Effect of Concentrate Supplementation on the Expression Profile of miRNA in the Ovaries of Yak during Non-Breeding Season
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Declaration
2.2. Animals and Management
2.3. Sample Collection
2.4. Blood Biochemistry and Hormonal Concentration
2.5. Extraction of RNA and Sequencing of Small RNA
2.6. Analysis through Nioinformatics for Sequencing of Small RNA
2.7. Quantification and Differential Analysis of MicroRNA
2.8. Target Gene Prediction, Gene Ontology, and Pathway Analysis
2.9. Validation of miRNA Expression by Quantitative Real-Time PCR
2.10. Statistical Analysis
3. Results
3.1. Effect of Mixed Diet Supplementation during the Cold Season on the Growth Performance of Yaks
3.2. Effect of Mix Diet Supplementation during the Non-Breeding Season on Serum Biochemical Parameters of the Yaks
3.3. Effect of Cold Season Mix Diet Supplementation on Serum Reproductive Hormone of Yaks
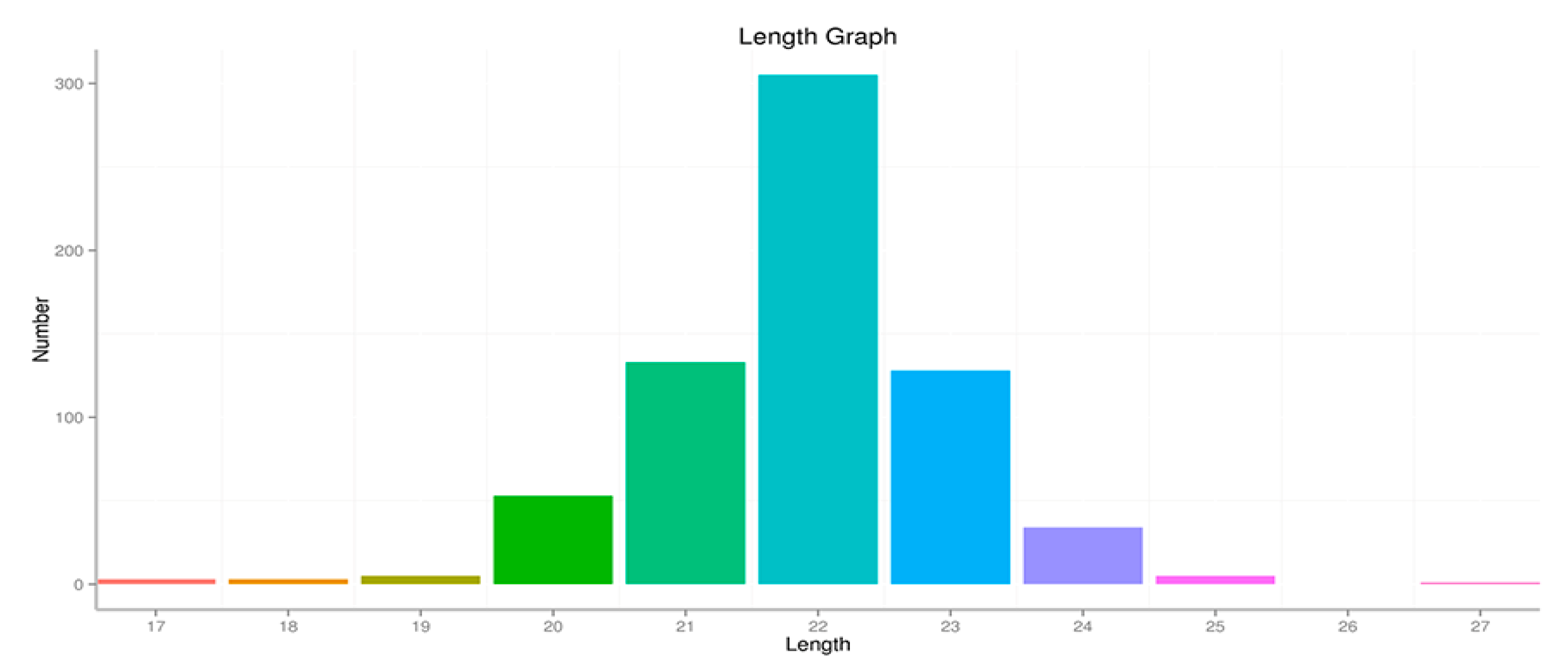
3.4. Solexa Sequencing of Ovary Small RNAs
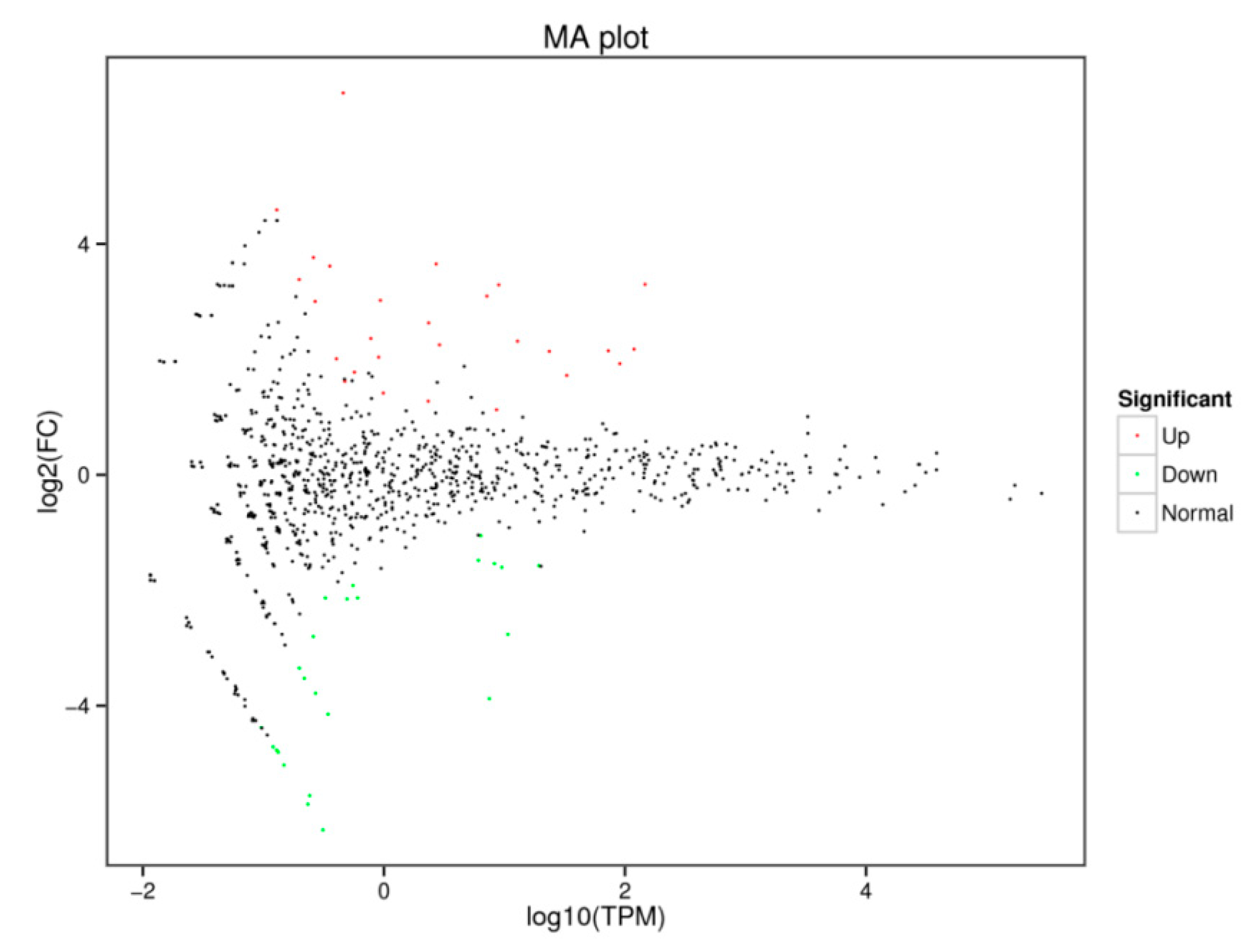
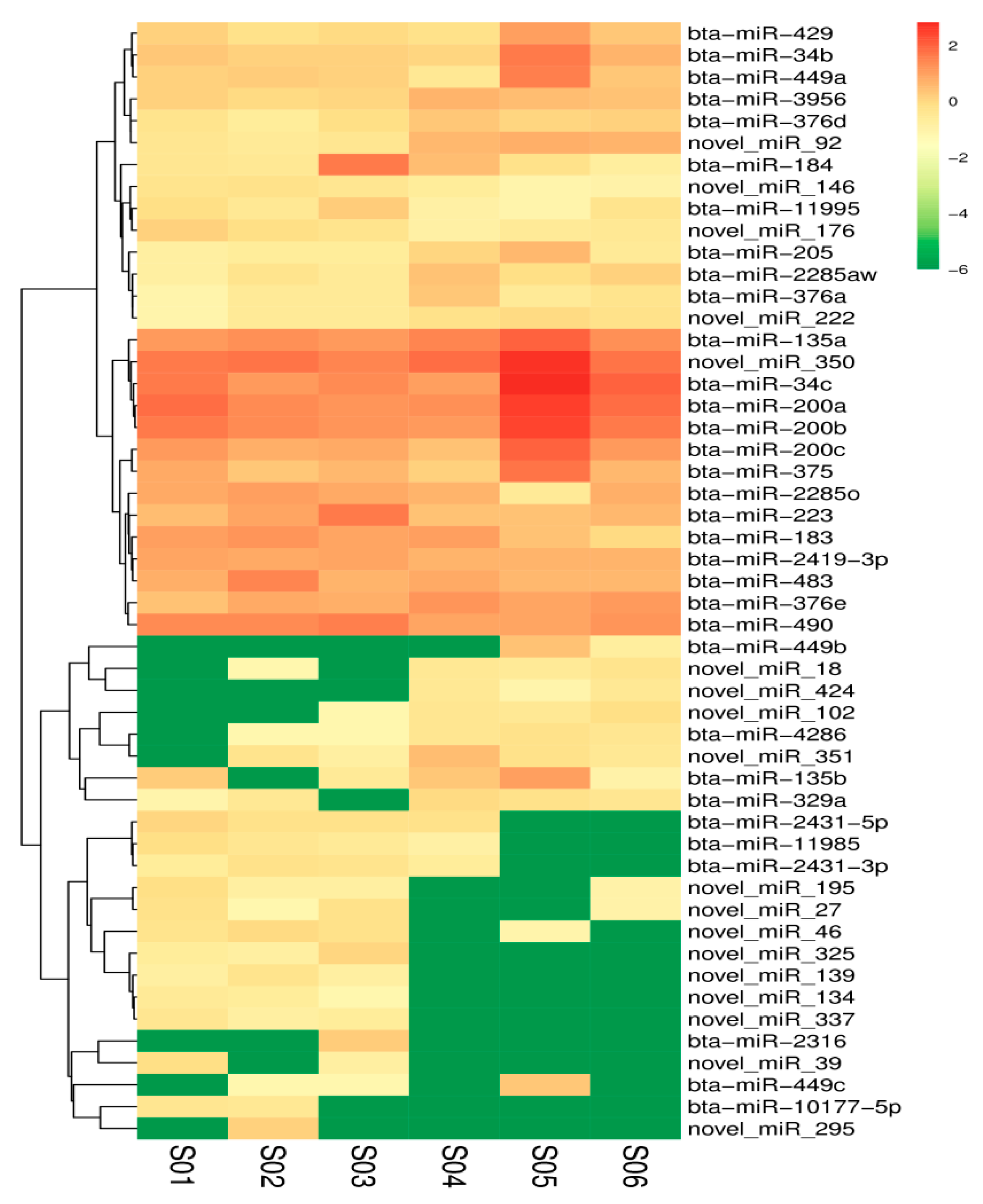
3.5. Identification of Differentially Expressed miRNAs in the Ovary
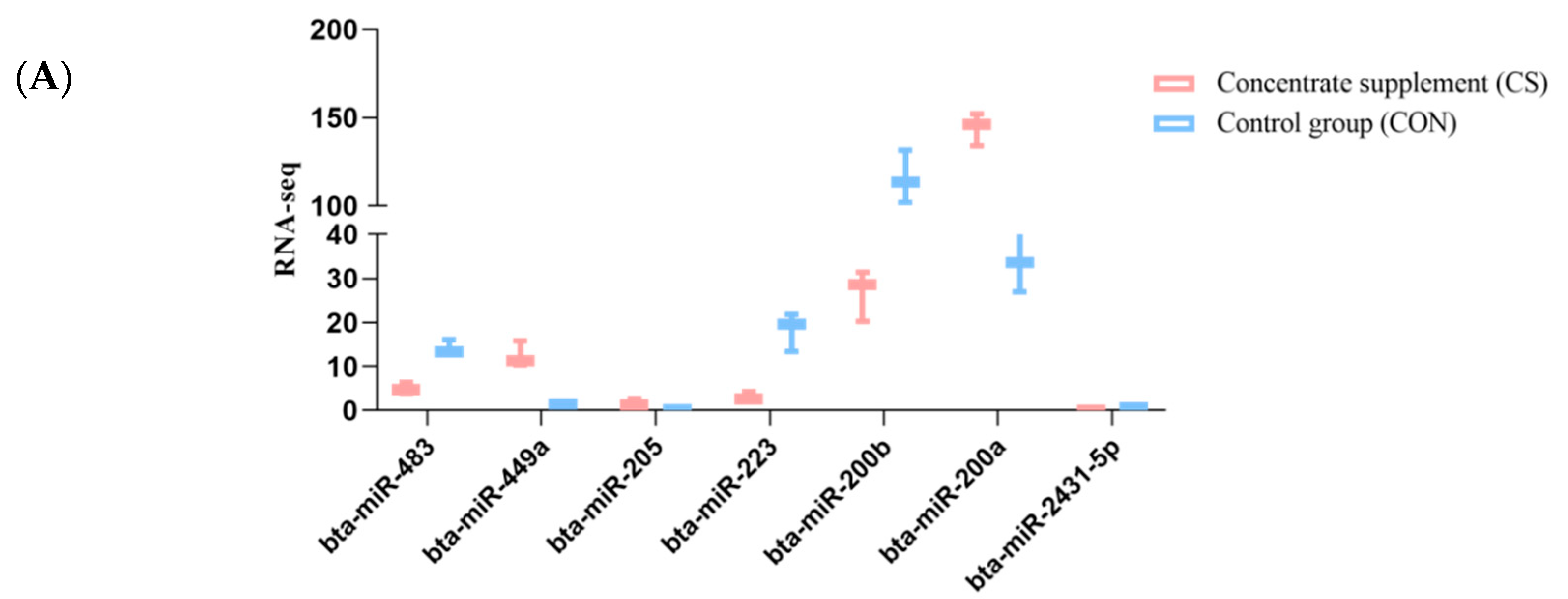
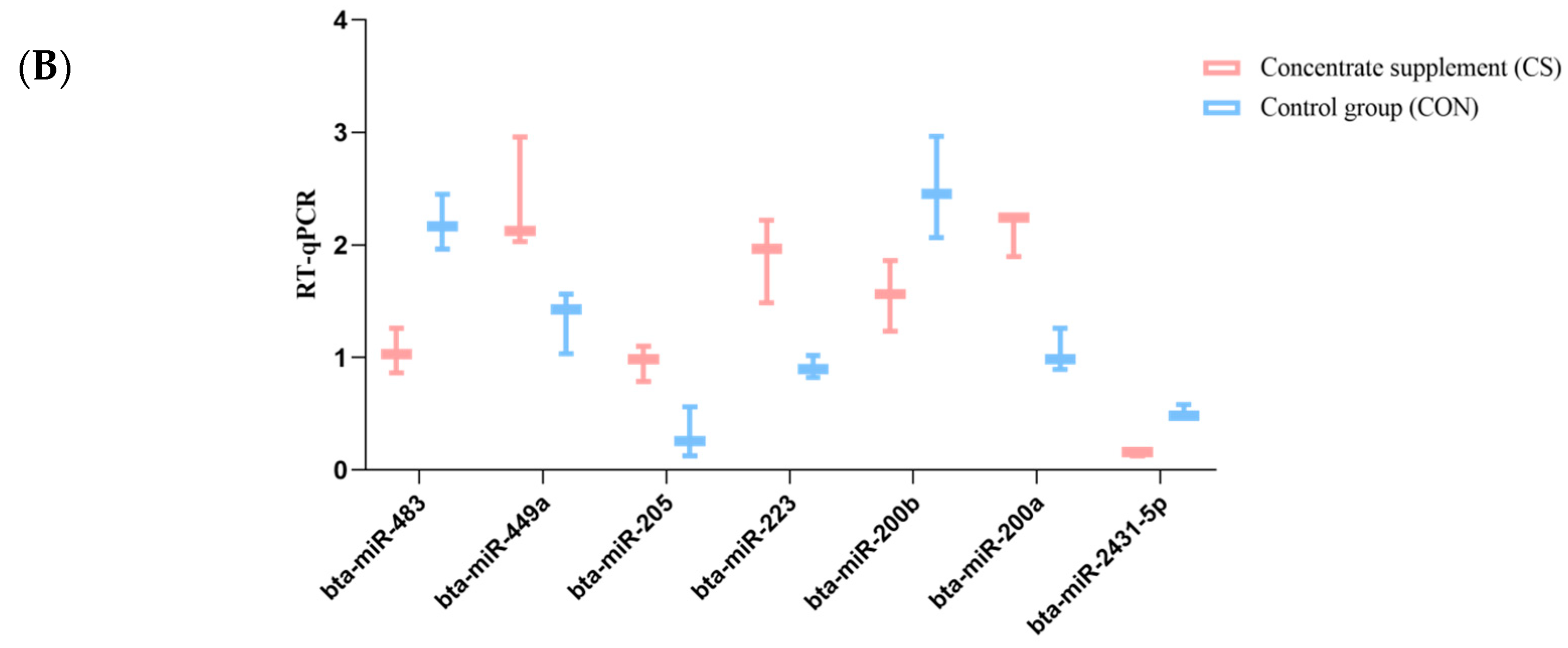
3.6. Validation of miRNA Expression with qRT-PCR
3.7. miRNA Target Gene Prediction, GO Enrichment, and KEGG Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Qiang, Q.; Guojie, Z.; Tao, M.; Wubin, Q.; Junyi, W.; Zhiqiang, Y.; Changchang, C.; Quanjun, H.; Jaebum, K.; Larkin, D.M. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Xiong, X.; Fu, M.; Lan, D.; Li, J.; Zi, X.; Zhong, J. Yak response to high-altitude hypoxic stress by altering mRNA expression and DNA methylation of hypoxia-inducible factors. Anim. Biotechnol. 2015, 26, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Miao, F.; Guo, Z.; Xue, R.; Wang, X.; Shen, Y. Effects of grazing and precipitation on herbage biomass, herbage nutritive value, and yak performance in an alpine meadow on the qinghai-tibetan plateau. PLoS ONE 2015, 10, e0127275. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Long, R.; Kreuzer, M.; Ding, L.; Shang, Z.; Zhang, Y.; Yang, Y.; Cui, G. Importance of functional ingredients in yak milk-derived food on health of Tibetan nomads living under high-altitude stress: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Mei, F.; Yabing, C.; Xianrong, X.; Daoliang, L.; Jian, L. Establishment of mammary gland model in vitro: Culture and evaluation of a yak mammary epithelial cell line. PLoS ONE 2014, 9, e113669. [Google Scholar] [CrossRef]
- Long, R.J.; Zhang, D.G.; Wang, X.; Hu, Z.Z.; Dong, S.K. Effect of strategic feed supplementation on productive and reproductive performance in yak cows. Prev. Vet. Med. 1999, 38, 195–206. [Google Scholar] [CrossRef]
- Prakash, B.S.; Sarkar, M.; Mondal, M. An update on reproduction in yak and mithun. Reprod. Domest. Anim. 2010, 43, 217–223. [Google Scholar] [CrossRef]
- Fu, M.; Xiong, X.R.; Lan, D.L.; Li, J. Molecular characterization and tissue distribution of estrogen receptor genes in domestic yak. Asian-Australas. J. Anim. Sci. 2014, 27, 1684–1690. [Google Scholar] [CrossRef]
- Xiao, X.; Zi, X.D.; Niu, H.R.; Xiong, X.R.; Zhong, J.C.; Li, J.; Wang, L.; Wang, Y. Effect of addition of FSH, LH and proteasome inhibitor MG132 to in vitro maturation medium on the developmental competence of yak (Bos grunniens) oocytes. Reprod. Biol. Endocrinol. 2014, 12, 30. [Google Scholar] [CrossRef]
- Mcgee, E.; Hsueh, A. Initial and cyclic recruitment of ovarian follicles. Endocr. Rev. 2000, 21, 200–214. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Y.; Wang, T.; Guan, J.; Luo, Z.; Chen, H.; Wang, X.; Chen, L.; Ma, J.; Mu, Z.; et al. Repertoire of porcine microRNAs in adult ovary and testis by deep sequencing. Int. J. Biol. Sci. 2011, 7, 1045–1055. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Ghanem, N.; Hoelker, M.; Rings, F.; Phatsara, C.; Tholen, E.; Schellander, K.; Tesfaye, D. Identification and characterization of miRNAs expressed in the bovine ovary. BMC Genom. 2009, 10, 443. [Google Scholar] [CrossRef]
- McBride, D.; Carre, W.; Sontakke, S.D.; Hogg, C.O.; Law, A.; Donadeu, F.X.; Clinton, M. Identification of miRNAs associated with the follicular-luteal transition in the ruminant ovary. Reproduction 2012, 144, 221–233. [Google Scholar] [CrossRef]
- Otsuka, M.; Jing, Q.; Georgel, P.; New, L.; Chen, J.; Mols, J.; Kang, Y.J.; Jiang, Z.; Du, X.; Cook, R.; et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 2007, 27, 123–134. [Google Scholar] [CrossRef]
- Otsuka, M.; Zheng, M.; Hayashi, M.; Lee, J.D.; Yoshino, O.; Lin, S.; Han, J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J. Clin. Investig. 2008, 118, 1944–1954. [Google Scholar] [CrossRef]
- Jing, X.; Peng, Q.; Hu, R.; Wang, H.; Yu, X.; Degen, A.; Zou, H.; Bao, S.; Zhao, S.; Wang, Z. Effect of supplements during the cold season on the reproductive system in prepubertal Tibetan sheep ewes. Anim. Sci. J. 2017, 88, 1269–1278. [Google Scholar] [CrossRef]
- Li, B.; Ruotti, V.; Stewart, R.M.; Thomson, J.A.; Dewey, C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 2010, 26, 493–500. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Roseler, D.K.; Ferguson, J.D.; Sniffen, C.J.; Herrema, J. Dietary Protein Degradability Effects on Plasma and Milk Urea Nitrogen and Milk Nonprotein Nitrogen in Holstein Cows. J. Dairy Sci. 1993, 76, 525–534. [Google Scholar] [CrossRef]
- Campanile, G.; Filippo, C.D.; Palo, R.D.; Taccone, W.; Zicarelli, L. Influence of dietary protein on urea levels in blood and milk of buffalo cows. Livest. Prod. Sci. 1998, 55, 135–143. [Google Scholar] [CrossRef]
- Law, R.A.; Young, F.J.; Patterson, D.C.; Kilpatrick, D.J.; Wylie, A.R.G.; Mayne, C.S. Effect of dietary protein content on animal production and blood metabolites of dairy cows during lactation. J. Dairy Sci. 2009, 92, 1001–1012. [Google Scholar] [CrossRef]
- Hazeleger, W.; Soede, N.M.; Kemp, B. The effect of feeding strategy during the pre-follicular phase on subsequent follicular development in the pig. Domest. Anim. Endocrinol. 2005, 29, 362–370. [Google Scholar] [CrossRef]
- Armstrong, D.G.; McEvoy, T.G.; Baxter, G.; Robinson, J.J.; Hogg, C.O.; Woad, K.J.; Webb, R.; Sinclair, K.D. Effect of dietary energy and protein on bovine follicular dynamics and embryo production in vitro: Associations with the ovarian insulin-like growth factor system. Biol. Reprod. 2001, 64, 1624–1632. [Google Scholar] [CrossRef] [PubMed]
- Meza-Herrera, C.A.; Hallford, D.M.; Ortiz, J.A.; Cuevas, R.A.; Sanchez, J.M.; Salinas, H.; Mellado, M.; Gonzalez-Bulnes, A. Body condition and protein supplementation positively affect periovulatory ovarian activity by non LH-mediated pathways in goats. Anim. Reprod. Sci. 2008, 106, 412–420. [Google Scholar] [CrossRef]
- O’Callaghan, D.; Yaakub, H.; Hyttel, P.; Spicer, L.J.; Boland, M.P. Effect of nutrition and superovulation on oocyte morphology, follicular fluid composition and systemic hormone concentrations in ewes. J. Reprod. Fertil. 2000, 118, 303–313. [Google Scholar] [CrossRef]
- Beam, S.W.; Butler, W.R. Effects of energy balance on follicular development and first ovulation in postpartum dairy cows. J. Reprod. Fertil. 1999, 54, 411–424. [Google Scholar] [CrossRef]
- Fitz-Rodríguez, G.; Santiago-Miramontes, M.A.D.; Scaramuzzi, R.J.; Malpaux, B.; Delgadillo, J.A. Nutritional supplementation improves ovulation and pregnancy rates in female goats managed under natural grazing conditions and exposed to the male effect. Anim. Reprod. Sci. 2009, 116, 85–94. [Google Scholar] [CrossRef]
- Ma, X.; Jia, C.; Chu, M.; Fu, D.; Lei, Q.; Ding, X.; Wu, X.; Guo, X.; Pei, J.; Bao, P.; et al. Transcriptome and DNA Methylation Analyses of the Molecular Mechanisms Underlying with Longissimus dorsi Muscles at Different Stages of Development in the Polled Yak. Genes 2019, 10, 970. [Google Scholar] [CrossRef] [PubMed]
- Dao-Liang, L.; Xian-Rong, X.; Zhi-Xin, C.; Yi, A.I.; Cai, H.; Jian, L.I. Comparative Transcriptome Analysis between Yak and Cattle Estrus Ovary. Chin. J. Anim. Vet. Sci. 2016, 47, 1830–1839. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Harrison, C.A.; O’Donnell, L.; Stanton, P.G. 148. Hormonal regulation of miRNA in the tistis. Reprod. Fertil. Dev. 2009, 21, 66. [Google Scholar] [CrossRef]
- Guelfi, G.; Stefanetti, V.; De Luca, S.; Giontella, A.; Barile, V.L.; Barbato, O. Serum microRNAs in buffalo cows: Potential biomarkers of pregnancy. Res. Vet. Sci. 2017, 115, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Liu, X.; Hu, G.; Xie, Y.; Lin, S.; Zhao, Z.; Chen, J. Identification and analysis of microRNAs-mRNAs pairs associated with nutritional status in seasonal sheep. Biochem. Biophys. Res. Commun. 2018, 499, 321–327. [Google Scholar] [CrossRef]
- Sohel, M.M.H.; Salilew-Wondim, D.; Rings, F.; Schellander, K.; Tesfaye, D. 207 circulatory microRNA signatures in follicular fluid in relation to the growth status of bovine oocytes. Reprod. Fertil. Dev. 2012, 25, 251–252. [Google Scholar] [CrossRef]
- Paczoska-Eliasiewicz, H.E.; Proszkowiec-Weglarz, M.; Proudman, J.; Jacek, T.; Mika, M.; Sechman, A.; Rzasa, J.; Gertler, A. Exogenous leptin advances puberty in domestic hen. Domest. Anim. Endocrinol. 2006, 31, 211–226. [Google Scholar] [CrossRef]
- Seeber, R.M.; Smith, J.T.; Waddell, B.J. Plasma Leptin-Binding Activity and Hypothalamic Leptin Receptor Expression During Pregnancy and Lactation in the Rat. Biol. Reprod. 2002, 66, 1762–1767. [Google Scholar] [CrossRef][Green Version]
- Sarkar, M.; Schilffarth, S.; Schams, D.; Meyer, H.H.; Berisha, B. The expression of leptin and its receptor during different physiological stages in the bovine ovary. Mol. Reprod. Dev. 2010, 77, 174–181. [Google Scholar] [CrossRef]
- Nicklin, L.T.; Robinson, R.S.; Marsters, P.; Campbell, B.K.; Mann, G.E.; Hunter, M.G. Leptin in the bovine corpus luteum: Receptor expression and effects on progesterone production. Mol. Reprod. Dev. 2007, 74, 724–729. [Google Scholar] [CrossRef]
- Wang, Y.H.; Xu, M.; Wang, F.N.; Yu, Z.P.; Yao, J.H.; Zan, L.S.; Yang, F.X. Effect of dietary starch on rumen and small intestine morphology and digesta pH in goats. Livest. Sci. 2009, 122, 48–52. [Google Scholar] [CrossRef]
- Meikle, A.; Kulcsar, M.; Chilliard, Y.; Febel, H.; Delavaud, C.; Cavestany, D.; Chilibroste, P. Effects of parity and body condition at parturition on endocrine and reproductive parameters of the cow. Reproduction 2004, 127, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Paula-Lopes, F.F.; Boelhauve, M.; Habermann, F.A.; Sinowatz, F.; Wolf, E. Leptin promotes meiotic progression and developmental capacity of bovine oocytes via cumulus cell-independent and -dependent mechanisms. Biol. Reprod. 2007, 76, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V. Growth factors controlling ovarian functions. J. Cell. Physiol. 2011, 226, 2222–2225. [Google Scholar] [CrossRef]
- Kaczmarek, M.M.; Schams, D.; Ziecik, A.J. Role of vascular endothelial growth factor in ovarian physiology—An overview. Reprod. Biol. 2005, 5, 111. [Google Scholar] [CrossRef]
- Kosaka, N.; Sudo, N.; Miyamoto, A.; Shimizu, T. Vascular endothelial growth factor (VEGF) suppresses ovarian granulosa cell apoptosis in vitro. Biochem. Biophys. Res. Commun. 2007, 363, 733–737. [Google Scholar] [CrossRef]
- Quintana, R.; Kopcow, L.; Sueldo, C.; Marconi, G.; Rueda, N.G.; Barañao, R.I. Direct injection of vascular endothelial growth factor into the ovary of mice promotes follicular development. Fertil. Steril. 2004, 82, 1101–1105. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, J.; Pan, Z.; Li, Q.; Liu, H. The distribution and expression of vascular endothelial growth factor A (VEGFA) during follicular development and atresia in the pig. Reprod. Fertil. Dev. 2019, 32. [Google Scholar] [CrossRef]
- Mizuno, N.; Itoh, H. Functions and regulatory mechanisms of Gq-signaling pathways. Neuro-Signals 2009, 17, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Mönkkönen, K.S.; Aflatoonian, R.; Lee, K.-F.; Yeung, W.S.B.; Tsao, S.-W.; Laitinen, J.T.; Fazeli, A. Hormonal regulation of Galphai2 and mPRalpha in immortalized human oviductal cell line OE-E6/E7. Mol. Hum. Reprod. 2007, 13, 845–851. [Google Scholar] [CrossRef][Green Version]
- Sánchez-Fernández, G.; Cabezudo, S.; García-Hoz, C.; Benincá, C.; Aragay, A.M.; Mayor, F.; Ribas, C. Gαq signalling: The new and the old. Cell Signal. 2014, 26, 833–848. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Ling, H.; Grimm, M.; Zhang, T.; Bers, D.M.; Brown, J.H. Cardiac hypertrophy and heart failure development through Gq and CaM kinase II signaling. J. Cardiovasc. Pharmacol. 2010, 56, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Markku, H.; Marina, E.; Malgorzata, B.; Rahman, N.A.; Naoko, N.; Huhtaniemi, I.T.; Tapanainen, J.S.; Wilson, D.B. Expression and hormonal regulation of transcription factors GATA-4 and GATA-6 in the mouse ovary. Endocrinology 1997, 138, 3505–3514. [Google Scholar] [CrossRef]
- Laitinen, M.P.; Anttonen, M.; Ketola, I.; Wilson, D.B.; Ritvos, O.; Butzow, R.; Heikinheimo, M. Transcription factors GATA-4 and GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord-derived ovarian tumors. J. Clin. Endocrinol. Metab. 2000, 85, 3476–3483. [Google Scholar] [CrossRef][Green Version]
- Gillio-Meina, C. GATA-4 and GATA-6 transcription factors: Expression, immunohistochemical localization, and possible function in the porcine ovary. Biol. Reprod. 2003, 68, 412–422. [Google Scholar] [CrossRef]
- Sicinski, P.; Donaher, J.L.; Geng, Y.; Parker, S.B.; Gardner, H.; Park, M.Y.; Robker, R.L.; Richards, J.S.; McGinnis, L.K.; Biggers, J.D.; et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 1996, 384, 470–474. [Google Scholar] [CrossRef]
- Shimizu, T.; Hirai, Y.; Miyamoto, A. Expression of cyclins and cyclin-dependent kinase inhibitors in granulosa cells from bovine ovary. Reprod. Domest. Anim. 2013, 48, e65–e69. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, H.; Jiang, Y.; Ding, L.; Wu, S.; Fang, T.; Yan, G.; Hu, Y. MicroRNA-181a suppresses mouse granulosa cell proliferation by targeting activin receptor IIA. PLoS ONE 2013, 8, e59667. [Google Scholar] [CrossRef]
- Haldar, S.M.; Ibrahim, O.A.; Jain, M.K. Kruppel-like Factors (KLFs) in muscle biology. J. Mol. Cell. Cardiol. 2007, 43, 1–10. [Google Scholar] [CrossRef]
- Moore, D.L.; Apara, A.; Goldberg, J.L. Krüppel-like transcription factors in the nervous system: Novel players in neurite outgrowth and axon regeneration. Mol. Cell. Neurosci. 2011, 47, 233–243. [Google Scholar] [CrossRef]
- Kanazawa, A.; Kawamura, Y.; Sekine, A.; Iida, A.; Tsunoda, T.; Kashiwagi, A.; Tanaka, Y.; Babazono, T.; Matsuda, M.; Kawai, K. Single nucleotide polymorphisms in the gene encoding Krüppel-like factor 7 are associated with type 2 diabetes. Diabetologia 2005, 48, 1315. [Google Scholar] [CrossRef] [PubMed]
- Yoshihiro, K.; Yasushi, T.; Ryuzo, K.; Shiro, M. Overexpression of Kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol. Endocrinol. 2006, 20, 844–856. [Google Scholar] [CrossRef]


| Constituent | Hay (%) | Concentrate Mixture (%) |
|---|---|---|
| Crude protein | 3.65 | 16.74 |
| Crude fat | 1.72 | 3.15 |
| Crude fiber | 68.3 | 6.00 |
| Crude ash | 7.53 | 5.27 |
| Calcium | 0.54 | 0.84 |
| Phosphorus | 0.47 | 0.50 |
| miRNA | miRNA Sequence (5′–3′) | Forward Primer Sequence (5′–3′) |
|---|---|---|
| bta-miR-483 | UCACUCCUCUCCUCCCGUCUU | TCACTCCTCTCCTCCCGTCTT |
| bta-miR-449a | UGGCAGUGUAUUGUUAGCUGGU | TGGCAGTGTATTGTTAGCTGGT |
| bta-miR-200b | UAAUACUGCCUGGUAAUGAUG | TAATACTGCCTGGTAATGATG |
| bta-miR-200a | UAACACUGUCUGGUAACGAUGUU | TAACACTGTCTGGTAACGATGTT |
| bta-miR-205 | UCCUUCAUUCCACCGGAGUCUG | TCCTTCATTCCACCGGAGTCT |
| bta-miR-223 | UGUCAGUUUGUCAAAUACCCCA | TGTCAGTTTGTCAAATACCCCA |
| bta-miR-2431-5p | CAGGUCAUAUAAGUGUGGAGUU | CAGGTCATATAAGTGTGGAGTT |
| Parameter | Groups | SEM | p-Value | |
|---|---|---|---|---|
| CON (kg) | CS (kg) | |||
| Initial Weight | 171.7 ± 24.83 a | 172.8 ± 31.32 a | 7.20 | 0.901 |
| Final Weight | 170.4 ± 23.88 a | 202.9 ± 31.75 b | 9.57 | 0.037 |
| Average Daily Gain | −0.008 ± 0.01 a | 0.283 ± 0.11 b | 0.034 | ≤0.01 |
| Parameter | Days | Groups | SEM | p-Value | |
|---|---|---|---|---|---|
| CON | CS | ||||
| GLU (mmol/L) | 0 | 2.84 ± 0.25 a | 2.87 ± 0.21 a | 0.06 | 0.761 |
| 106 | 2.69 ± 0.28 a | 3.63 ± 0.31 b | 0.10 | ≤0.01 | |
| TP (g/L) | 0 | 69.60 ± 4.37 a | 70.40 ± 3.27 a | 1.34 | 0.649 |
| 106 | 66.50 ± 3.86 a | 84.00 ± 5.85 b | 1.85 | ≤0.01 | |
| ALB (g/L) | 0 | 22.20 ± 1.03 a | 22.60 ± 1.65 b | 0.32 | 0.523 |
| 106 | 21.00 ± 1.30 a | 32.80 ± 2.57 b | 0.55 | ≤0.01 | |
| TRIG (mmol/L) | 0 | 0.15 ± 0.06 a | 0.13 ± 0.03 a | 0.02 | 0.483 |
| 106 | 0.11 ± 0.03 a | 0.22 ± 0.06 b | 0.01 | ≤0.01 | |
| CHOL (mmol/L) | 0 | 2.27 ± 0.37 a | 2.24 ± 0.31 a | 0.12 | 0.855 |
| 106 | 2.11 ± 0.16 a | 3.59 ± 0.24 b | 0.08 | ≤0.01 | |
| Parameter | Days | Groups | SEM | p-Value | |
|---|---|---|---|---|---|
| CON | CS | ||||
| GnRH (mIU/mL) | 0 | 8.1875 ± 1.3287 a | 8.5225 ± 1.2386 a | 0.42 | 0.567 |
| 106 | 7.8782 ± 1.8952 a | 15.9439 ± 2.8643 b | 0.59 | ≤0.01 | |
| FSH (mIU/mL) | 0 | 1.1149 ± 0.2541 a | 1.1788 ± 0.1697 a | 0.08 | 0.517 |
| 106 | 0.9870 ± 0.2471 a | 2.6079 ± 0.1806 b | 0.07 | ≤0.01 | |
| LH (mIU/mL) | 0 | 1.0563 ± 0.2505 a | 1.1102 ± 0.2677 a | 0.07 | 0.648 |
| 106 | 0.9431 ± 0.2780 a | 2.6328 ± 0.2877 b | 0.09 | ≤0.01 | |
| E2 (pg/mL) | 0 | 10.2806 ± 4.1495 a | 10.9653 ± 4.2415 a | 0.68 | 0.719 |
| 106 | 6.2496 ± 1.4354 a | 16.1687 ± 2.0088 b | 0.63 | ≤0.01 | |
| P (pmol/L) | 0 | 450.4571 ± 50.2930 a | 458.0429 ± 29.6429 a | 9.37 | 0.686 |
| 106 | 471.1857 ± 44.5153 a | 715.0143 ± 780,343 b | 14.0 | ≤0.01 | |
| Types | CS | CON | ||
|---|---|---|---|---|
| Number | Percentage | Number | Percentage | |
| rRNA | 1425029 | 3.46% | 1800091 | 3.41% |
| snRNA | 130 | 0.00% | 142 | 0.00% |
| scRNA | 0 | 0.00% | 0 | 0.00% |
| snoRNA | 145152 | 0.35% | 112544 | 0.21% |
| tRNA | 89338 | 0.22% | 72260 | 0.14% |
| Repbase | 131696 | 0.32% | 174194 | 0.33% |
| Unannotated | 59020 | 95.65% | 50568470 | 95.91% |
| Total | 16362355 | 100% | 52727701 | 100% |
| MiRNA-mRNA | Predicted Consequential Pairing of Target Region (Top) and miRNA (Bottom) | Site Type | Context Score | Context Score Percentile | Weighted Context Score | Conserved Branch Length | PCT |
|---|---|---|---|---|---|---|---|
| Position 1903-1909 of LEP 3′ UTR bta-miR-483 | 5′ ....AGUAGGGCAGAGGGCAGGAGUGU... | | | | | | | | | | | 3′ UUCUGCCCUCCUCUCCUCACU | 7mer-m8 | −0.16 | 86 | −0.16 | 0.748 | N/A |
| Position 190-196 of NOTCH1 3′ UTR bta-miR-449a | 5′ ...UAUUUUACAUGGAAACACUGCCU... | | | | | | | 3′ UGGUCGAUUGUUAUGUGACGGU | 7mer-m8 | −0.49 | 98 | −0.49 | 6.934 | 0.79 |
| Position 2688-2694 of KLF7 3′ UTR bta-miR-223 | 5′ ...UUAAUUUAAUUUUUG-AACUGACC... | | | | | | | | | | | 3′ ACCCCAUAAACUGUUUGACUGU | 7mer-m8 | −0.21 | 87 | −0.21 | 3.082 | 0.33 |
| Position 1328-1334 of VEGFA 3′ UTR bta-miR-200b | 5′ ...GAGUAGGGUUUUUUUCAGUAUUC... | | | | | | | 3′ GUAGUAAUGGUCCGUCAUAAU | 7mer-m8 | −0.23 | 95 | −0.19 | 4.184 | 0.62 |
| Position 4877-4883 of GNAQ 3′ UTR bta-miR-200b | 5′ ...AACAUUUUUAACUUGCAGUAUUU... | | | | | | | | | | 3′ GUAGUAAUGGUCCGUCAUAAU | 7mer-m8 | −0.18 | 91 | −0.15 | 3.766 | 0.53 |
| Position 736-743 of GATA6 3′ UTR bta-miR-200a | 5′ ....UUGCGUUGCAGCAAUCAGUGUUA... | | | | | | | | | | | 3′ .UUGUAGCAAUGGUCU--GUCACAAU | 8mer | −0.43 | 99 | −0.42 | 10.634 | 0.86 |
| Position5142-5148 of CCND2 3′ UTR bta-miR-2431-5p | 5′ ....ACAAUAAACUCACCUUGACCUAA... | | | | | | 3′ UUGAGGUGUGAAUAUACUGGAC | 7mer-A1 | −0.19 | 91 | 0.00 | 0 | N/A |
| miRNAs | Genes |
|---|---|
| bta-miR-483 | LEP |
| bta-miR-449a | NOTCH1 |
| bta-miR-223 | KLF7 |
| bta-miR-200b | VEGFA |
| GNAQ | |
| bta-miR-200a | GTAT6 |
| bta-miR-2431-5p | CCND2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, J.; Kalwar, Q.; Yan, P.; Guo, X. Effect of Concentrate Supplementation on the Expression Profile of miRNA in the Ovaries of Yak during Non-Breeding Season. Animals 2020, 10, 1640. https://doi.org/10.3390/ani10091640
Xie J, Kalwar Q, Yan P, Guo X. Effect of Concentrate Supplementation on the Expression Profile of miRNA in the Ovaries of Yak during Non-Breeding Season. Animals. 2020; 10(9):1640. https://doi.org/10.3390/ani10091640
Chicago/Turabian StyleXie, Jianpeng, Qudratullah Kalwar, Ping Yan, and Xian Guo. 2020. "Effect of Concentrate Supplementation on the Expression Profile of miRNA in the Ovaries of Yak during Non-Breeding Season" Animals 10, no. 9: 1640. https://doi.org/10.3390/ani10091640
APA StyleXie, J., Kalwar, Q., Yan, P., & Guo, X. (2020). Effect of Concentrate Supplementation on the Expression Profile of miRNA in the Ovaries of Yak during Non-Breeding Season. Animals, 10(9), 1640. https://doi.org/10.3390/ani10091640





