The Milk Microbiota of the Spanish Churra Sheep Breed: New Insights into the Complexity of the Milk Microbiome of Dairy Species
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
Churra Milk Sampling and Bioinformatic Data Analysis
3. Results
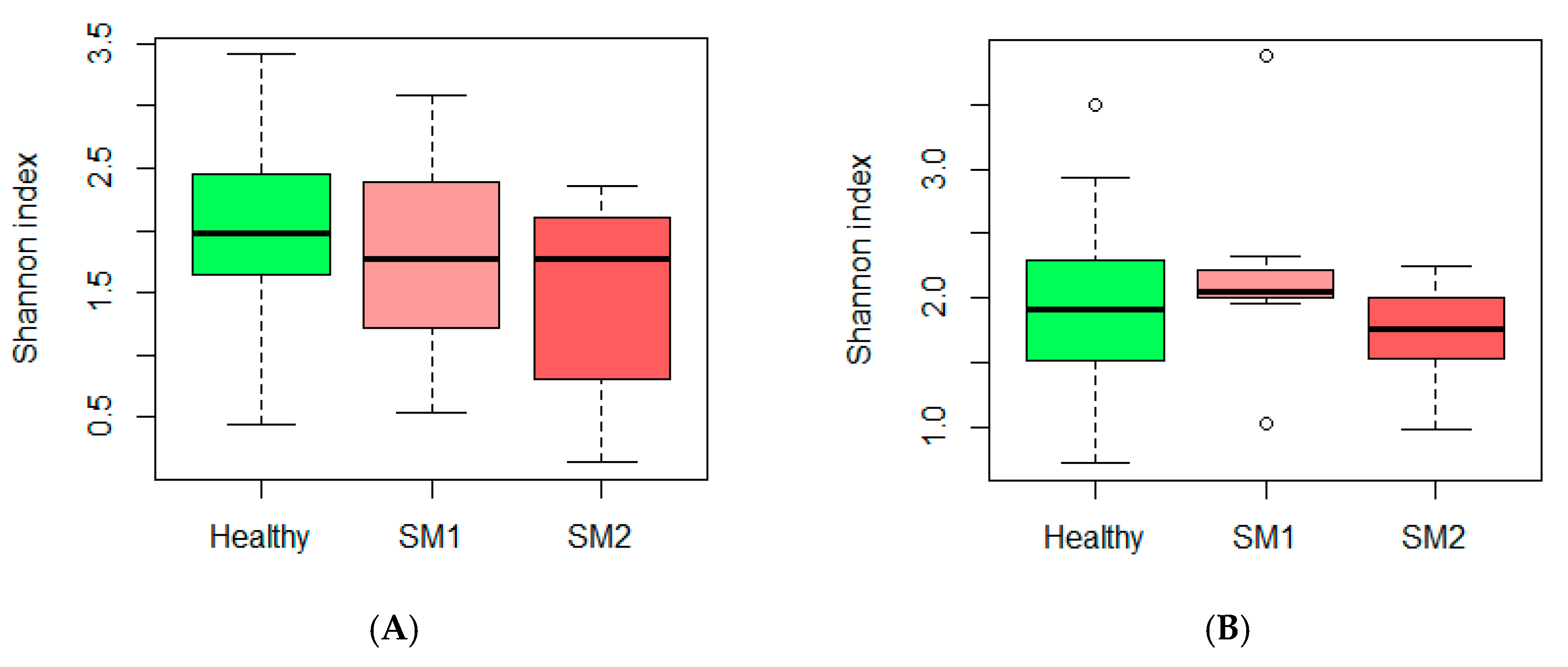
3.1. Microbiota and Diversity Profile in Churra Samples
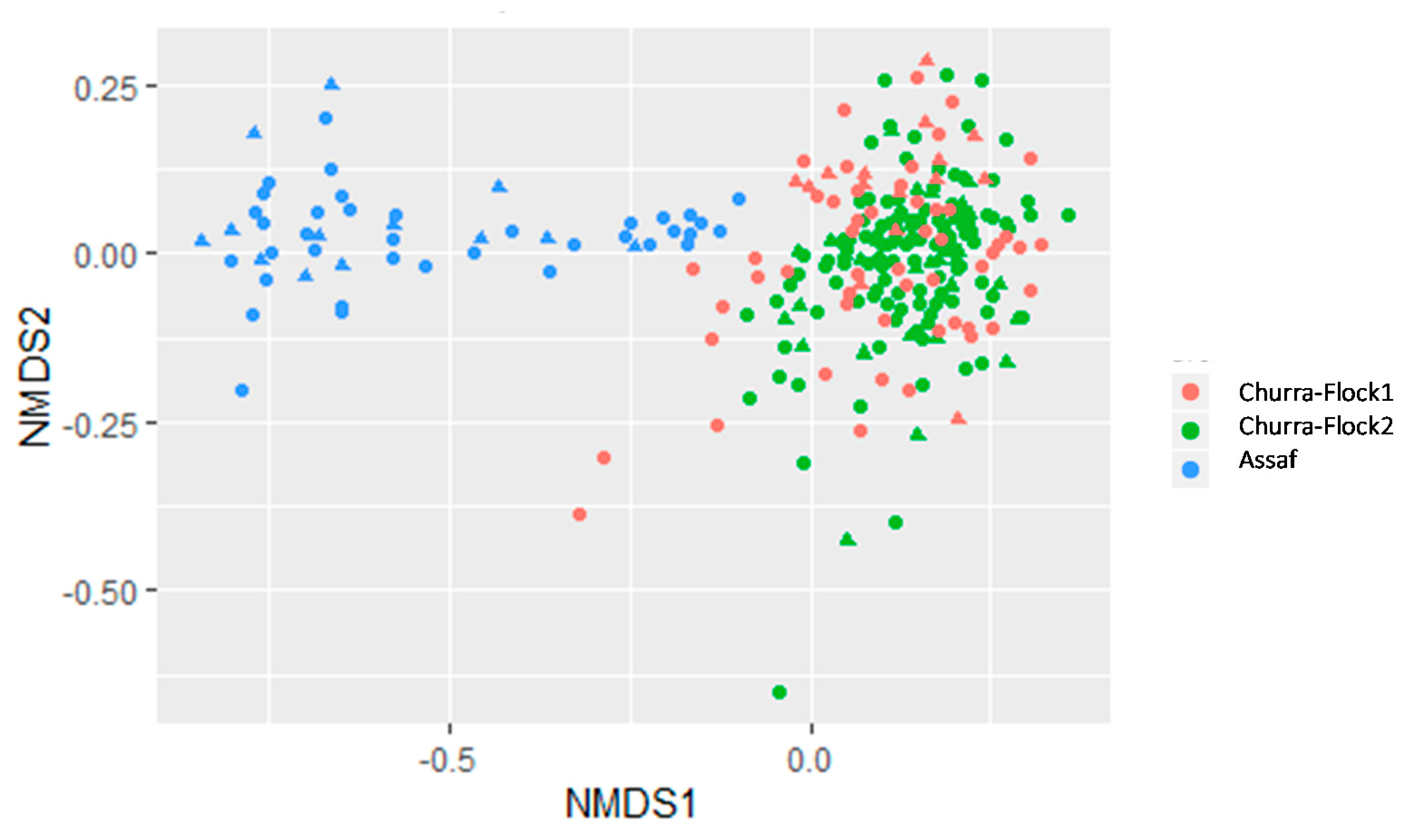
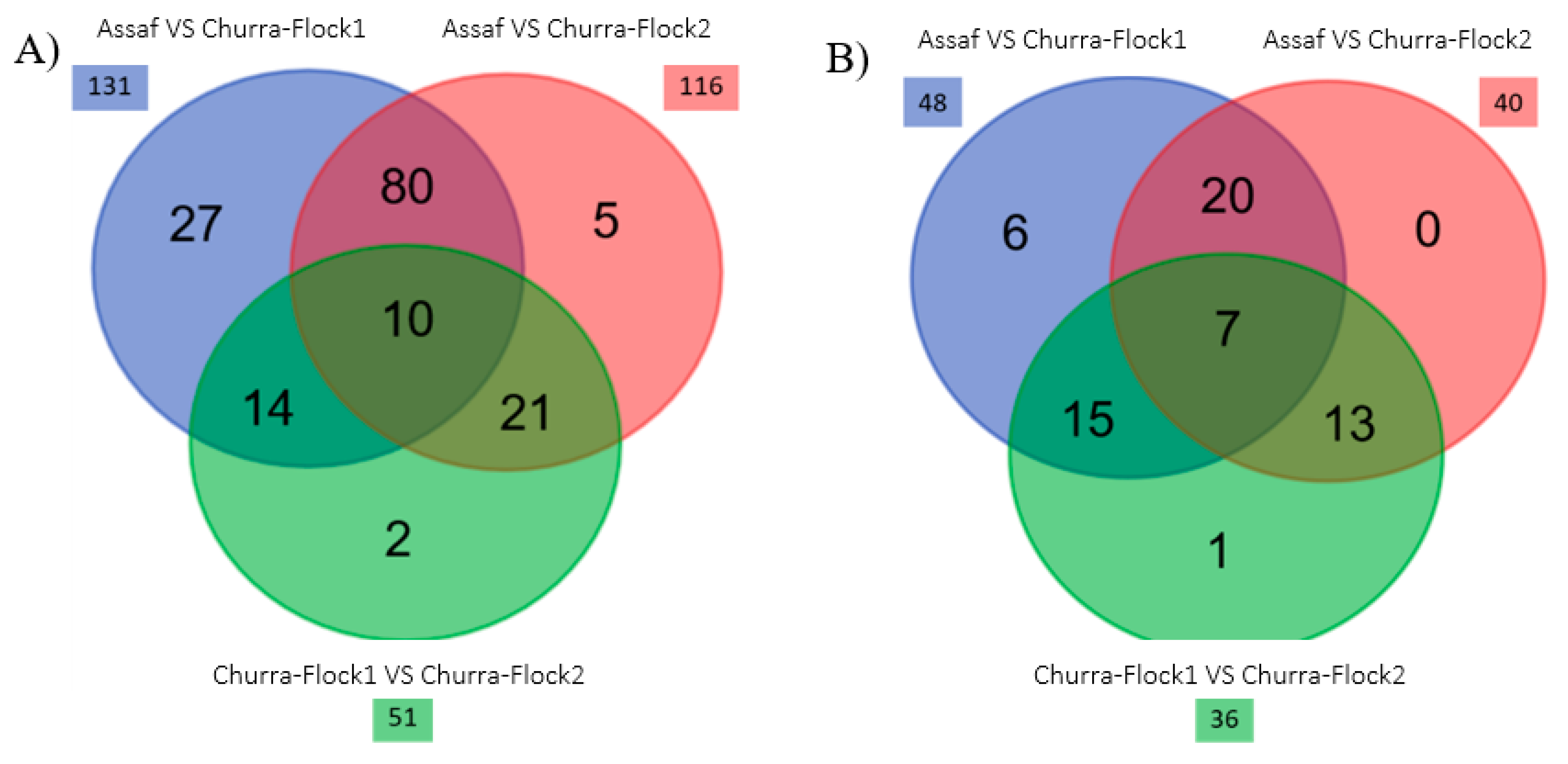
3.2. Comparative Study of the Milk Microbiota in Churra and Assaf Flocks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oikonomou, G.; Machado, V.S.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbial Diversity of Bovine Mastitic Milk as Described by Pyrosequencing of Metagenomic 16s rDNA. PLoS ONE 2012, 7, e47671. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, J.S.; Gorden, P.J.; Munro, D.; Rong, R.; Dong, Q.; Plummer, P.J.; Wang, C.; Phillips, G.J. Bacterial Community Profiling of Milk Samples as a Means to Understand Culture-Negative Bovine Clinical Mastitis. PLoS ONE 2013, 8, e61959. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef] [PubMed]
- Catozzi, C.; Sanchez Bonastre, A.; Francino, O.; Lecchi, C.; De Carlo, E.; Vecchio, D.; Martucciello, A.; Fraulo, P.; Bronzo, V.; Cusco, A.; et al. The microbiota of water buffalo milk during mastitis. PLoS ONE 2017, 12, e0184710. [Google Scholar] [CrossRef] [PubMed]
- Esteban-Blanco, C.; Gutierrez-Gil, B.; Puente-Sanchez, F.; Marina, H.; Tamames, J.; Acedo, A.; Arranz, J.J. Microbiota characterization of sheep milk and its association with somatic cell count using 16s rRNA gene sequencing. J. Anim. Breed. Genet. 2019. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef]
- Oikonomou, G.; Bicalho, M.L.; Meira, E.; Rossi, R.E.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.V.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbiota of Cow’s Milk; Distinguishing Healthy, Sub-Clinically and Clinically Diseased Quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef]
- Pang, M.; Xie, X.; Bao, H.; Sun, L.; He, T.; Zhao, H.; Zhou, Y.; Zhang, L.; Zhang, H.; Wei, R.; et al. Insights Into the Bovine Milk Microbiota in Dairy Farms With Different Incidence Rates of Subclinical Mastitis. Front. Microbiol. 2018, 9, 2379. [Google Scholar] [CrossRef]
- Taponen, S.; McGuinness, D.; Hiitiö, H.; Simojoki, H.; Zadoks, R.; Pyörälä, S. Bovine milk microbiome: A more complex issue than expected. Vet. Res. 2019, 50, 44. [Google Scholar] [CrossRef]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What Are We Exactly Talking About? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente, L.-F.; Fernández, G.; San Primitivo, F. Breeding programme for the Spnaish Churra sheep breed. Cah. Options Méditerr. 1995, 11, 165–172. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rodriguez, M.C.; Gonzalo, C.; San Primitivo, F.; Carmenes, P. Relationship between somatic cell count and intramammary infection of the half udder in dairy ewes. J. Dairy Sci. 1995, 78, 2753–2759. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Hiergeist, A.; Gläsner, J.; Reischl, U.; Gessner, A. Analyses of intestinal microbiota: Culture versus sequencing. ILAR J. 2015, 56, 228–240. [Google Scholar] [CrossRef]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef]
- Derakhshani, H.; Fehr, K.B.; Sepehri, S.; Francoz, D.; De Buck, J.; Barkema, H.W.; Plaizier, J.C.; Khafipour, E. Invited review: Microbiota of the bovine udder: Contributing factors and potential implications for udder health and mastitis susceptibility. J. Dairy Sci. 2018, 101, 10605–10625. [Google Scholar] [CrossRef]
- Dario, C.G.B. Investigation of Mastitis Occurrence in Purebred and Crossbred Ewes; Owen, J.B., Axford, R.F.E., Eds.; Breeding for Disease Resistance in Farm Animals: Wallingford, UK, 1991. [Google Scholar]
- Gonzalo, C.; Carriedo, J.A.; Blanco, M.A.; Beneitez, E.; Juárez, M.T.; De La Fuente, L.F.; Primitivo, F.S. Factors of Variation Influencing Bulk Tank Somatic Cell Count in Dairy Sheep. J. Dairy Sci. 2005, 88, 969–974. [Google Scholar] [CrossRef]
- Gonzalo, C. Control de las Condiciones Higiénico-Sanitarias de la Leche en Ganaderías de Ovino y Caprino; Jornada del programa de formación agraria, El Centro Regional de Selección y Reproducción Animal de Valdepeñas (CERSYRA): Valdepeñas, Spain, 2008. [Google Scholar]
- Brüggemann, H.; Henne, A.; Hoster, F.; Liesegang, H.; Wiezer, A.; Strittmatter, A.; Hujer, S.; Dürre, P.; Gottschalk, G. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science 2004, 305, 671–673. [Google Scholar] [CrossRef] [PubMed]
- Postollec, F.; Mathot, A.-G.; Bernard, M.; Divanac’h, M.-L.; Pavan, S.; Sohier, D. Tracking spore-forming bacteria in food: From natural biodiversity to selection by processes. Int. J. Food Microbiol. 2012, 158, 1–8. [Google Scholar] [CrossRef]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef] [PubMed]
- Bobbo, T.; Ruegg, P.L.; Stocco, G.; Fiore, E.; Gianesella, M.; Morgante, M.; Pasotto, D.; Bittante, G.; Cecchinato, A. Associations between pathogen-specific cases of subclinical mastitis and milk yield, quality, protein composition, and cheese-making traits in dairy cows. J. Dairy Sci. 2017, 100, 4868–4883. [Google Scholar] [CrossRef] [PubMed]
- Mahmmod, Y.S.; Klaas, I.C.; Svennesen, L.; Pedersen, K.; Ingmer, H. Communications of Staphylococcus aureus and non-aureus Staphylococcus species from bovine intramammary infections and teat apex colonization. J. Dairy Sci. 2018, 101, 7322–7333. [Google Scholar] [CrossRef] [PubMed]
- Jara, S.; Sanchez, M.; Vera, R.; Cofre, J.; Castro, E. The inhibitory activity of Lactobacillus spp. isolated from breast milk on gastrointestinal pathogenic bacteria of nosocomial origin. Anaerobe 2011, 17, 474–477. [Google Scholar] [CrossRef]
- Coeuret, V.; Dubernet, S.; Bernardeau, M.; Gueguen, M.; Vernoux, J.P. Isolation, characterisation and identification of lactobacilli focusing mainly on cheeses and other dairy products. Lait 2003, 83, 269–306. [Google Scholar] [CrossRef]
- Khalid, N.M.; Marth, E.H. Lactobacilli—Their Enzymes and Role in Ripening and Spoilage of Cheese: A Review. J. Dairy Sci. 1990, 73, 2669–2684. [Google Scholar] [CrossRef]
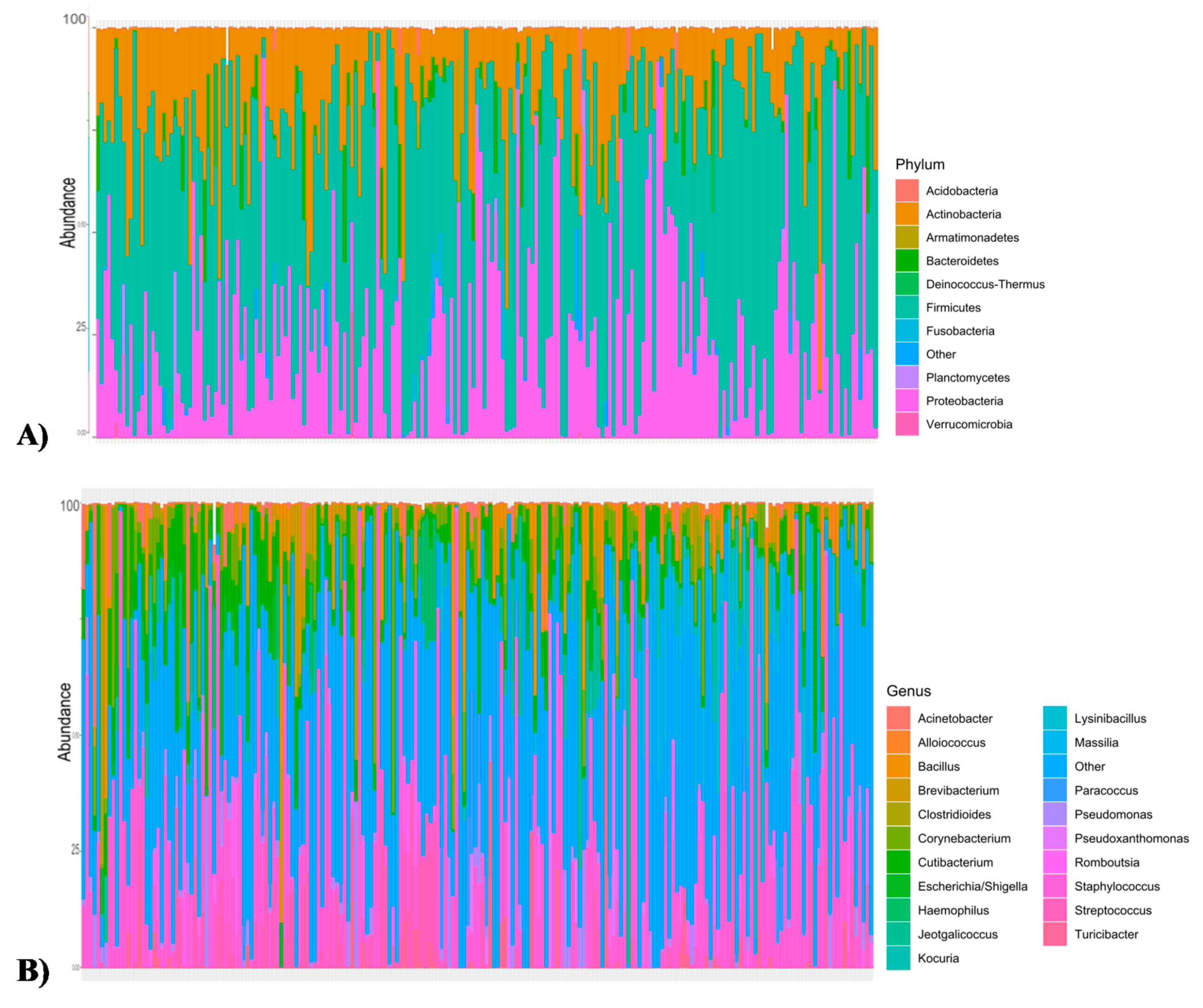
| Level | Churra | Assaf (Esteban-Blanco et al. [5]) |
|---|---|---|
| At phylum level | Firmicutes (50.28%) Proteobacteria (25.5%) Actinobacteria (18.9%) Bacteroidetes (2.6%) | Firmicutes (64.44%) Actinobacteria (14.25%) Proteobacteria (9.08%) Acidobacteria (2.7%) Bacteroidetes (2.3%) |
| At genus level | Staphylococcus (20.29%) Cutibacterium (6.27%) Corynebacterium (4.34%) Streptococcus (4.1%) Massilia (3.5%) Bacillus (3.2%) | Staphylococcus (16.8%) Lactobacillus (14.1%) Corynebacterium (8.8%) Alloiococcus (6.8%) Streptococcus (4%) Romboutsia (3%) |
| All Samples (n = 212) | Healthy (n = 166) | SM (n = 46) |
|---|---|---|
| Staphylococcus (20.3%) | Staphylococcus (14.1%) | Staphylococcus (42.6%) |
| Cutibacterium (6.3%) | Cutibacterium (7.1%) | Cutibacterium (3.4%) |
| Corynebacterium (4.3%) | Corynebacterium (4.3%) | Corynebacterium (4.5%) |
| Streptococcus (4.1%) | Streptococcus (4.3%) | Streptococcus (3.5%) |
| Massilia (3.5%) | Massilia (3.6%) | Massilia (3.1%) |
| Bacillus (3.2%) | Bacillus (3.7%) | Enterococcus (2.4%) |
| Romboutsia (2.6%) | Romboutsia (3.1%) | Escherichia/Shigella (2.1%) |
| Pseudomonas (2.4%) | Pseudomonas (2.6%) | Mannheimia (1.9%) |
| Jeotgalicoccus (2.2%) | Jeotgalicoccus (2.5%) | Pseudomonas (1.7%) |
| Escherichia/Shigella (2.2%) | Lysinibacillus (2.5%) | Alloiococcus (1.7%) |
| Pairwiswe Comparations | Genus | Base Mean | log2FC | lfcSE | Stat | p-Value | p-Adj |
|---|---|---|---|---|---|---|---|
| Assaf vs. Churra-Flock1 | Cutibacterium | 5597.47 | −11.55 | 0.43 | −26.92 | 1.35 × 10−159 | 1.19 × 10−157 |
| Bacillus | 1864.60 | −4.96 | 0.62 | −8.05 | 8.31 × 10−16 | 9.14 × 10−15 | |
| Staphylococcus | 27,203.72 | −2.43 | 0.45 | −5.45 | 5.17 × 10−8 | 3.03 × 10−7 | |
| Lactobacillus | 4126.47 | 5.85 | 1.14 | 5.15 | 2.65 × 10−7 | 1.23 × 10−6 | |
| Assaf vs. Churra-Flock2 | Cutibacterium | 5597.47 | −10.07 | 0.48 | −20.75 | 1.29 × 10−95 | 1.13 × 10−93 |
| Bacillus | 1864.60 | −4.65 | 0.70 | −6.63 | 3.42 × 10−11 | 3.01 × 10−10 | |
| Staphylococcus | 27,203.72 | −1.82 | 0.51 | −3.58 | 3.48 × 10−4 | 1.13 × 10−3 | |
| Lactobacillus | 4126.47 | 4.15 | 1.29 | 3.20 | 1.35 × 10−3 | 4.25 × 10−3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esteban-Blanco, C.; Gutiérrez-Gil, B.; Marina, H.; Pelayo, R.; Suárez-Vega, A.; Acedo, A.; Arranz, J.-J. The Milk Microbiota of the Spanish Churra Sheep Breed: New Insights into the Complexity of the Milk Microbiome of Dairy Species. Animals 2020, 10, 1463. https://doi.org/10.3390/ani10091463
Esteban-Blanco C, Gutiérrez-Gil B, Marina H, Pelayo R, Suárez-Vega A, Acedo A, Arranz J-J. The Milk Microbiota of the Spanish Churra Sheep Breed: New Insights into the Complexity of the Milk Microbiome of Dairy Species. Animals. 2020; 10(9):1463. https://doi.org/10.3390/ani10091463
Chicago/Turabian StyleEsteban-Blanco, Cristina, Beatriz Gutiérrez-Gil, Héctor Marina, Rocío Pelayo, Aroa Suárez-Vega, Alberto Acedo, and Juan-José Arranz. 2020. "The Milk Microbiota of the Spanish Churra Sheep Breed: New Insights into the Complexity of the Milk Microbiome of Dairy Species" Animals 10, no. 9: 1463. https://doi.org/10.3390/ani10091463
APA StyleEsteban-Blanco, C., Gutiérrez-Gil, B., Marina, H., Pelayo, R., Suárez-Vega, A., Acedo, A., & Arranz, J.-J. (2020). The Milk Microbiota of the Spanish Churra Sheep Breed: New Insights into the Complexity of the Milk Microbiome of Dairy Species. Animals, 10(9), 1463. https://doi.org/10.3390/ani10091463







