Transcriptome Analysis Provides Insights into Hepatic Responses to Trichloroisocyanuric Acid Exposure in Goldfish (Carassius auratus)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Chemicals
2.2. Experimental Design and Sampling
2.3. RNA Extraction and Transcriptome Sequencing
2.4. Bioinformatics Analysis
2.5. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.6. Determination of Antioxidant Enzyme Activities
2.7. Histopathological Analysis
3. Results
3.1. Transcriptome Sequence and Reads Mapping
3.2. Differential Expression Analysis and Functional Annotation
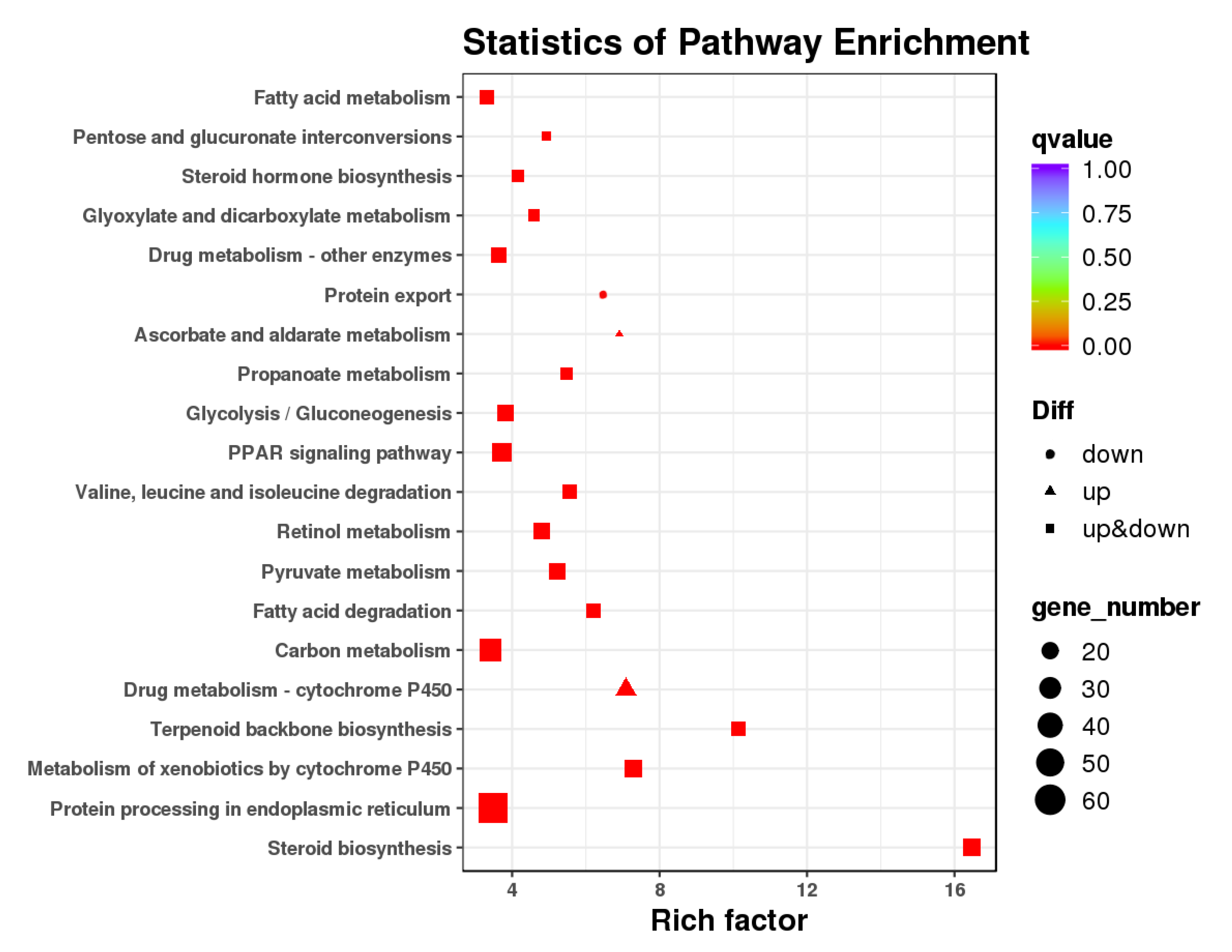
3.3. GO and KEGG Pathway Enrichment Analysis
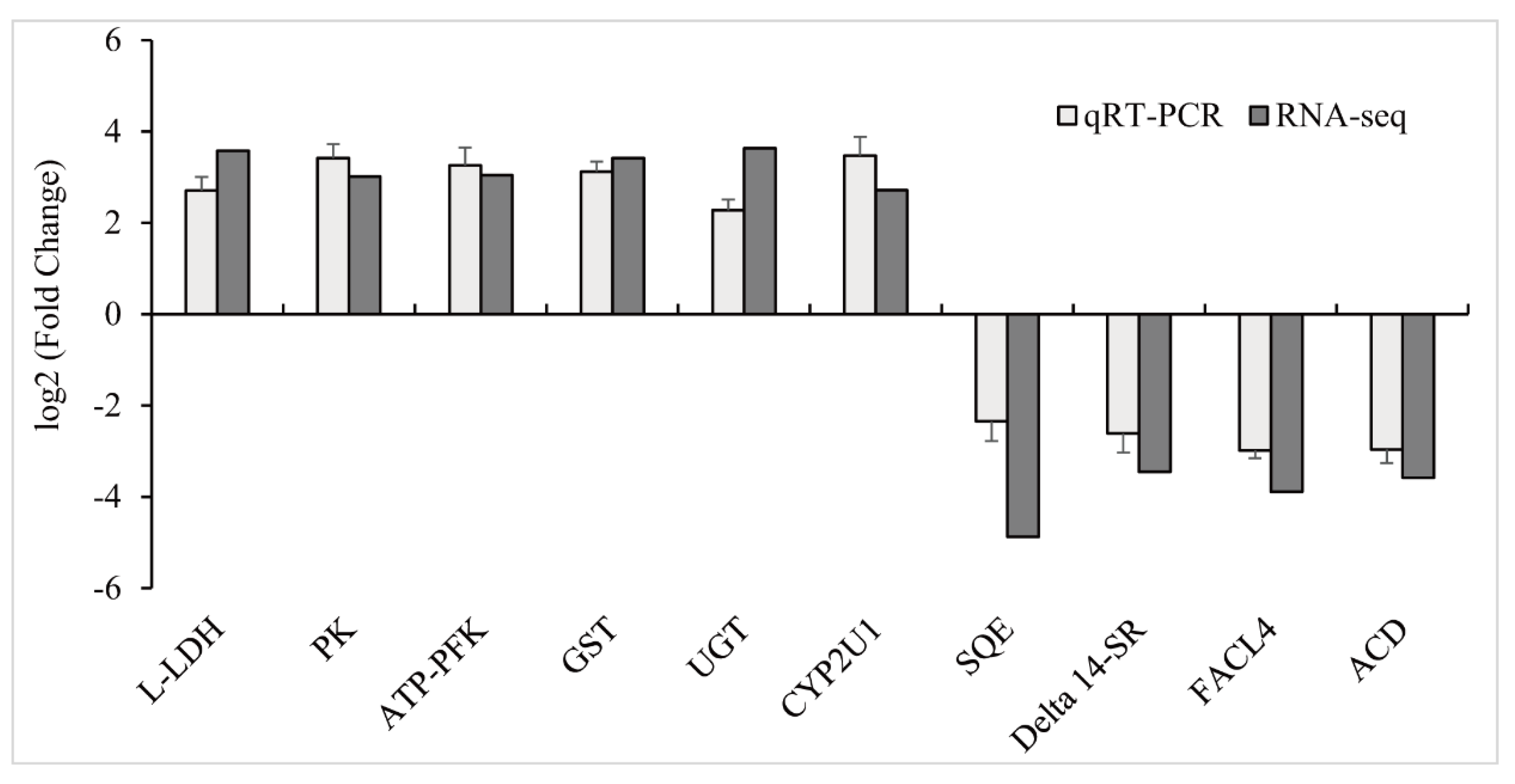
3.4. Validation of RNA-Seq Results with qPCR
3.5. Analysis of Antioxidant Enzyme Activities
3.6. Histopathological Analysis after Exposures to TCCA
4. Discussion
4.1. Effects of TCCA Exposure on Energy Metabolism
4.2. Effects of TCCA Exposure on Detoxification Responses
4.3. TCCA Exposure Induced Oxidative Damage in Liver
4.4. TCCA Exposure Induced Histopathological Damage in Gill
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ben-Asher, R.; Ravid, S.; Ucko, M.; Smirnov, M.; Lahav, O. Chlorine-based disinfection for controlling horizontal transmission of VNN in a seawater recirculating aquaculture system growing European seabass. Aquaculture 2019, 510, 329–336. [Google Scholar] [CrossRef]
- Tkachenko, H.; Kurhaluk, N.; Grudniewska, J. Biomarkers of oxidative stress and antioxidant defences as indicators of different disinfectants exposure in the heart of rainbow trout (Oncorhynchus Mykiss Walbaum). Aquac. Res. 2015, 46, 679–689. [Google Scholar] [CrossRef]
- Tilstam, U.; Weinmann, H. Trichloroisocyanuric Acid: A Safe and Efficient Oxidant. Org. Process. Res. Dev. 2002, 6, 384–393. [Google Scholar] [CrossRef]
- Pullar, J.M.; Vissers, M.C.M.; Winterbourn, C.C. Living with a killer: The effects of hypochlorous acid on mammalian cells. Iubmb Life 2000, 50, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, N.; Nakada, A.; Itoh, Y.; Watanabe, A.; Niki, E. Formation of Active Oxygen Species and Lipid Peroxidation Induced by Hypochlorite. Arch. Biochem. Biophys. 2002, 397, 440–447. [Google Scholar] [CrossRef]
- Nie, X.; Wang, X.; Chen, J.; Zitko, V.; An, T. Response of the freshwater alga chlorella vulgaris to trichloroisocyanuric acid and ciprofloxacin. Environ. Toxicol. Chem. 2008, 27, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Fortún, S.; Llorente, M.T.; Castaño, A. Cytotoxic and genotoxic effect in RTG-2 cell line exposed to selected biocides used in the disinfection of cooling towers. Ecotoxicology 2008, 17, 273–279. [Google Scholar] [CrossRef]
- López-Galindo, C.; Vargas-Chacoff, L.; Nebot, E.; Casanueva, J.F.; Rubio, D.; Sole, M.; Mancera, J.M. Biomarker responses in Solea senegalensis exposed to sodium hypochlorite used as antifouling. Chemosphere 2010, 78, 885–893. [Google Scholar] [CrossRef]
- López-Galindo, C.; Vargas-Chacoff, L.; Nebot, E.; Casanueva, J.F.; Rubio, D.; Mancera, J.M.; Solé, M. Sublethal responses of the common mussel (Mytilus galloprovincialis) exposed to sodium hypochlorite and Mexel®432 used as antifoulants. Ecotoxicol. Environ. Saf. 2010, 73, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Blanco, A.M.; Sundarrajan, L.; Bertucci, J.I.; Unniappan, S. Why goldfish? Merits and challenges in employing goldfish as a model organism in comparative endocrinology research. Gen. Comp. Endocrinol. 2017, 257, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Omori, Y.; Koren, S.; Shirokiya, T.; Kuroda, T.; Miyamoto, A.; Wada, H.; Fujiyama, A.; Toyoda, A.; Zhang, S.; et al. De novo assembly of the goldfish (Carassius auratus) genome and the evolution of genes after whole-genome duplication. Sci. Adv. 2019, 5, eaav0547. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; Von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef]
- Zhou, S.; Li, W.X.; Wang, Y.Q.; Zou, H.; Wu, S.G.; Wang, G.T. Anthelmintic efficacies of three common disinfectants and extracts of four traditional Chinese medicinal plants against Gyrodactylus kobayashii (Monogenea) in goldfish (Carassius auratus). Aquaculture 2017, 466, 72–77. [Google Scholar] [CrossRef]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Peskin, A.; Winterbourn, C.C. A microtiter plate assay for superoxide dismutase using a water-soluble tetrazolium salt (WST-1). Clin. Chim. Acta 2000, 293, 157–166. [Google Scholar] [CrossRef]
- Góth, L. A simple method for determination of serum catalase activity and revision of reference range. Clin. Chim. Acta 1991, 196, 143–151. [Google Scholar] [CrossRef]
- Mannervik, B.; Guthenberg, C. Glutathione transferase (human placenta). Methods Enzymol. 1981, 77, 231–235. [Google Scholar]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Torrecillas, S.; Makol, A.; Caballero, M.J.; Montero, D.; Robaina, L.; Real, F.; Sweetman, J.; Tort, L.; Izquierdo, M.S. Immune stimulation and improved infection resistance in European sea bass (Dicentrarchus labrax) fed mannan oligosaccharides. Fish Shellfish. Immunol. 2007, 23, 969–981. [Google Scholar] [CrossRef] [PubMed]
- Manju, M.; Suresh, S.; Vivekanand, P.; Gunasekaran, S.; Srinivasan, S.; Biju, C. Vibrational spectroscopic investigation and antibacterial activity studies on Trichloroisocyanuric acid. Mater. Today: Proc. 2020, 36, 857–862. [Google Scholar] [CrossRef]
- Rye, P.T.; LaMarr, W.A. Measurement of glycolysis reactants by high-throughput solid phase extraction with tandem mass spectrometry: Characterization of pyrophosphate-dependent phosphofructokinase as a case study. Anal. Biochem. 2015, 482, 40–47. [Google Scholar] [CrossRef]
- Hoffmann, K.H.; Mustafa, T.; Jørgensen, J.B. Role of pyruvate kinase, phosphoenolpyruvate carboxykinase, malic enzyme and lactate dehydrogenase in anaerobic energy metabolism of Tubifex spec. J. Comp. Physiol. 1979, 130, 337–345. [Google Scholar] [CrossRef]
- Cuninghame, S.; Jackson, R.; Lees, S.J.; Zehbe, I. Two common variants of human papillomavirus type 16 E6 differentially deregulate sugar metabolism and hypoxia signalling in permissive human keratinocytes. J. Gen. Virol. 2017, 98, 2310–2319. [Google Scholar] [CrossRef]
- Jamal, N.; Allen, J.I.; Moore, M.N.; Dyke, P. Role for autophagy and programmed cell death (PCD) in maintenance of fish liver function during stress: A modelling study. Mar. Environ. Res. 2006, 62, S439–S440. [Google Scholar]
- Hu, J.; Le, Q.; Wang, Y.; Yu, N.; Cao, X.; Kuang, S.; Zhang, M.; Gu, W.; Sun, Y.; Yang, Y.; et al. Effects of formaldehyde on detoxification and immune responses in silver pomfret (Pampus argenteus). Fish Shellfish. Immunol. 2019, 88, 449–457. [Google Scholar] [CrossRef]
- Lv, X.-M.; Yang, X.-L.; Xie, X.-Y.; Yang, Z.-Y.; Hu, K.; Wu, Y.-J.; Jiang, Y.-y.; Liu, T.-F.; Fang, W.-H.; Huang, X.-Y. Comparative transcriptome analysis of Anguilla japonica livers following exposure to methylene blue. Molecules 2018, 49, 1232–1241. [Google Scholar] [CrossRef]
- Özaslan, M.S.; Demir, Y.; Aksoy, M.; Küfrevioğlu, Ö.I.; Beydemir, Ş. Inhibition effects of pesticides on glutathione-S -transferase enzyme activity of Van Lake fish liver. J. Biochem. Mol. Toxicol. 2018, 32, e22196. [Google Scholar] [CrossRef] [PubMed]
- Testa, B.; Pedretti, A.; Vistoli, G. Reactions and enzymes in the metabolism of drugs and other xenobiotics. Drug Discov. Today 2012, 17, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Gurung, A.B.; Mayengbam, B.; Bhattacharjee, A. Discovery of novel drug candidates for inhibition of soluble epoxide hydrolase of arachidonic acid cascade pathway implicated in atherosclerosis. Comput. Biol. Chem. 2018, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sapone, A.; Gustavino, B.; Monfrinotti, M.; Canistro, D.; Broccoli, M.; Pozzetti, L.; Affatato, A.; Valgimigli, L.; Forti, G.C.; Pedulli, G.F.; et al. Perturbation of cytochrome P450, generation of oxidative stress and induction of DNA damage in Cyprinus carpio exposed in situ to potable surface water. Mutat. Res. Genet. Toxicol. Environ. Mutagenesis 2007, 626, 143–154. [Google Scholar] [CrossRef]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular Mechanisms of Metabolic Resistance to Synthetic and Natural Xenobiotics. Annu. Rev. Èntomol. 2007, 52, 231–253. [Google Scholar] [CrossRef]
- Yin, P.; Xie, S.; Huo, Y.; Guo, T.; Fang, H.; Zhang, Y.; Liu, Y.; Tian, L.; Niu, J. Effects of dietary oxidized fish oil on growth performance, antioxidant defense system, apoptosis and mitochondrial function of juvenile largemouth bass (Micropterus salmoides). Aquaculture 2018, 500, 347–358. [Google Scholar] [CrossRef]
- Awoyemi, O.M.; Kumar, N.; Schmitt, C.; Subbiah, S.; Crago, J. Behavioral, molecular and physiological responses of embryo-larval zebrafish exposed to types I and II pyrethroids. Chemosphere 2019, 219, 526–537. [Google Scholar] [CrossRef]
- Koh, Y.H.; Park, Y.S.; Takahashi, M.; Suzuki, K.; Taniguchi, N. Aldehyde reductase gene expression by lipid peroxidation end products, MDA and HNE. Free Radic. Res. 2000, 33, 739–746. [Google Scholar] [CrossRef]
- Kandimalla, R.; Dash, S.; Kalita, S.; Choudhury, B.; Malampati, S.; Kalita, K.; Kalita, B.; Devi, R.; Kotoky, J. Protective Effect of Bioactivity Guided Fractions of Ziziphus jujuba Mill. Root Bark against Hepatic Injury and Chronic Inflammation via Inhibiting Inflammatory Markers and Oxidative Stress. Front. Pharmacol. 2016, 7, 298. [Google Scholar] [CrossRef]
- Elia, A.C.; Anastasi, V.; Dörr, A.J.M. Hepatic antioxidant enzymes and total glutathione of Cyprinus carpio exposed to three disinfectants, chlorine dioxide, sodium hypochlorite and peracetic acid, for superficial water potabilization. Chemosphere 2006, 64, 1633–1641. [Google Scholar] [CrossRef]
- Yonkos, L.; Fisher, D.; Wright, D.; Kane, A. Pathology of fathead minnows (Pimephales promelas) exposed to chlorine dioxide and chlorite. Mar. Environ. Res. 2000, 50, 267–271. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, S.; Dong, J.; Liu, Y.; Yang, Q.; Xu, N.; Yang, Y.; Ai, X. Transcriptome Analysis Provides Insights into Hepatic Responses to Trichloroisocyanuric Acid Exposure in Goldfish (Carassius auratus). Animals 2021, 11, 2775. https://doi.org/10.3390/ani11102775
Zhou S, Dong J, Liu Y, Yang Q, Xu N, Yang Y, Ai X. Transcriptome Analysis Provides Insights into Hepatic Responses to Trichloroisocyanuric Acid Exposure in Goldfish (Carassius auratus). Animals. 2021; 11(10):2775. https://doi.org/10.3390/ani11102775
Chicago/Turabian StyleZhou, Shun, Jing Dong, Yongtao Liu, Qiuhong Yang, Ning Xu, Yibin Yang, and Xiaohui Ai. 2021. "Transcriptome Analysis Provides Insights into Hepatic Responses to Trichloroisocyanuric Acid Exposure in Goldfish (Carassius auratus)" Animals 11, no. 10: 2775. https://doi.org/10.3390/ani11102775
APA StyleZhou, S., Dong, J., Liu, Y., Yang, Q., Xu, N., Yang, Y., & Ai, X. (2021). Transcriptome Analysis Provides Insights into Hepatic Responses to Trichloroisocyanuric Acid Exposure in Goldfish (Carassius auratus). Animals, 11(10), 2775. https://doi.org/10.3390/ani11102775




