Genetic Population Structure of Wild Pigs in Southern Texas
Abstract
Simple Summary
Abstract
1. Introduction
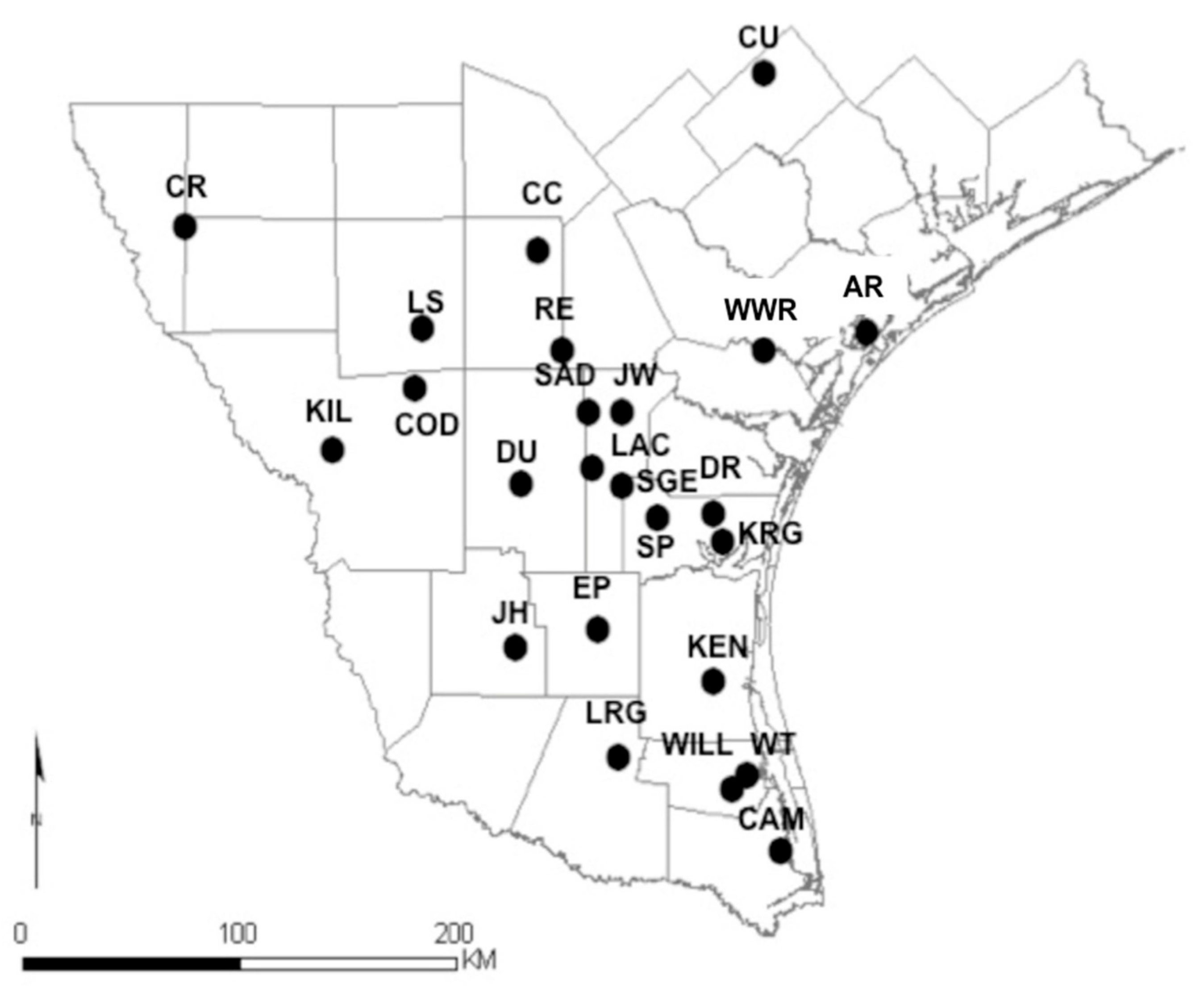
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Amplification
2.2. Locus Properties and Genetic Differentiation among Populations
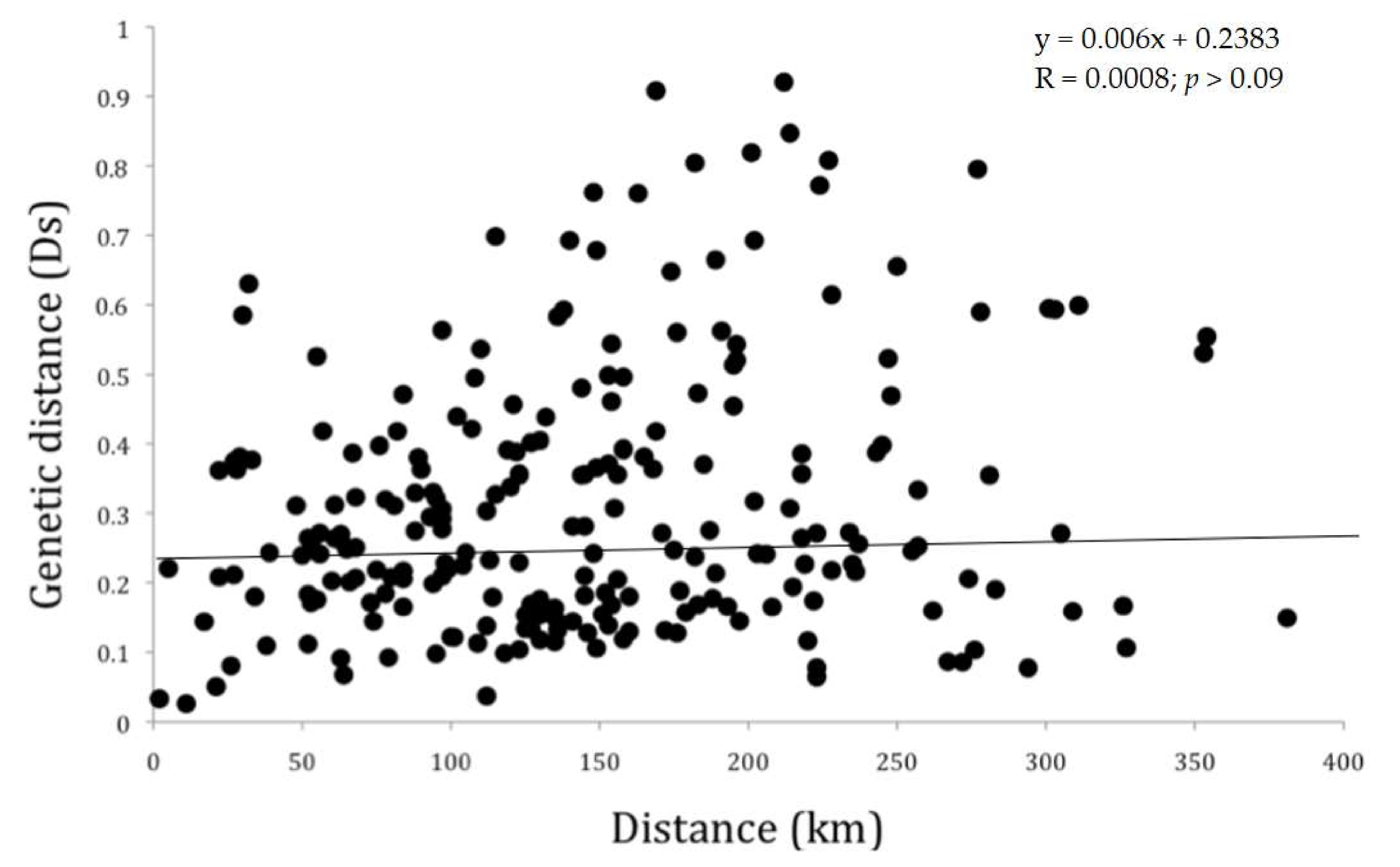
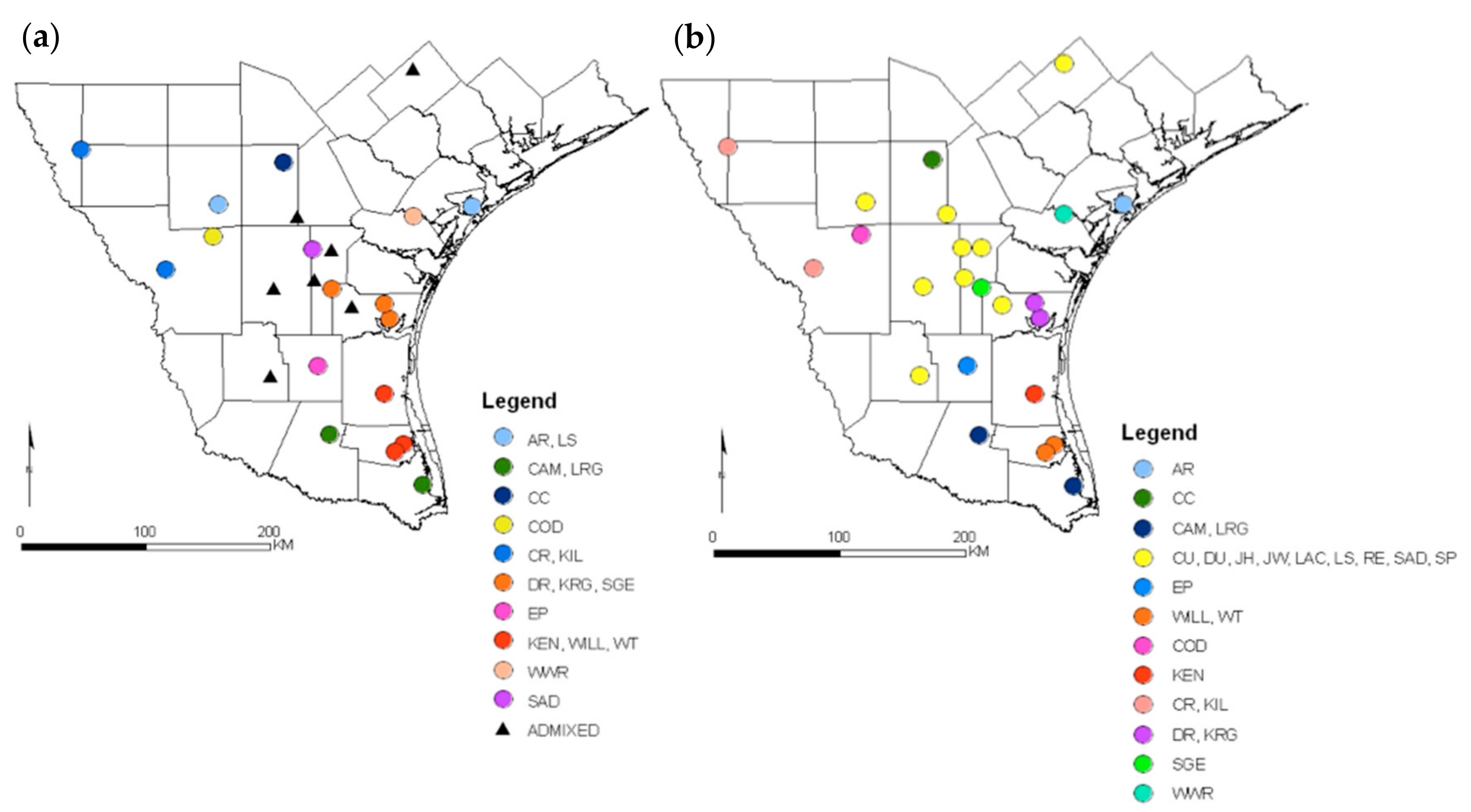
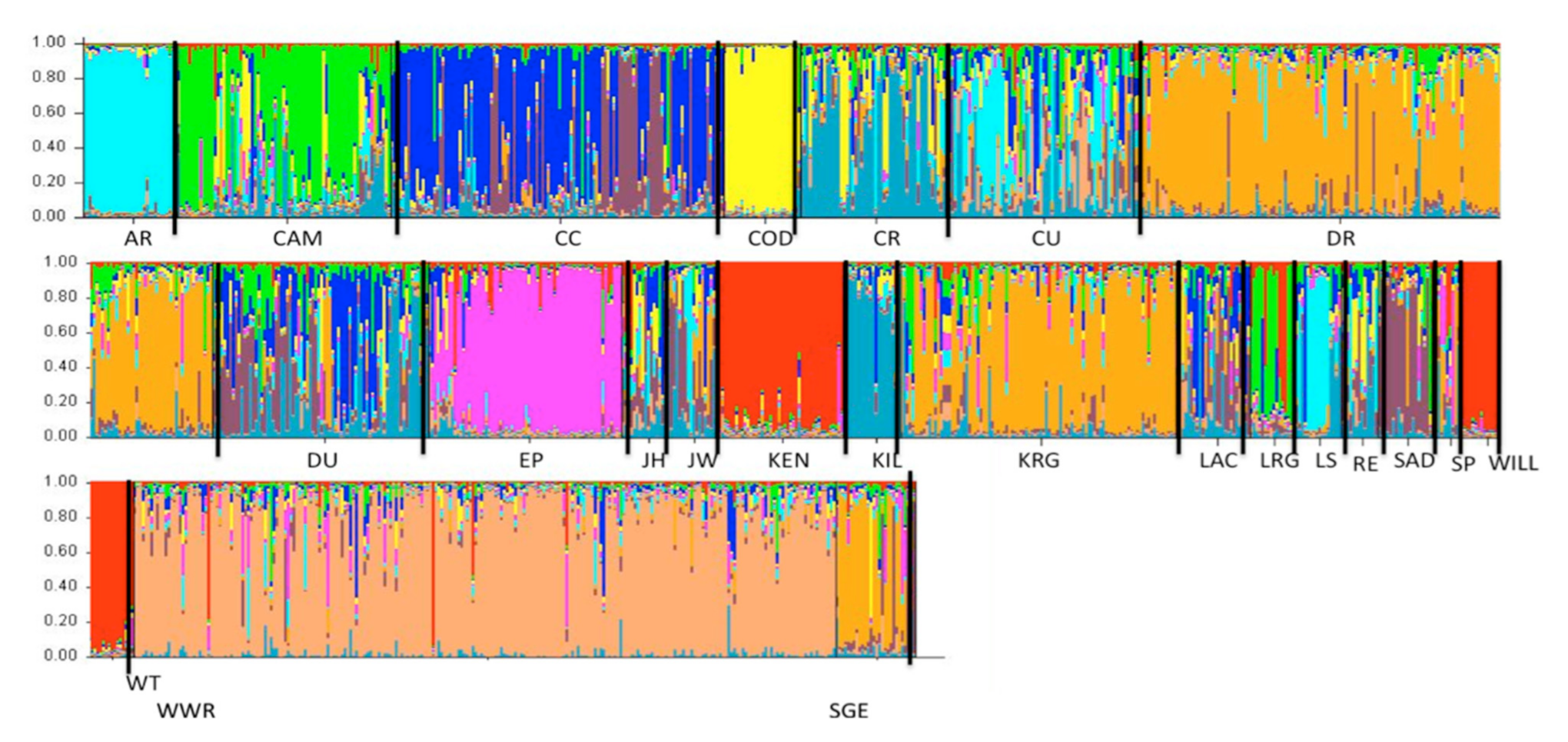
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Linders, T.E.W.; Schaffner, U.; Eschen, R.; Abebe, A.; Choge, S.K.; Nigatu, L.; Mbaabu, P.R.; Allan, R. Direct and indirect effects of invasive species: Biodiversity loss is a major mechanism by which an invasive tree affects ecosystem functioning. J. Ecol. 2019, 107, 2660–2672. [Google Scholar] [CrossRef]
- Pejchar, L.; Mooney, H.A. Invasive species, ecosystem services, and human well-being. Trends Ecol. Evol. 2009, 24, 497–504. [Google Scholar] [CrossRef]
- Crooks, J.A. Characterizing ecosystem-level consequences of biological invasions: The role of ecosystem engineers. Oikos 2002, 97, 153–166. [Google Scholar] [CrossRef]
- Mihailović, D.T.; Petrić, D.; Petrović, T.; Hrnjaković-Cvjetković, I.; Djurdjevic, V.; Nikolić-Dorić, E.; Arsenić, I.; Petrić, M.; Mimić, G.; Ignjatović-Cupina, A. Assessment of climate change impact on the malaria vector Anopheles hyrcanus, west Nile disease, and incidence of melanoma in the Vojvodina Province (Serbia) using data from a regional climate model. PLoS ONE 2020, 15, e0227679. [Google Scholar] [CrossRef]
- Capitani, C.; van Soesbergen, A.; Mukama, K.; Malugu, I. Scenarios of land use and land cover change and their multiple impacts on natural capital in Tanzania. Environ. Conserv. 2019, 46, 17–24. [Google Scholar] [CrossRef]
- He, X.; Liang, J.; Zeng, G.; Youan, Y.; Li, X. The effects of interaction between climate change and land-use/cover change on biodiversity-related ecosystem services. Glob. Chall. 2019, 3, 1800095. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, J.R.; Sweeney, J.M.; Sweeney, S.W. Feral hog. In Wild Mammals of North. America; Feldhamer, G., Thomson, B., Chapman, J., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2003; pp. 1164–1179. [Google Scholar]
- APHIS. Available online: https://www.aphis.usda.gov/aphis/ourfocus/wildlifedamage/operational-activities/feral-swine/sa-fs-history (accessed on 30 October 2020).
- Lewis, J.S.; Corn, J.L.; Mayer, J.J.; Jordan, T.R.; Farnsworth, M.L.; Burdett, C.L.; VerCauteren, K.C.; Sweeney, S.J.; Miller, R.S. Historical, current, and potential population size estimates of invasive wild pigs (Sus Scrofa) in the United States. Biol. Invasions 2019, 21, 2373–2384. [Google Scholar] [CrossRef]
- Timmons, J.B.; Higginbotham, B.; Lopez, R.; Cathey, J.C.; Mellish, J.; Griffin, J.; Sumrall, A.; Skow, K. Feral Hog Population, Growth, Density, and Harvest in Texas; AgriLife Extension Service Texas A&M University: College Station, TX, USA, 2012. [Google Scholar]
- Mellish, J.; Sumrall, A.; Campbell, T.; Collier, B.; Neill, W.; Higginbotham, B.; Lopez, R. Simulating potential population growth of wild pig, Sus scrofa, in Texas. Southeast. Nat. 2014, 13, 367–376. [Google Scholar] [CrossRef]
- Dondina, O.; Orioli, V.; Toretta, E.; Merli, F.; Bani, L.; Meriggi, A. Combining ensemble models and connectivity analyses to predict wolf expected dispersal routes through a lowland corridor. PLoS ONE 2020, 15, e0229261. [Google Scholar] [CrossRef] [PubMed]
- Palsbøll, P.J.; Bérubé, M.; Allendorf, F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007, 22, 11–16. [Google Scholar] [CrossRef]
- Wood, D.A.; Rose, J.P.; Halstead, B.J.; Stoelting, R.E.; Swaim, K.E.; Vandergast, A.G. Combining genetic and demographic monitoring better informs conservation of an endangered urban snake. PLoS ONE 2020, 15, e0231744. [Google Scholar] [CrossRef] [PubMed]
- Beacham, T.D.; Wallace, C.; Jonsen, K.; McIntosh, B.; Candy, J.R.; Willis, D.; Lynch, C.; Moore, J.S.; Bernatchez, L.; Withler, R.E. Comparison of coded-wire tagging with parentage-based tagging and genetic stock identification in a large-scale coho salmon fisheries application in British Columbia, Canada. Evol. Appl. 2019, 12, 230–254. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, R.W.; Zamorano, A.; Mesenbrink, B.T.; Campbell, T.A.; Leland, B.R.; Moore, G.M.; Honeycutt, R.L.; Root, J.J. Landscape-genetic analysis of population structure in the Texas gray fox (Urocyon cinereoargenteus) oral rabies vaccination zone. J. Wildl. Manag. 2009, 73, 1292–1299. [Google Scholar] [CrossRef]
- Epps, C.W.; Wehausen, J.D.; Bleich, V.C.; Torres, S.G.; Brashares, J.S. Optimizing dispersal and corridor models using landscape genetics. J. Appl. Ecol. 2007, 44, 714–724. [Google Scholar] [CrossRef]
- Frantz, A.C.; Cellina, S.; Scheley, L.; Burke, T. Using spatial Bayesian methods to determine the genetic structure of a continuously distributed population: Clusters or isolation by distance? J. Appl. Ecol. 2009, 46, 493–505. [Google Scholar] [CrossRef]
- Hampton, J.O.; Spencer, P.B.S.; Alpers, D.L.; Twigg, L.E.; Woolnough, A.P.; Doust, J.; Higgs, T.; Pluske, J. Molecular techniques, wildlife management and the importance of genetic population structure and dispersal: A case study with feral pigs. J. Appl. Ecol. 2004, 41, 735–743. [Google Scholar] [CrossRef]
- El Mousadik, A.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree Argania spinosa (Skeels) endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef]
- Goudet, J.; Perrin, N.; Waser, P. Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol. Ecol. 2002, 11, 1103–1114. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Wright, S. Isolation by distance. Genetics 1942, 28, 114–138. [Google Scholar]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Hardy, O.J.; Vekemans, X. SPAGeDi: A versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol. Ecol. Notes 2002, 2, 618–620. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Corander, J.; Waldmann, P.; Sillanpää, M.J. Bayesian analysis of genetic differentiation between populations. Genetics 2004, 163, 367–374. [Google Scholar]
- Corander, J.; Waldmann, P.; Marttinen, P.; Sillanpää, M.J. BAPS 4.2: Enhanced possibilities for the analysis of genetic population structure. Bioinformatics 2003, 20, 2363–2369. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Dudley, J.; Nei, M.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. An Introduction to Adegenet 2.1.0.; Imperial College London, MEC Center for Outbreak Analysis and Modeling: London, UK, 2007; pp. 1–79. [Google Scholar]
- Jombart, T. Adenet: An R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed]
- Mapston, M.E. Feral Hogs in Texas; Publication B-6149; Texas Cooperative Extension, Texas A&M University: College Station, TX, USA, 2004. [Google Scholar]
- Maes, J.; Van Oosten, A.R.; Van Houtte, N.; Matthysen, E. Genetic structure of natterjack toad (Epidalea calamita) populations in Flanders, Belgium, and its implications for conservation. Amphib. Reptile 2019, 40, 193–205. [Google Scholar] [CrossRef]
- Kheng, V.; Zichello, J.M.; Lumbantobing, D.N.; Lawalata, S.Z.S.; Andayani, N.; Melnick, D.J. Phylogeography, population structure, and conservation of the Javan gibbon (Hylobates moloch). Int. J. Primatol. 2018, 39, 5–26. [Google Scholar] [CrossRef]
- Latch, E.K.; Scognamillo, D.G.; Fike, J.A.; Chamberlain, M.J.; Rhodes, O.E. Deciphering ecological barriers to North America river otter (Lontra canadensis) gene flow in the Louisiana landscape. J. Hered. 2008, 99, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Rowe, G.; Beebee, T.J.C. Defining population boundaries: Use of three Bayesian approaches with microsatellite data from British natterjack toads (Bufo calamita). Mol. Ecol. 2007, 16, 785–796. [Google Scholar] [CrossRef]
- Hernández, F.A.; Parker, B.M.; Pylant, C.L.; Smyser, T.J.; Piaggio, A.J.; Lance, S.L.; Milleson, M.P.; Austin, J.D.; Wisely, S.M. Invasion ecology of wild pigs (Sus scrofa) in Florida, USA: The role of humans in the expansion and colonization of an invasive wild ungulate. Biol. Invasions 2018, 20, 1865–1880. [Google Scholar] [CrossRef]
- DeYoung, R.W.; Demarais, S.; Honeycutt, R.L.; Rooney, A.P.; Gonzales, R.A.; Gee, K.L. Genetic consequences of white-tailed deer (Odocoileus virginianus) restoration in Mississippi. Mol. Ecol. 2003, 12, 3237–3252. [Google Scholar] [CrossRef]
- Kolbe, J.J.; Glor, R.E.; Schettino, L.R.; Lara, A.C.; Larson, A.; Losos, J.B. Multiple sources, admixture, and genetic variation in introduced Anolis lizard populations. Cons. Biol. 2007, 21, 1612–1625. [Google Scholar] [CrossRef]
- McCan, B.E.; Smyser, T.J.; Schmidt, B.S.; Newman, R.A.; Piaggio, A.J.; Malek, M.J.; Swafford, S.R.; Sweitzer, R.A.; Simmons, R.B. Molecular population structure of feral swine in the United States. J. Wildl. Manag. 2018, 82, 821–832. [Google Scholar] [CrossRef]
- Smyser, T.J.; Tabak, M.A.; Slootmaker, C.; Robeson II., M.S.; Miller, R.S.; Bosse, M.; Megens, H.J.; Groenen, M.A.M.; Rezende Paiva, S.; Asis de Faria, D.; et al. Mixed ancestry from wild and domestic lineages contributes to the rapid expansion of invasive feral swine. Mol. Ecol. 2020, 29, 1103–1119. [Google Scholar] [CrossRef] [PubMed]
- Beasley, J.C.; Ditchkoff, S.S.; Mayer, J.J.; Smith, M.D.; VerCauteren, K.C. Research priorities for managing invasive wild pigs in North America: Research priorities for wild pigs. J. Wildl. Manag. 2018, 82, 674–681. [Google Scholar] [CrossRef]
- Campbell, T.A.; Long, D.B. Feral swine damage and damage management in forested ecosystems. For. Ecol. Manag. 2009, 257, 2319–2326. [Google Scholar] [CrossRef]
- Snow, N.P.; Halseth, J.M.; Lavelle, M.J.; Hanson, T.E.; Blass, C.R.; Foster, J.A.; Humphrys, S.T.; Staples, L.D.; Hewitt, D.G.; VerCauteren, K.C. Bait preference of free-ranging wild pig for delivery of a novel toxicant. PLoS ONE 2016, 11, e0146712. [Google Scholar] [CrossRef]
- Cowled, B.D.; Elsworth, P.; Lapidge, S.J. Additional toxins for feral pig (Sus scrofa) control: Identifying and testing Achilles’ heels. Wildl. Res. 2008, 35, 651–662. [Google Scholar] [CrossRef]
- Campbell, T.A.; Long, D.B. Mammalian visitation to candidate feral swine attractants. J. Wild. Manag. 2008, 71, 305–309. [Google Scholar] [CrossRef]
- Campbell, T.A.; Garcia, M.R.; Miller, L.A.; Ramirez, M.A.; Long, D.B.; Marchand, J.; Hill, F. Immunocontraception of male feral swine with a recombinant GnRH vaccine. J. Swine Health Prod. 2010, 18, 118–124. [Google Scholar] [CrossRef]
- Massei, G.; Cowan, D.P.; Coats, J.; Gladweel, F.; Lane, J.E.; Miller, L.A. Effect of the GnRH vaccine GonaCon on the fertility, physiology and behaviour of wild boar. Wildl. Res. 2008, 35, 540–547. [Google Scholar] [CrossRef]
- Ballesteros, C.; Gortázar, C.; Canales, M.; Vicente, J.; Lasagna, A.; Gamarra, J.A.; Carrasco-García, R.; de la Fuente, J. Evaluation of baits for oral vaccination of European wild boar piglets. Res. Vet. Sci. 2009, 86, 388–393. [Google Scholar] [CrossRef]
- Lavelle, M.J.; Snow, N.P.; Halseth, J.M.; Kinsey, J.C.; Foster, J.A.; VerCauteren, K.C. Development and evaluation of a bait station for selectively dispensing bait to invasive wild pigs. Wildl. Soc. Bull. 2018, 42, 102–110. [Google Scholar] [CrossRef]
- Campbell, T.A.; Lapidge, S.J.; Long, D.B. Using baits to deliver pharmaceuticals to feral swine in southern Texas. Wildl. Soc. Bull. 2006, 34, 1184–1189. [Google Scholar] [CrossRef]
- Long, D.B.; Campbell, T.A.; Massei, G. Evaluation of feral swine-specific feeder systems. Rangelands 2010, 32, 8–13. [Google Scholar] [CrossRef]
- Massei, G.; Coats, J.; Quy, R.; Storer, K.; Cowan, D.P. The Boar-Operated-System: A novel method to deliver baits to wild pigs. J. Wildl. Manag. 2010, 74, 333–336. [Google Scholar] [CrossRef]
- Lavoie, C.; Donlan, C.J.; Campbell, K.; Cruz, F.; Carrion, G.V. Geographic tools for eradication programs of insular non-native mammals. Biol. Invasions 2007, 9, 139–148. [Google Scholar] [CrossRef]
- Blanchong, J.A.; Scribner, K.T.; Winterstein, S.R. Assignment of individuals to populations: Bayesian methods and multilocus genotypes. J. Wildl. Manag. 2002, 66, 321–329. [Google Scholar] [CrossRef]
- Funk, W.C.; Garcia, T.S.; Cortina, G.A.; Hill, R.H. Population genetics of introduced bullfrogs, Rana (Lithobates) catesbeianus, in the Willamette Valley, Oregon, USA. Biol. Invasions 2011, 13, 651–658. [Google Scholar] [CrossRef]
- Smith, A.L. Wild pig policy and legislation. In Invasive Wild Pigs in North America—Ecology, Impacts and Management; VerCauteren, K.C., Beasley, J.C., Ditchkoff, S.S., Mayer, J.J., Roloff, G.J., Strickland, B.K., Eds.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2019; pp. 245–265. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delgado-Acevedo, J.; Zamorano, A.; DeYoung, R.W.; Campbell, T.A. Genetic Population Structure of Wild Pigs in Southern Texas. Animals 2021, 11, 168. https://doi.org/10.3390/ani11010168
Delgado-Acevedo J, Zamorano A, DeYoung RW, Campbell TA. Genetic Population Structure of Wild Pigs in Southern Texas. Animals. 2021; 11(1):168. https://doi.org/10.3390/ani11010168
Chicago/Turabian StyleDelgado-Acevedo, Johanna, Angeline Zamorano, Randy W. DeYoung, and Tyler A. Campbell. 2021. "Genetic Population Structure of Wild Pigs in Southern Texas" Animals 11, no. 1: 168. https://doi.org/10.3390/ani11010168
APA StyleDelgado-Acevedo, J., Zamorano, A., DeYoung, R. W., & Campbell, T. A. (2021). Genetic Population Structure of Wild Pigs in Southern Texas. Animals, 11(1), 168. https://doi.org/10.3390/ani11010168




