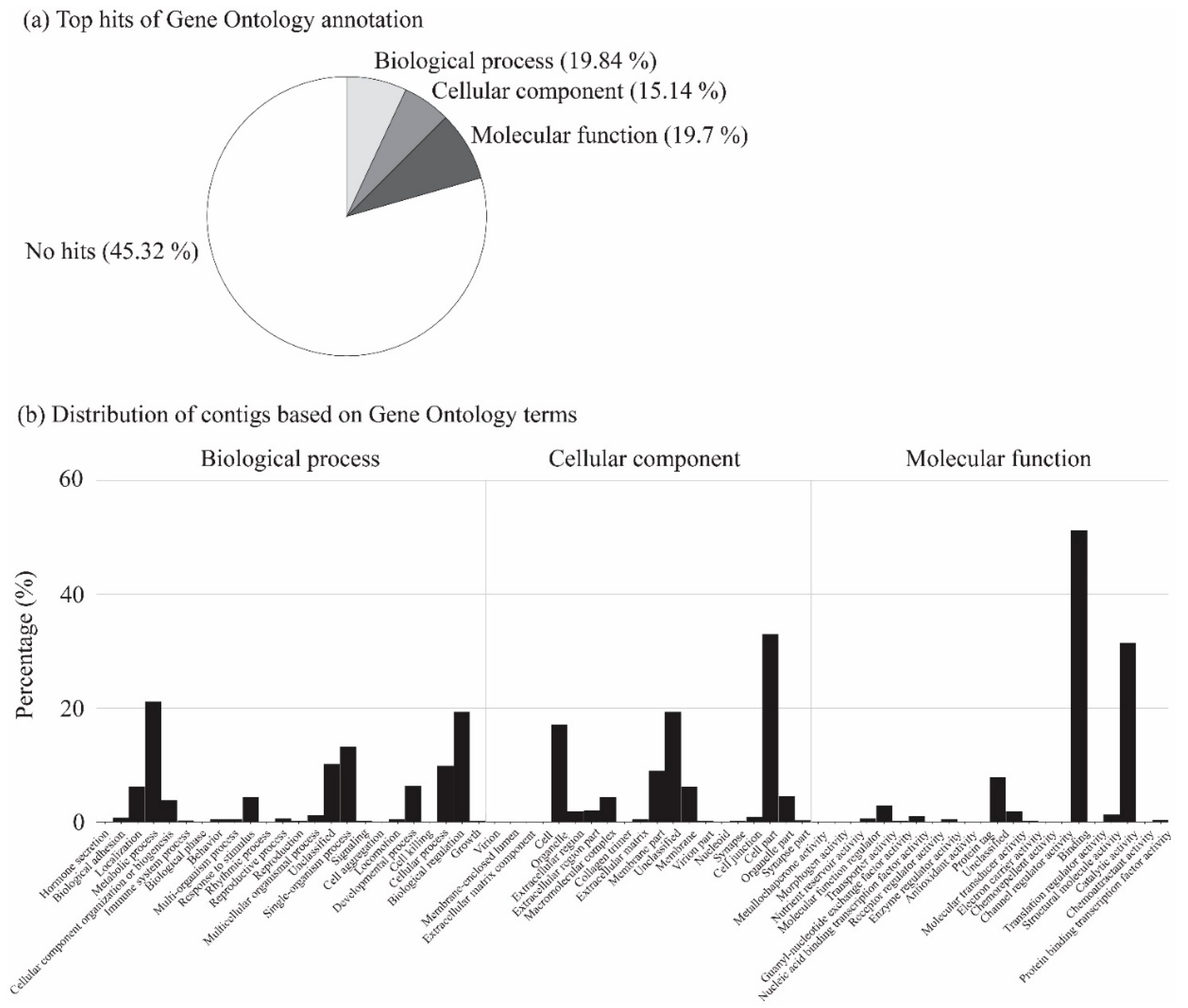
2.5. BLAST and Gene Ontology Annotations
3. Results
3.1. The Heat Stress Experiment
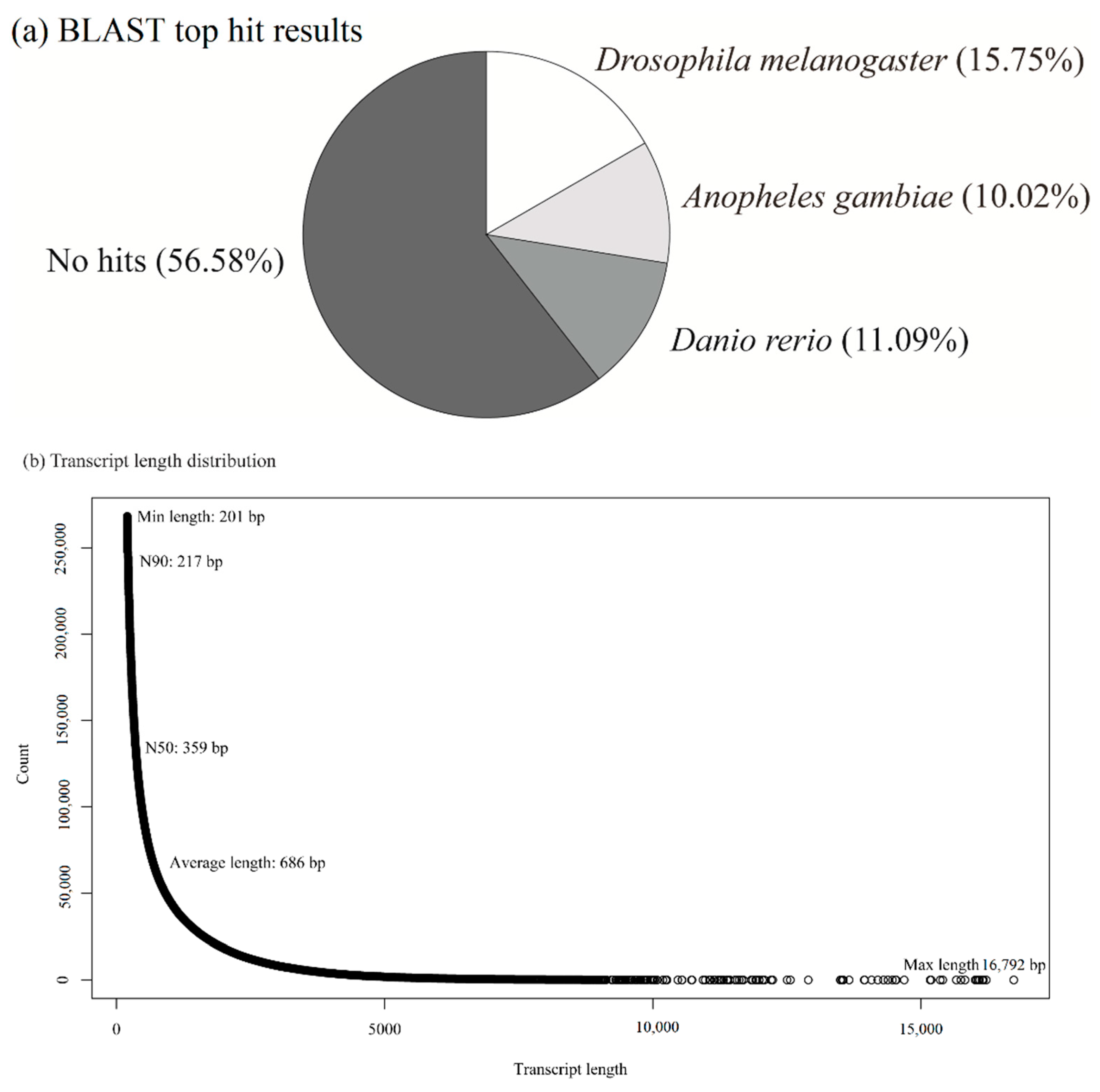
3.2. Illumina Sequencing, De Novo Assembly, and RNA-Seq Mapping
3.3. Responses at Normal Conditions
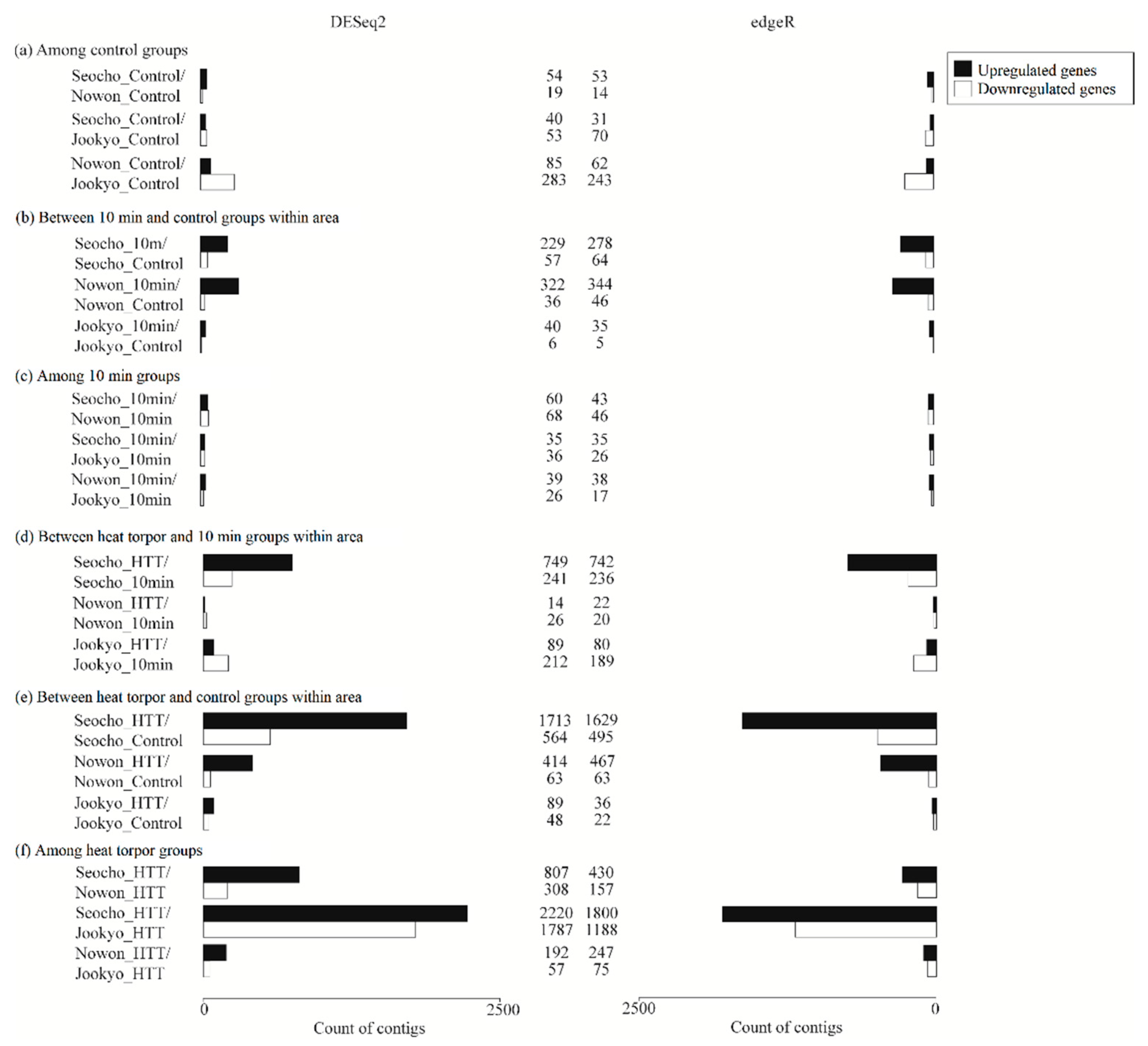
Among Control Groups
3.4. Acute Heat Stress Responses
3.4.1. Between 10 min and Control Groups from the Same Area
3.4.2. Among 10 min Groups
3.5. Heat Torpor Responses
3.5.1. Between Heat Torpor and 10 min Groups from the same Area
3.5.2. Between Heat Torpor and Control Groups from the Same Area
3.5.3. Among Heat Torpor Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denlinger, D.L.; Yocum, G.D. Physiology of heat sensitivity. In Temperature Sensitivity in Insects and Application in Integrated Pest Management; Hallman, G.J., Denlinger, D.L., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 7–53. [Google Scholar]
- Moriyama, M.; Numata, H. A cicada that ensures its fitness during climate warming by synchronizing its hatching time with the rainy season. Zool. Sci. 2011, 28, 875–881. [Google Scholar] [CrossRef]
- Sato, Y.; Sato, S. Spring temperature predicts the long-term molting phenology of two cicadas, Cryptotympana facialis and Graptopsaltria nigrofuscata (Hemiptera: Cicadidae). Ann. Entomol. Soc. Am. 2015, 108, 494–500. [Google Scholar] [CrossRef]
- Toolson, E.C. Comparative thermal physiological ecology of syntopic populations of Cacama valvata and Tibicen bifidus (Homoptera: Cicadidae): Modeling fitness consequences of temperature variation. Am. Zool. 1998, 38, 568–582. [Google Scholar] [CrossRef]
- Sanborn, A.F.; Phillips, P.K. Thermal responses of the Diceroprocta cinctifera species group (Homoptera: Cicadidae). Southwest. Nat. 1996, 41, 136–139. [Google Scholar]
- Schowalter, T.D. Insect Ecology: An Ecosystem Approach; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Gleason, L.U.; Burton, R.S. RNA-seq reveals regional differences in transcriptome response to heat stress in the marine snail Chlorostoma Funebralis. Mol. Ecol. 2015, 24, 610–627. [Google Scholar] [CrossRef]
- Jesus, T.F.; Grosso, A.R.; Almeida-Val, V.M.F.; Coelho, M.M. Transcriptome profiling of two Iberian freshwater fish exposed to thermal stress. J. Therm. Biol. 2016, 55, 54–61. [Google Scholar] [CrossRef]
- Lang, R.P.; Bayne, C.J.; Camara, M.D.; Cunningham, C.; Jenny, M.J.; Langdon, C.J. Transcriptome profiling of selectively bred Pacific oyster Crassostrea gigas families that differ in tolerance of heat shock. Mar. Biotechnol. 2009, 11, 650–668. [Google Scholar] [CrossRef]
- Liu, Y.; Su, H.; Li, R.; Li, X.; Xu, Y.; Dai, X.; Zhou, Y.; Wang, H. Comparative transcriptome analysis of Glyphodes pyloalis Walker (Lepidoptera: Pyralidae) reveals novel insights into heat stress tolerance in insects. BMC Genom. 2017, 18, 974. [Google Scholar] [CrossRef]
- Wang, W.; Hui, J.H.; Chan, T.F.; Chu, K.H. De Novo transcriptome sequencing of the snail Echinolittorina malaccana: Identification of genes responsive to thermal stress and development of genetic markers for population studies. Mar. Biotechnol. 2014, 16, 547–559. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Chen, B.; Wei, J.-N.; Liu, T.-X. Roles of thermal adaptation and chemical ecology in Liriomyza distribution and control. Annu. Rev. Entomol. 2009, 54, 127–145. [Google Scholar] [CrossRef]
- Klepsatel, P.; Gáliková, M.; Xu, Y.; Kühnlein, R.P. Thermal stress depletes energy reserves in Drosophila. Sci. Rep. 2016, 6, 33667. [Google Scholar] [CrossRef]
- Lindquist, S. The heat-shock response. Annu. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef]
- Chown, S.L.; Nicolson, S. Insect Physiological Ecology: Mechanisms and Patterns; Oxford University Press: Oxford, UK, 2004. [Google Scholar]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Parsell, D.; Lindquist, S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu. Rev. Genet. 1993, 27, 437–496. [Google Scholar] [CrossRef]
- Wang, L.; Yang, S.; Zhao, K.; Han, L. Expression profiles of the heat shock protein 70 gene in response to heat stress in Agrotis c-nigrum (Lepidoptera: Noctuidae). J. Insect Sci. 2015, 15, 9. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bukau, B.; Horwich, A.L. The Hsp70 and Hsp60 chaperone machines. Cell 1998, 92, 351–366. [Google Scholar] [CrossRef]
- Bukau, B.; Deuerling, E.; Pfund, C.; Craig, E.A. Getting newly synthesized proteins into shape. Cell 2000, 101, 119–122. [Google Scholar] [CrossRef]
- Sørensen, J.G. Application of heat shock protein expression for detecting natural adaptation and exposure to stress in natural populations. Curr. Zool. 2010, 56, 703–713. [Google Scholar] [CrossRef]
- Chidawanyika, F.; Terblanche, J.S. Rapid thermal responses and thermal tolerance in adult codling moth Cydia pomonella (Lepidoptera: Tortricidae). J. Insect Physiol. 2011, 57, 108–117. [Google Scholar] [CrossRef]
- Gilchrist, G.W.; Huey, R.B.; Partridge, L. Thermal sensitivity of Drosophila melanogaster: Evolutionary responses of adults and eggs to laboratory natural selection at different temperatures. Physiol. Zool. 1997, 70, 403–414. [Google Scholar] [CrossRef]
- Krebs, R.A.; Feder, M.E. Natural variation in the expression of the heat-shock protein HSP70 in a population of Drosophila melanogaster and its correlation with tolerance of ecologically relevant thermal stress. Evolution 1997, 51, 173–179. [Google Scholar] [CrossRef]
- McColl, G.; Hoffmann, A.A.; McKechnie, S.W. Response of two heat shock genes to selection for knockdown heat resistance in Drosophila melanogaster. Genetics 1996, 143, 1615–1627. [Google Scholar] [CrossRef]
- Gehring, W.J.; Wehner, R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara Desert. Proc. Natl. Acad. Sci. USA 1995, 92, 2994–2998. [Google Scholar] [CrossRef]
- Wang, H.; Dong, S.-Z.; Li, K.; Hu, C.; Ye, G.-Y. A heat shock cognate 70 gene in the endoparasitoid, Pteromalus puparum, and its expression in relation to thermal stress. BMB Rep. 2008, 41, 388–393. [Google Scholar] [CrossRef]
- Joanisse, D.; Storey, K. Oxidative stress and antioxidants in overwintering larvae of cold-hardy goldenrod gall insects. J. Exp. Biol. 1996, 199, 1483–1491. [Google Scholar] [CrossRef]
- Lalouette, L.; Williams, C.; Hervant, F.; Sinclair, B.J.; Renault, D. Metabolic rate and oxidative stress in insects exposed to low temperature thermal fluctuations. Comp. Biochem. Physiol. Mol. Integr. Physiol. 2011, 158, 229–234. [Google Scholar] [CrossRef]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef]
- Monaghan, P.; Metcalfe, N.B.; Torres, R. Oxidative stress as a mediator of life history trade-offs: Mechanisms, measurements and interpretation. Ecol. Lett. 2009, 12, 75–92. [Google Scholar] [CrossRef]
- Howe, G.A.; Schilmiller, A.L. Oxylipin metabolism in response to stress. Curr. Opin. Plant. Biol. 2002, 5, 230–236. [Google Scholar] [CrossRef]
- Gelain, D.P.; Dalmolin, R.J.S.; Belau, V.L.; Moreira, J.C.F.; Klamt, F.; Castro, M.A. A systematic review of human antioxidant genes. Front. Biosci. 2009, 14, 4457–4463. [Google Scholar] [CrossRef]
- Cui, Y.; Du, Y.; Lu, M.; Qiang, C. Antioxidant responses of Chilo suppressalis (Lepidoptera: Pyralidae) larvae exposed to thermal stress. J. Therm. Biol. 2011, 36, 292–297. [Google Scholar] [CrossRef]
- Sokolova, I.M. Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr. Comp. Biol. 2013, 53, 597–608. [Google Scholar] [CrossRef]
- Calow, P. Physiological costs of combating chemical toxicants: Ecological implications. Comp. Biochem. Physiol. Comp. Pharmacol. 1991, 100, 3–6. [Google Scholar] [CrossRef]
- Harada, E.; Goto, S.G. Upregulation of heat-shock proteins in larvae, but not adults, of the flesh fly during hot summer days. Cell Stress Chaperones 2017, 22, 823–831. [Google Scholar] [CrossRef]
- Hoffmann, A.A. Acclimation: Increasing survival at a cost. Trends Ecol. Evol. 1995, 10, 1–2. [Google Scholar]
- Koehn, R.K.; Bayne, B.L. Towards a physiological and genetical understanding of the energetics of the stress response. Biol. J. Linn. Soc. 1989, 37, 157–171. [Google Scholar] [CrossRef]
- Kültz, D. Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 2005, 67, 225–257. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 2007, 87, 99–163. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Andersen, D.K.; Kim, Y.; Jang, Y. Urban heat island effect on cicada densities in metropolitan Seoul. PeerJ 2018, 6, e4238. [Google Scholar] [CrossRef] [PubMed]
- Oke, T.R. City size and the urban heat island. Atmos. Environ. 1973, 7, 769–779. [Google Scholar] [CrossRef]
- Nguyen, H.Q.; Serret, H.; Bae, Y.; Ji, S.; Chae, S.; Kim, Y.I.; Ha, J.; Jang, Y. Not all cicadas increase thermal tolerance in response to a temperature gradient in metropolitan Seoul. Sci. Rep. 2020, 10, 1–12. [Google Scholar]
- Sanborn, A.F.; Heath, J.E.; Heath, M.S.; Phillips, P.K. Thermal adaptation in North American cicadas (Hemiptera: Cicadidae). J. Therm. Biol. 2017, 69, V-XVIII. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Da Lage, J.-L.; Renard, E.; Chartois, F.; Lemeunier, F.; Cariou, M.-L. Amyrel, a paralogous gene of the amylase gene family in Drosophila melanogaster and the Sophophora subgenus. Proc. Natl. Acad. Sci. USA 1998, 95, 6848–6853. [Google Scholar] [CrossRef] [PubMed]
- Maczkowiak, F.; Da Lage, J.-L. Origin and evolution of the Amyrel gene in the α-amylase multigene family of Diptera. Genetica 2006, 128, 145–158. [Google Scholar] [CrossRef]
- Hull-Thompson, J.; Muffat, J.; Sanchez, D.; Walker, D.W.; Benzer, S.; Ganfornina, M.D.; Jasper, H. Control of metabolic homeostasis by stress signaling is mediated by the lipocalin NLaz. PLoS Genet. 2009, 5, e1000460. [Google Scholar] [CrossRef]
- Ruiz, M.; Wicker-Thomas, C.; Sanchez, D.; Ganfornina, M.D. Grasshopper Lazarillo, a GPI-anchored Lipocalin, increases Drosophila longevity and stress resistance, and functionally replaces its secreted homolog NLaz. Insect Biochem. Mol. Biol. 2012, 42, 776–789. [Google Scholar] [CrossRef]
- Ruiz, M.; Sanchez, D.; Correnti, C.; Strong, R.K.; Ganfornina, M.D. Lipid-binding properties of human ApoD and Lazarillo-related lipocalins: Functional implications for cell differentiation. FEBS J. 2013, 280, 3928–3943. [Google Scholar] [CrossRef]
- Ruiz, M.; Ganfornina, M.D.; Correnti, C.; Strong, R.K.; Sanchez, D. Ligand binding-dependent functions of the lipocalin NLaz: An in vivo study in Drosophila. FASEB J. 2014, 28, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Baudry, J.; Berenbaum, M.R.; Schuler, M.A. Structural and functional divergence of insect CYP6B proteins: From specialist to generalist cytochrome P450. Proc. Natl. Acad. Sci. USA 2004, 101, 2939–2944. [Google Scholar] [CrossRef]
- Radyuk, S.N.; Klichko, V.I.; Spinola, B.; Sohal, R.S.; Orr, W.C. The peroxiredoxin gene family in Drosophila melanogaster. Free Radic. Biol. Med. 2001, 31, 1090–1100. [Google Scholar] [CrossRef]
- Schulte, J. Peroxiredoxin 4: A multifunctional biomarker worthy of further exploration. BMC Med. 2011, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Schulte, J.; Struck, J.; Köhrle, J.; Müller, B. Circulating levels of peroxiredoxin 4 as a novel biomarker of oxidative stress in patients with sepsis. Shock 2011, 35, 460–465. [Google Scholar] [CrossRef]
- Enayati, A.A.; Ranson, H.; Hemingway, J. Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 2005, 14, 3–8. [Google Scholar] [CrossRef]
- Usuki, K.; Saras, J.; Waltenberger, J.; Miyazono, K.; Pierce, G.; Thomason, A.; Heldin, C.-H. Platelet-derived endothelial cell growth factor has thymidine phosphorylase activity. Biochem. Biophys. Res. Commun. 1992, 184, 1311–1316. [Google Scholar] [CrossRef]
- Wheeler, M.M.; Robinson, G.E. Diet-dependent gene expression in honey bees: Honey vs. sucrose or high fructose corn syrup. Sci. Rep. 2014, 4, 5726. [Google Scholar] [CrossRef]
- Goodsell, D.S. The molecular perspective: The ras oncogene. Stem Cells 1999, 17, 235–236. [Google Scholar] [CrossRef]
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 1–23. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- Vlahopoulos, S.; Zoumpourlis, V. JNK: A key modulator of intracellular signaling. Biochemistry 2004, 69, 844–854. [Google Scholar] [CrossRef]
- Miao, Y.; Tenor, J.L.; Toffaletti, D.L.; Maskarinec, S.A.; Liu, J.; Lee, R.E.; Perfect, J.R.; Brennan, R.G. Structural and in vivo studies on Trehalose-6-phosphate synthase from pathogenic fungi provide insights into its catalytic mechanism, biological necessity, and potential for novel antifungal drug design. mBio 2017, 8, e00643-17. [Google Scholar] [CrossRef]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant. Sci. 2011, 2, 70. [Google Scholar] [CrossRef]
- Yang, H.-L.; Liu, Y.-J.; Wang, C.-L.; Zeng, Q.-Y. Molecular evolution of trehalose-6-phosphate synthase (TPS) gene family in Populus, Arabidopsis and rice. PLoS ONE 2012, 7, e42438. [Google Scholar] [CrossRef]
- Kultz, D. Evolution of the cellular stress proteome: From monophyletic origin to ubiquitous function. J. Exp. Biol. 2003, 206, 3119–3124. [Google Scholar] [CrossRef] [PubMed]
- Denlinger, D.L.; Rinehart, J.P.; Yocum, G.D. Stress proteins: A role in insect diapause? In Insect Timing: Circadian Rhythmicity to Seasonality; Elsevier: Amsterdam, The Netherlands, 2001; pp. 155–171. [Google Scholar]
- Harrison, J.F.; Woods, H.A.; Roberts, S.P. Ecological and Environmental Physiology of Insects; Oxford University Press: Oxford, UK, 2012; Volume 3. [Google Scholar]
- Qiao, L.; Wu, J.X.; Qin, D.Z.; Liu, X.C.; Lu, Z.C.; Lv, L.Z.; Pan, Z.L.; Chen, H.; Li, G.W. Gene expression profiles of heat shock proteins 70 and 90 from Empoasca onukii (Hemiptera: Cicadellidae) in response to temperature stress. J. Insect Sci. 2015, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Lu, M. Cloning of the heat shock protein 70 gene from Chilo suppressalis and the analysis of its expression characteristics under heat stress. Acta Entomol. Sin. 2010, 53, 841–848. [Google Scholar]
- Genevaux, P.; Georgopoulos, C.; Kelley, W.L. The Hsp70 chaperone machines of Escherichia coli: A paradigm for the repartition of chaperone functions. Mol. Microbiol. 2007, 66, 840–857. [Google Scholar] [CrossRef]
- Bimston, D.; Song, J.; Winchester, D.; Takayama, S.; Reed, J.C.; Morimoto, R.I. BAG-1, a negative regulator of Hsp70 chaperone activity, uncouples nucleotide hydrolysis from substrate release. EMBO J. 1998, 17, 6871–6878. [Google Scholar] [CrossRef]
- Gebauer, M.; Zeiner, M.; Gehring, U. Proteins interacting with the molecular chaperone hsp70/hsc70: Physical associations and effects on refolding activity. FEBS Lett. 1997, 417, 109–113. [Google Scholar] [CrossRef]
- Argaman, M.; Aly, R.; Shapira, M. Expression of heat shock protein 83 in Leishmania is regulated post-transcriptionally. Mol. Biochem. Parasitol. 1994, 64, 95–110. [Google Scholar] [CrossRef]
- van der Straten, A.; Rommel, C.; Dickson, B.; Hafen, E. The heat shock protein 83 (Hsp83) is required for Raf-mediated signalling in Drosophila. EMBO J. 1997, 16, 1961–1969. [Google Scholar] [CrossRef] [PubMed]
- Erlejman, A.G.; Lagadari, M.; Toneatto, J.; Piwien-Pilipuk, G.; Galigniana, M.D. Regulatory role of the 90-kDa-heat-shock protein (Hsp90) and associated factors on gene expression. Biochim. Biophys. Acta 2014, 1839, 71–87. [Google Scholar] [CrossRef]
- Knorr, E.; Vilcinskas, A. Post-embryonic functions of HSP90 in Tribolium castaneum include the regulation of compound eye development. Dev. Genes Evol. 2011, 221, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Parkhurst, S.M.; Halsell, S.R.; Lipshitz, H.D. Dynamic Hsp83 RNA localization during Drosophila oogenesis and embryogenesis. Mol. Cell. Biol. 1993, 13, 3773–3781. [Google Scholar] [CrossRef][Green Version]
- Pisa, V.; Cozzolino, M.; Gargiulo, S.; Ottone, C.; Piccioni, F.; Monti, M.; Gigliotti, S.; Talamo, F.; Graziani, F.; Pucci, P. The molecular chaperone Hsp90 is a component of the cap-binding complex and interacts with the translational repressor Cup during Drosophila oogenesis. Gene 2009, 432, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Fee, L.; Lee, T.H.; Wharton, R.P. The molecular chaperone Hsp90 is required for mRNA localization in Drosophila melanogaster embryos. Genetics 2007, 176, 2213–2222. [Google Scholar] [CrossRef]
- Yue, L.; Karr, T.L.; Nathan, D.F.; Swift, H.; Srinivasan, S.; Lindquist, S. Genetic analysis of viable Hsp90 alleles reveals a critical role in Drosophila spermatogenesis. Genetics 1999, 151, 1065–1079. [Google Scholar] [CrossRef]
- Atkinson, B. Changes in Eukaryotic Gene Expression in Response to Environmental Stress; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Krebs, R.A.; Feder, M.E. Hsp70 and larval thermotolerance in Drosophila melanogaster: How much is enough and when is more too much? J. Insect Physiol. 1998, 44, 1091–1101. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Sørensen, J.G.; Loeschcke, V. Adaptation of Drosophila to temperature extremes: Bringing together quantitative and molecular approaches. J. Therm. Biol. 2003, 28, 175–216. [Google Scholar] [CrossRef]
- Feder, J.H.; Rossi, J.M.; Solomon, J.; Solomon, N.; Lindquist, S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes Dev. 1992, 6, 1402–1413. [Google Scholar] [CrossRef]
- Heise, K.; Puntarulo, S.; Pörtner, H.-O.; Abele, D. Production of reactive oxygen species by isolated mitochondria of the Antarctic bivalve Laternula elliptica (King and Broderip) under heat stress. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2003, 134, 79–90. [Google Scholar] [CrossRef]
- Ryter, S.W.; Kim, H.P.; Hoetzel, A.; Park, J.W.; Nakahira, K.; Wang, X.; Choi, A.M. Mechanisms of cell death in oxidative stress. Antioxid. Redox Signal. 2007, 9, 49–89. [Google Scholar] [CrossRef]
- Dubovskiy, I.; Martemyanov, V.; Vorontsova, Y.; Rantala, M.; Gryzanova, E.; Glupov, V. Effect of bacterial infection on antioxidant activity and lipid peroxidation in the midgut of Galleria mellonella L. larvae (Lepidoptera, Pyralidae). Comp. Biochem. Physiol. Toxicol. Pharmacol. 2008, 148, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Felton, G.W.; Summers, C.B. Antioxidant systems in insects. Arch. Insect Biochem. Physiol. 1995, 29, 187–197. [Google Scholar] [CrossRef]
- Wang, Y.; Oberley, L.W.; Murhammer, D.W. Antioxidant defense systems of two lipidopteran insect cell lines. Free Radic. Biol. Med. 2001, 30, 1254–1262. [Google Scholar] [CrossRef]
- Forman, H.J.; Fisher, A.B. Antioxidant defenses. In Oxygen and Living Processes; Springer: New York, NY, USA, 1981; pp. 235–249. [Google Scholar]
- Storey, K.B. Oxidative stress: Animal adaptations in nature. Braz. J. Med. Biol. Res. 1996, 29, 1715–1733. [Google Scholar]
- Young, I.; Woodside, J. Antioxidants in health and disease. J. Clin. Pathol. 2001, 54, 176–186. [Google Scholar] [CrossRef]
- Jena, K.; Kar, P.K.; Kausar, Z.; Babu, C.S. Effects of temperature on modulation of oxidative stress and antioxidant defenses in testes of tropical tasar silkworm Antheraea mylitta. J. Therm. Biol. 2013, 38, 199–204. [Google Scholar] [CrossRef]
- Meng, J.-Y.; Zhang, C.-Y.; Zhu, F.; Wang, X.-P.; Lei, C.-L. Ultraviolet light-induced oxidative stress: Effects on antioxidant response of Helicoverpa armigera adults. J. Insect Physiol. 2009, 55, 588–592. [Google Scholar] [CrossRef]
- Manevich, Y.; Fisher, A.B. Peroxiredoxin 6, a 1-Cys peroxiredoxin, functions in antioxidant defense and lung phospholipid metabolism. Free Radic. Biol. Med. 2005, 38, 1422–1432. [Google Scholar] [CrossRef]
- Yang, S.; Luo, A.; Hao, X.; Lai, Z.; Ding, T.; Ma, X.; Mayinuer, M.; Shen, W.; Wang, X.; Lu, Y. Peroxiredoxin 2 inhibits granulosa cell apoptosis during follicle atresia through the NFKB pathway in mice. Biol. Reprod. 2011, 84, 1182–1189. [Google Scholar] [CrossRef]
- Walker, D.W.; Muffat, J.; Rundel, C.; Benzer, S. Overexpression of a Drosophila homolog of apolipoprotein D leads to increased stress resistance and extended lifespan. Curr. Biol. 2006, 16, 674–679. [Google Scholar] [CrossRef]
- Zhang, Y.; Cong, Y.; Wang, S.; Zhang, S. Antioxidant activities of recombinant amphioxus (Branchiostoma belcheri) apolipoprotein D. Mol. Biol. Rep. 2011, 38, 1847–1851. [Google Scholar] [CrossRef]
- Ganfornina, M.D.; Do Carmo, S.; Lora, J.M.; Torres-Schumann, S.; Vogel, M.; Allhorn, M.; González, C.; Bastiani, M.J.; Rassart, E.; Sanchez, D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 2008, 7, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Creagh, E.; Carmody, R.; Cotter, T. Heat shock protein 70 inhibits caspase-dependent and-independent apoptosis in Jurkat T cells. Exp. Cell Res. 2000, 257, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.S.; Csermely, P. Heat shock proteins in the regulation of apoptosis: New strategies in tumor therapy: A comprehensive review. Pharmacol. Ther. 2004, 101, 227–257. [Google Scholar] [CrossRef]
- Hill, J.O.; Wyatt, H.R.; Peters, J.C. Energy balance and obesity. Circulation 2012, 126, 126–132. [Google Scholar] [CrossRef]
- Hawkins, A. Protein turnover: A functional appraisal. Funct. Ecol. 1991, 5, 222–233. [Google Scholar] [CrossRef]
- Hawkins, A.J.; Day, A.J. The metabolic basis of genetic differences in growth efficiency among marine animals. J. Exp. Mar. Biol. Ecol. 1996, 203, 93–115. [Google Scholar] [CrossRef]
- Hawkins, A.; Wilson, I.; Bayne, B. Thermal responses reflect protein turnover in Mytilus edulis L. Funct. Ecol. 1987, 1, 339–351. [Google Scholar] [CrossRef]
- Hawkins, A.J.; Rusin, J.; Bayne, B.L.; Day, A.J. The metabolic/physiological basis of genotype-dependent mortality during copper exposure in Mytilus Edulis. Mar. Environ. Res. 1989, 28, 253–257. [Google Scholar] [CrossRef]
- López-Maury, L.; Marguerat, S.; Bähler, J. Tuning gene expression to changing environments: From rapid responses to evolutionary adaptation. Nat. Rev. Genet. 2008, 9, 583–593. [Google Scholar] [CrossRef]
- Buckley, B.A.; Gracey, A.Y.; Somero, G.N. The cellular response to heat stress in the goby Gillichthys mirabilis: A cDNA microarray and protein-level analysis. J. Exp. Biol. 2006, 209, 2660–2677. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.V. Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 2000, 12, 13–19. [Google Scholar] [CrossRef]
- Blum, J.S.; Wearsch, P.A.; Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef]
- Boullier, A.; Bird, D.A.; Chang, M.-K.; Dennis, E.A.; Friedman, P.; Gillotte-Taylor, K.; Hörkkö, S.; Palinski, W.; Quehenberger, O.; Shaw, P. Scavenger receptors, oxidized LDL, and atherosclerosis. Ann. N. Y. Acad. Sci. 2001, 947, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.S.; Pardhasaradhi, B.V.V.; Khar, A.; Srinivas, U.K. A cross talk between cellular signalling and cellular redox state during heat-induced apoptosis in a rat histiocytoma. Free Radic. Biol. Med. 2002, 32, 221–227. [Google Scholar] [CrossRef]
- Yi, S.-X.; Lee, R.E. Rapid cold-hardening blocks cold-induced apoptosis by inhibiting the activation of pro-caspases in the flesh fly Sarcophaga crassipalpis. Apoptosis 2011, 16, 249–255. [Google Scholar] [CrossRef]
- Verheij, M.; Ruiter, G.A.; Zerp, S.F.; van Blitterswijk, W.J.; Fuks, Z.; Haimovitz-Friedman, A.; Bartelink, H. The role of the stress-activated protein kinase (SAPK/JNK) signaling pathway in radiation-induced apoptosis. Radiother. Oncol. 1998, 47, 225–232. [Google Scholar] [CrossRef]
- Cross, M.; Rajan, S.; Chekaiban, J.; Saunders, J.; Hamilton, C.; Kim, J.-S.; Coster, M.J.; Gasser, R.B.; Hofmann, A. Enzyme characteristics of pathogen-specific trehalose-6-phosphate phosphatases. Sci. Rep. 2017, 7, 2015. [Google Scholar] [CrossRef]
- Soto, T.; Fernández, J.; Vicente-Soler, J.; Cansado, J.; Gacto, M. Accumulation of trehalose by overexpression oftps1, coding for trehalose-6-phosphate synthase, causes increased resistance to multiple stresses in the fission yeast Schizosaccharomyces pombe. Appl. Environ. Microbiol. 1999, 65, 2020–2024. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, M.; Numata, H. Diapause and prolonged development in the embryo and their ecological significance in two cicadas, Cryptotympana facialis and Graptopsaltria nigrofuscata. J. Insect Physiol. 2008, 54, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- White, J.A.; Lloyd, M. Growth rates of 17 and 13-year periodical cicadas. Am. Midl. Nat. 1975, 94, 127–143. [Google Scholar] [CrossRef]
| Total Number of Common Genes | Heat Shock Protein | Cell Repair | Metabolism | Antioxidant and Detoxification | Immune Response | Signal Transduction Pathway | Protein Turnover | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | Up | Down | |
| (a) Among control groups | ||||||||||||||||
| SC_control vs. NW_control | 46 | 13 | 0 | 0 | 0 | 0 | 3 | 1 | 2 | 2 | 2 | 1 | 0 | 0 | 1 | 0 |
| SC_control vs. JK_control | 27 | 27 | 0 | 1 | 0 | 0 | 4 | 2 | 0 | 5 | 0 | 2 | 3 | 0 | 1 | 0 |
| NW_control vs. JK_control | 60 | 192 | 0 | 0 | 0 | 1 | 5 | 6 | 2 | 7 | 1 | 1 | 1 | 2 | 0 | 6 |
| (b) Between 10 min and control groups within an area | ||||||||||||||||
| SC_10 min vs. SC_control | 209 | 53 | 0 | 1 | 0 | 0 | 18 | 5 | 12 | 1 | 2 | 1 | 0 | 0 | 1 | 0 |
| NW_10 min vs. NW_control | 291 | 29 | 0 | 0 | 0 | 1 | 38 | 1 | 10 | 0 | 2 | 0 | 6 | 1 | 4 | 0 |
| JK_10 min vs. JK_control | 30 | 4 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| (c) Among 10 min groups | ||||||||||||||||
| SC_10 min vs. NW_10min | 38 | 42 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 4 |
| SC_10 min vs. JK_10min | 28 | 24 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| NW_10 min vs. JK_10min | 33 | 14 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| (d) Between heat torpor and 10 min groups within an area | ||||||||||||||||
| SC_HTT vs. SC_10 min | 683 | 205 | 6 | 0 | 0 | 1 | 48 | 4 | 17 | 2 | 2 | 3 | 5 | 6 | 11 | 2 |
| NW_HTT vs. NW_10 min | 12 | 17 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| JK_HTT vs. JK_10 min | 54 | 168 | 0 | 1 | 2 | 1 | 5 | 9 | 0 | 5 | 1 | 0 | 0 | 2 | 1 | 1 |
| (e) Between heat torpor and control groups within an area | ||||||||||||||||
| SC_HTT vs. SC_control | 1466 | 426 | 10 | 0 | 2 | 1 | 128 | 11 | 62 | 8 | 14 | 5 | 72 | 25 | 42 | 13 |
| NW_HTT vs. NW_control | 355 | 53 | 0 | 0 | 1 | 0 | 57 | 1 | 23 | 0 | 1 | 0 | 6 | 0 | 3 | 0 |
| JK_HTT vs. JK_control | 32 | 19 | 0 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 |
| (f) Among heat torpor groups | ||||||||||||||||
| SC_HTT vs. NW_HTT | 385 | 116 | 0 | 0 | 0 | 0 | 7 | 6 | 10 | 4 | 2 | 3 | 3 | 13 | 4 | 7 |
| SC_HTT vs. JK_HTT | 1693 | 1088 | 2 | 3 | 0 | 0 | 99 | 45 | 64 | 18 | 3 | 19 | 17 | 62 | 12 | 50 |
| NW_HTT vs. JK_HTT | 168 | 38 | 0 | 0 | 0 | 2 | 7 | 2 | 12 | 0 | 1 | 1 | 1 | 2 | 3 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, H.Q.; Kim, Y.; Jang, Y. De Novo Transcriptome Analysis Reveals Potential Thermal Adaptation Mechanisms in the Cicada Hyalessa fuscata. Animals 2021, 11, 2785. https://doi.org/10.3390/ani11102785
Nguyen HQ, Kim Y, Jang Y. De Novo Transcriptome Analysis Reveals Potential Thermal Adaptation Mechanisms in the Cicada Hyalessa fuscata. Animals. 2021; 11(10):2785. https://doi.org/10.3390/ani11102785
Chicago/Turabian StyleNguyen, Hoa Quynh, Yuseob Kim, and Yikweon Jang. 2021. "De Novo Transcriptome Analysis Reveals Potential Thermal Adaptation Mechanisms in the Cicada Hyalessa fuscata" Animals 11, no. 10: 2785. https://doi.org/10.3390/ani11102785
APA StyleNguyen, H. Q., Kim, Y., & Jang, Y. (2021). De Novo Transcriptome Analysis Reveals Potential Thermal Adaptation Mechanisms in the Cicada Hyalessa fuscata. Animals, 11(10), 2785. https://doi.org/10.3390/ani11102785







