Mitochondrial DNA Diversity in Large White Pigs in Russia
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. PCR and Sequencing
2.3. Editing and Alignment of Sequences
2.4. Genetic Diversity
2.5. Haplogroups of MtDNA
2.6. Differences in the Distribution of Haplotypes and Haplogroups of MtDNA
3. Results
3.1. Genetic Diversity of Large White and Yorkshire Pig Populations in Russia
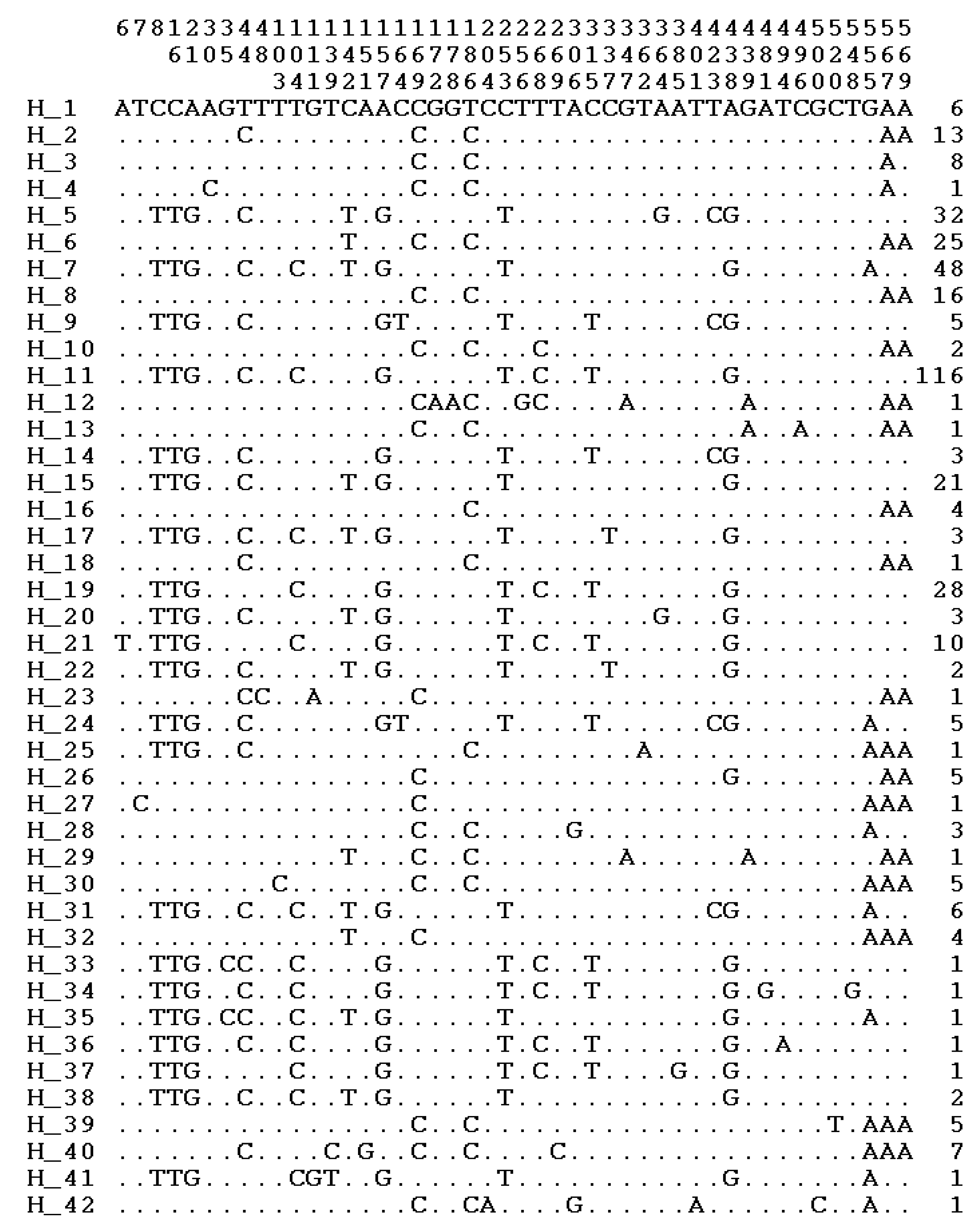
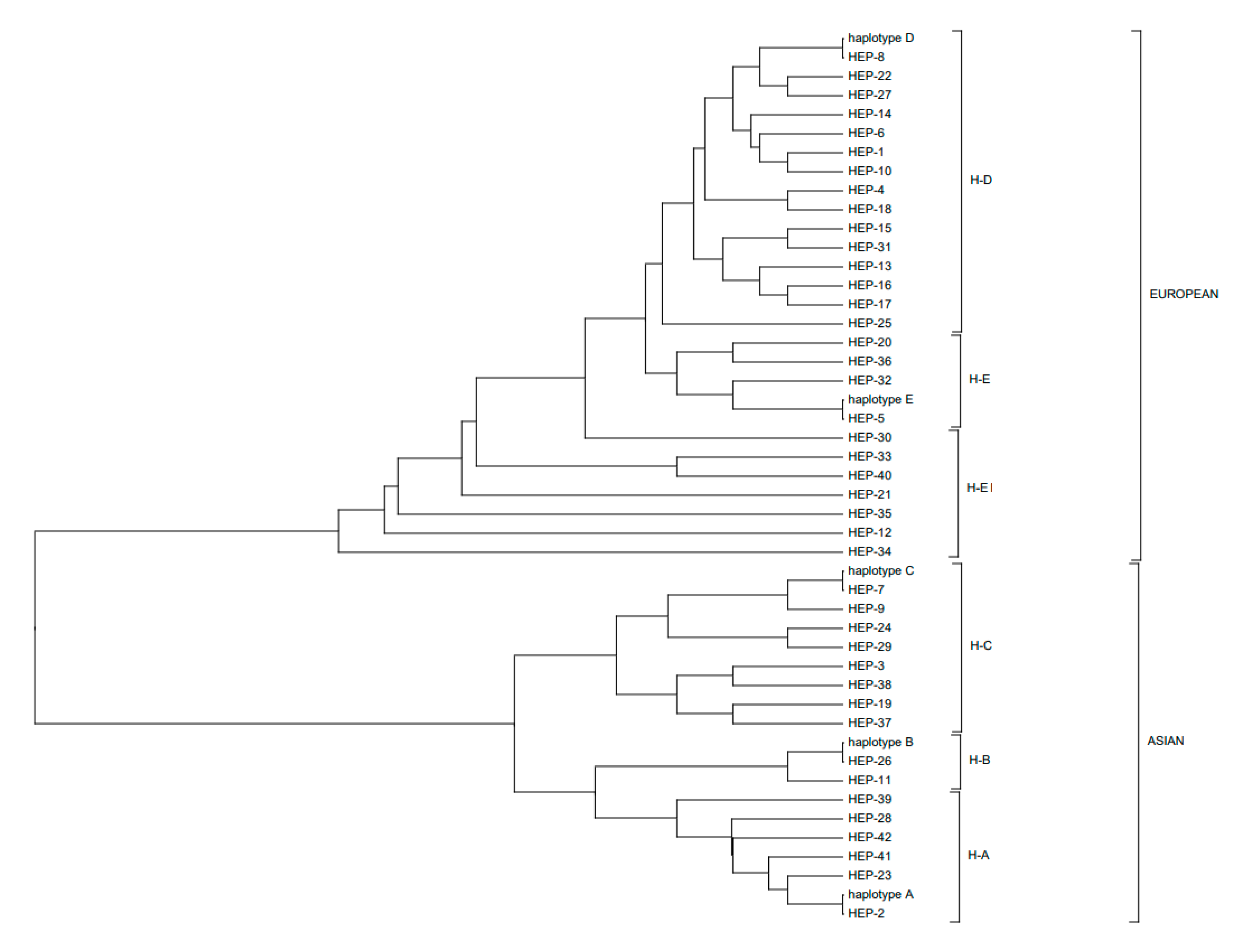
3.2. Polymorphic Sites, Haplotypes, and Haplogroups
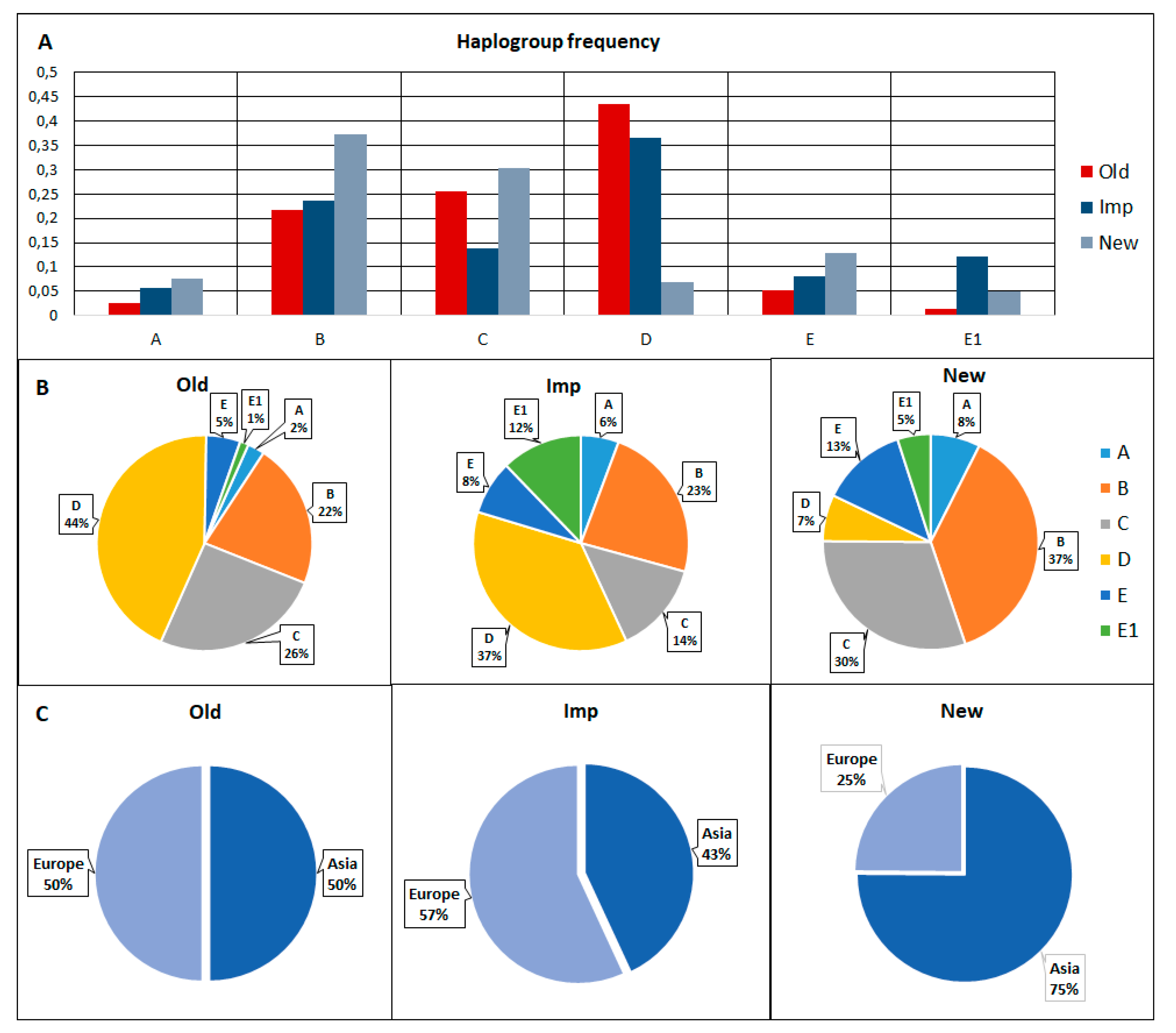
3.3. Haplotype and Haplogroup Frequencies between the Old, Imp, and New Groups of Pigs
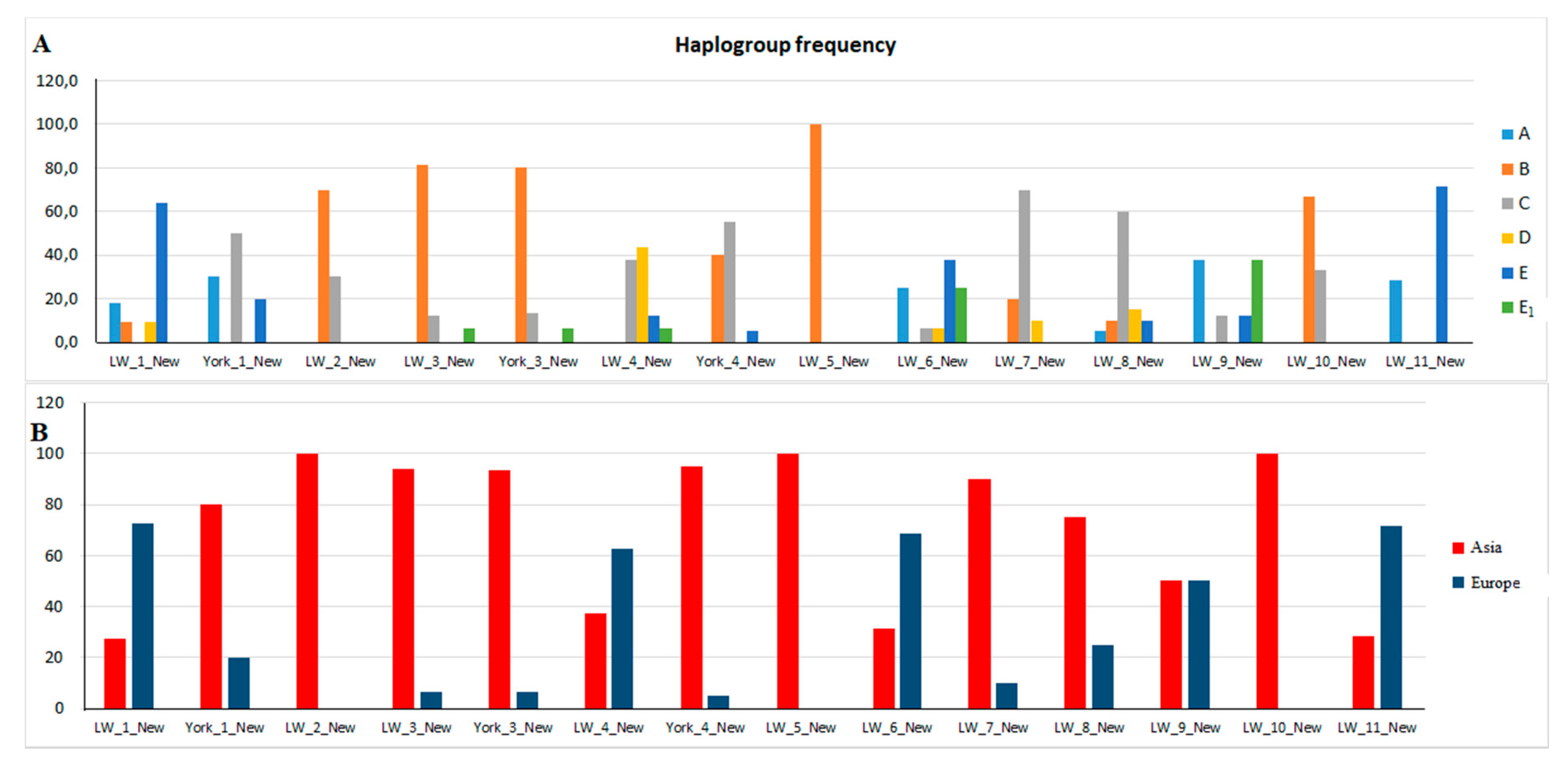
3.4. Haplotypes and Haplogroups in Pigs of the New Group
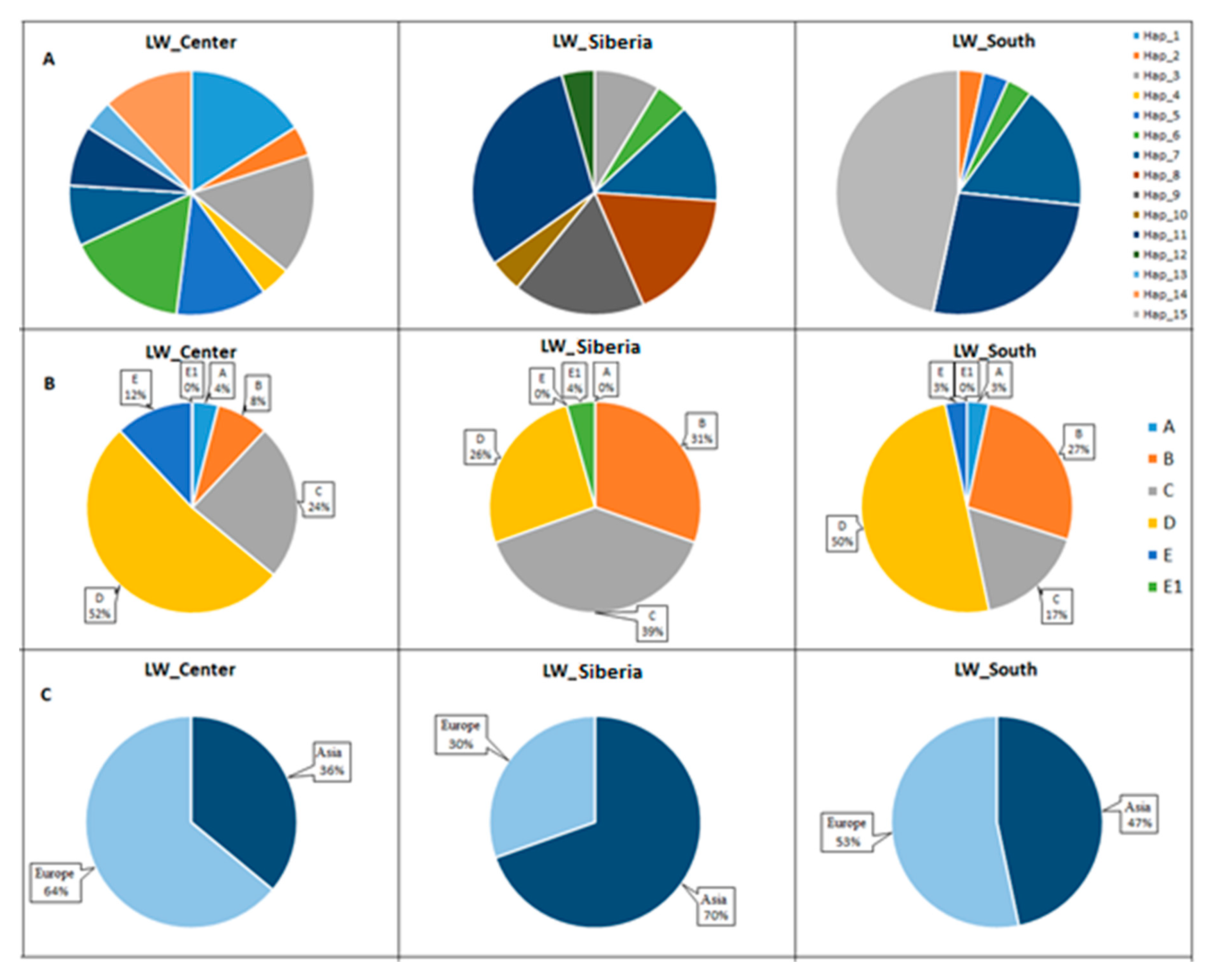
3.5. Frequencies of Haplotypes and Haplogroups in Old_Center, Old_Siberia, and Old_South Groups of Pigs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Amer, P.; Allain, D.; Avendano, S.; Baselga, M.; Boettcher, P.; Dürr, J.; Garreau, H.; Gootwine, E.; Gutierrez, G.; Knap, P.; et al. “Breeding strategies and programmes” Animal Science White Papers, Technical Reports and Fact Sheets. Anim. Sci. 2015, 6, 449–495. [Google Scholar]
- Wilkinson, S.; Lu, Z.H.; Megens, H.J.; Archibald, A.L.; Haley, C.; Jackson, I.J.; Groenen, M.A.; Crooijmans, R.P.; Ogden, R.; Wiener, P. Signatures of diversifying selection in European pig breeds. PLoS Genet. 2013, 9, e1003453. [Google Scholar] [CrossRef] [PubMed]
- Traspov, A.; Deng, W.; Kostyunina, O.; Ji, J.; Shatokhin, K.; Lugovoy, S.; Zinovieva, N.; Yang, B.; Huang, L. Population structure and genome characterization of local pig breeds in Russia, Belorussia, Kazakhstan and Ukraine. Genet. Sel. Evol. 2016, 1, 16. [Google Scholar] [CrossRef] [PubMed]
- Galtier, N.; Enard, D.; Radondy, Y.; Bazin, E.; Belkhir, K. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 2006, 16, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.M.; George, M.; Wilson, A.C. Rapid evolution of animal mitochondrial-DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef] [PubMed]
- Hill, G.E. Mitonuclear Ecology. Mol. Biol. Evol. 2015, 32, 1917–1927. [Google Scholar] [CrossRef]
- Tsai, T.S.; Rajasekar, S.; St John, J.C. The relationship between mitochondrial DNA haplotype and the reproductive capacity of domestic pigs (Sus scrofa domesticus). BMC Genet. 2016, 17, 67. [Google Scholar] [CrossRef]
- Toro, M.A.; Rodrigáñez, J.J.; Silió, L.; Rodr’ıguez, M.C. Genealogical analysis of a closed herd of black hairless Iberian pigs, cons. Conserv. Biol. 2000, 14, 1843–1851. [Google Scholar] [CrossRef]
- Srivastava, G.K.; Rajput, N.; Jadav, K.K.; Shrivastav, A.B.; Joshi, H.R. Single nucleotide markers of D-loop for identification of Indian wild pig (Sus scrofa cristatus). Vet. World. 2015, 8, 532–536. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, B.; Wen, X.; Sun, G. Genetic variation and relationships in the mitochondrial DNA D-loop region of Qinghai indigenous and commercial pig breeds. Cell. Mol. Biol. Lett. 2018, 23, 31. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.; Han, J.; Mo, D.; Li, X.; Liu, X. Mitochondrial DNA diversity and origin of indigenous pigs in South China and their contribution to western modern pig breeds. J. Integr. Agric. 2019, 18, 2338–2350. [Google Scholar] [CrossRef]
- Available online: https://www.vij.ru/infrastruktura/ckp (accessed on 21 February 2020).
- Available online: https://www.vij.ru/infrastruktura/46-infrastruktura/286-unikalnaya-nauchnaya-ustanovka-unu (accessed on 21 February 2020).
- Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 21 February 2020).
- Available online: https://bioedit.software.informer.com/7.2/ (accessed on 21 March 2020).
- Available online: https://mega.software.informer.com/7.2/ (accessed on 21 March 2020).
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Jiao, T.; Zhao, S. Genetic diversity in the mitochondrial DNA D-loop region of global swine (Sus scrofa) populations. Biochem. Biophys. Res. Commun. 2016, 473, 814–820. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. 2018. Available online: https://www.R-project.org (accessed on 21 April 2020).
- Mori, S.; Matsunami, M. Signature of positive selection in mitochondrial DNA in Cetartiodactyla. Genes Genet. Syst. 2018, 93, 65–73. [Google Scholar] [CrossRef]
- Li, X.-D.; Jiang, G.-F.; Yan, L.-Y.; Li, R.; Mu, Y.; Deng, W.-A. Positive Selection Drove the Adaptation of Mitochondrial Genes to the Demands of Flight and High-Altitude Environments in Grasshoppers. Front. Genet. 2017, 17, 46. [Google Scholar] [CrossRef]
- Lajbner, Z.; Pnini, R.; Camus, M.F.; Miller, J.; Dowling, D.K. Experimental evidence that thermal selection shapes mitochondrial genome evolution. Sci. Rep. 2018, 8, 9500. [Google Scholar] [CrossRef]
- Niu, L.; Chen, X.; Xiao, P.; Zhao, Q.; Zhou, J.; Hu, J.; Sun, H.; Guo, J.; Li, L.; Wang, L.; et al. Detecting signatures of selection within the Tibetan sheep mitochondrial genome. Mitochondrial DNA Part A 2017, 28, 801–809. [Google Scholar] [CrossRef]
- Slimen, H.B.; Schaschl, H.; Knauer, F.; Suchentrunk, F. Selection on the mitochondrial ATP synthase 6 and the NADH dehydrogenase 2 genes in hares (Lepus capensis L., 1758) from a steep ecological gradient in Siberia Africa. BMC Evol. Biol. 2017, 17, 46. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Z.; Zhang, Y.; Gao, T.; Zou, J.; Zhang, X. Evidence for intra-mitochondrial variation in population genetic structure of Platycephalus sp.1 In the Siberiawestern Pacific. Mitochondrial DNA Part A 2019, 30, 281–288. [Google Scholar] [CrossRef]
- Oliviero, C.; Junnikkala, S.; Peltoniemi, O. The challenge of large litters on the immune system of the sow and the piglets. Reprod. Domest. Anim. 2019, 54, 12–21. [Google Scholar] [CrossRef]
- Bakoev, S.; Getmantseva, L.; Bakoev, F.; Kolosova, M.; Gabova, V.; Kolosov, A.; Kostyunina, O. Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig. Genes 2020, 11, 491. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Da Silva, C.L.A.; Soede, N.M. Recent advances in pig reproduction: Focus on impact of genetic selection for female fertility. Reprod. Domest. Anim. 2018, 53, 28–36. [Google Scholar] [CrossRef] [PubMed]
- White, S. From globalized pig breeds to capitalist pigs: A study in animal cultures and evolutionary history. Environ. Hist. 2011, 16, 94–120. [Google Scholar] [CrossRef]
- Jones, G.; Rothschild, M.; Ruvinsky, A. Genetic aspects of domestication, common breeds and their origin. In The Genetics of the Pig; Rothschild, M.F., Ruvinsky, A., Eds.; CAB International: Wallingford, UK, 1998. [Google Scholar]
- Chinnery, P.F.; Hudson, G. Mitochondrial genetics. Br. Med. Bull. 2013, 106, 135–159. [Google Scholar] [CrossRef]
- Sutarno Cummins, J.M.; Greeff, J.; Lymbery, A.J. Mitochondrial DNA polymorphisms and fertility in beef cattle. Theriogenology 2002, 57, 1603–1610. [Google Scholar] [CrossRef]
- Mannen, H.; Morimoto, M.L.; Oyamat, K.; Mukai, F.; Tsuji, S. Identification of mitochondrial DNA substitutions related to meat quality in Japanese black cattle. J. Anim. Sci. 2003, 81, 68–73. [Google Scholar] [CrossRef]
- Yen, N.T.; Lin, C.S.; Ju, C.C.; Wang, S.C.; Huang, M.C. Mitochondrial DNA polymorphism and determination of effects on reproductive trait in pigs. Reprod. Domest. Anim. 2007, 42, 387–392. [Google Scholar] [CrossRef]
- Fernandez, A.I.; Alves, E.; Fernandez, A.; de Pedro, E.; Lopez-Garcia, M.A.; Ovilo, C.; Rodriguez, M.C.; Silio, L. Mitochondrial genome polymorphisms associated with longissimus muscle composition in Iberian pigs. J. Anim. Sci. 2008, 86, 1283–1290. [Google Scholar] [CrossRef]
- St John, J.C.; Tsai, T. The association of mitochondrial DNA haplotypes and phenotypic traits in pigs. BMC Genet. 2018, 19, 41. [Google Scholar] [CrossRef]
- Nisztuk-Pacek, S.; Slaska, B.; Zieba, G.; Rozempolska-Rucinska, I. Two Mitochondrial Genes Are Associated with Performance Traits in Farmed Raccoon Dogs (Nyctereutes procyonoides). Czech J. Anim. Sci. 2018, 63, 110–118. [Google Scholar] [CrossRef]
- Kolosova, M.A.; Getmantseva, L.V.; Bakoev, S.Y.; Kolosov, A.Y.; Bakoev, N.F.; Shevtsova, V.S.; Lyashenko, E.N.; Kostyunina, O.V. Associations of mtDNA haplotypes with productive traits in pigs. Rend. Fis. Acc. Lincei 2019, 30, 807–813. [Google Scholar] [CrossRef]
- Hulsegge, I.; Calus, M.; Hoving-Bolink, R.; Lopes, M.; Megens, H.-J.; Oldenbroek, K. Impact of merging commercial breeding lines on the genetic diversity of Landrace pigs. Genet. Sel. Evol. 2018, 51, 60. [Google Scholar] [CrossRef] [PubMed]
| Group | Farm | n | Year of Sampling | Place of Sampling |
|---|---|---|---|---|
| Old, n = 78 | LW_1_Old | 12 | 2006 | Central District (Center) |
| LW_2_Old | 12 | 2011 | Siberia (Siberia) | |
| LW_3_Old | 11 | 2005 | Siberia (Siberia) | |
| LW_4_Old | 13 | 2003 | Central District (Center) | |
| LW_5_Old | 16 | 2009 | Southern District (South) | |
| LW_6_Old | 14 | 2011 | Southern District (South) | |
| Imp, n = 123 | York_1_Imp | 20 | 2008 | |
| York_2_Imp | 13 | 2010 | ||
| York_3_Imp | 16 | 2013 | ||
| York_4_Imp | 16 | 2010 | ||
| York_5_Imp | 15 | 2011 | ||
| York_6_Imp | 16 | 2014 | ||
| LW_1_Imp | 13 | 2011 | ||
| LW_2_Imp | 7 | 2010 | ||
| New, n = 201 | LW_1_New | 11 | 2017 | |
| LW_2_New | 10 | 2018 | ||
| LW_3_New | 16 | 2017 | ||
| LW_4_New | 16 | 2018 | ||
| LW_5_New | 20 | 2018 | ||
| LW_6_New | 16 | 2018 | ||
| LW_7_New | 20 | 2018 | ||
| LW_8_New | 20 | 2017 | ||
| LW_9_New | 8 | 2017 | ||
| LW_10_New | 12 | 2018 | ||
| LW_11_New | 7 | 2018 | ||
| York_1_New | 10 | 2017 | ||
| York_3_New | 15 | 2018 | ||
| York_4_New | 20 | 2018 |
| Group and Farm | N | S | H | Hd | K | Pi |
|---|---|---|---|---|---|---|
| LW_1_Old | 12 | 17 | 7 | 0.879 | 6.424 | 0.00891 |
| LW_2_Old | 12 | 23 | 8 | 0.894 | 9.500 | 0.01318 |
| LW_3_Old | 11 | 16 | 4 | 0.745 | 7.782 | 0.01079 |
| LW_4_Old | 13 | 20 | 7 | 0.897 | 8.615 | 0.01195 |
| LW_5_Old | 16 | 17 | 5 | 0.767 | 3.875 | 0.00537 |
| LW_6_Old | 14 | 16 | 4 | 0.582 | 3.066 | 0.00425 |
| LW_1_Imp | 20 | 18 | 5 | 0.763 | 8.058 | 0.01118 |
| York_1_Imp | 20 | 17 | 7 | 0.800 | 5.179 | 0.00718 |
| LW_2_Imp | 7 | 18 | 4 | 0.810 | 7.333 | 0.01017 |
| York_2_Imp | 13 | 18 | 3 | 0.410 | 4.179 | 0.00580 |
| York_3_Imp | 16 | 15 | 2 | 0.125 | 1.875 | 0.00260 |
| York_4_Imp | 16 | 18 | 8 | 0.875 | 5.483 | 0.00761 |
| York_5_Imp | 15 | 19 | 6 | 0.819 | 7.867 | 0.01091 |
| York_6_Imp | 16 | 18 | 4 | 0.742 | 8.183 | 0.01135 |
| LW_1_New | 11 | 18 | 5 | 0.818 | 8.145 | 0.01130 |
| York_1_New | 10 | 19 | 4 | 0.733 | 8.111 | 0.01125 |
| LW_2_New | 10 | 4 | 2 | 0.467 | 1.867 | 0.00259 |
| LW_3_New | 16 | 5 | 3 | 0.342 | 1.058 | 0.00147 |
| York_3_New | 15 | 6 | 3 | 0.362 | 1.257 | 0.00174 |
| LW_4_New | 16 | 17 | 5 | 0.792 | 5.542 | 0.00769 |
| York_4_New | 20 | 6 | 5 | 0.663 | 1.026 | 0.00142 |
| LW_5_New | 20 | 0 | 1 | 0.000 | 0.000 | 0.00000 |
| LW_6_New | 16 | 21 | 5 | 0.775 | 9.617 | 0.01334 |
| LW_7_New | 20 | 16 | 5 | 0.558 | 2.368 | 0.00328 |
| LW_8_New | 20 | 8 | 5 | 0.626 | 1.921 | 0.00266 |
| LW_9_New | 8 | 25 | 6 | 0.893 | 10.143 | 0.01407 |
| LW_10_New | 12 | 4 | 2 | 0.485 | 1.939 | 0.00269 |
| LW_11_New | 7 | 13 | 2 | 0.476 | 6.190 | 0.00859 |
| Old | 78 | 26 | 15 | 0.890 | 7.452 | 0.01034 |
| Imp | 123 | 29 | 25 | 0.915 | 8.442 | 0.01171 |
| New | 201 | 33 | 23 | 0.810 | 5.971 | 0.00828 |
| ALL | 402 | 46 | 42 | 0.881 | 7.431 | 0.01031 |
| Item | Hd | Pi | K |
|---|---|---|---|
| Hd | 1.0 | 0.771 | 0.771 |
| Pi | 0.771 | 1.000 | 1.000 |
| K | 0.771 | 1.000 | 1.000 |
| Breed | F1 | F2 | Fz |
|---|---|---|---|
| LW_1_Old | −0.469869 | 0.397603 | −0.323178 |
| LW_2_Old | −0.216537 | −0.792395 | −0.393317 |
| LW_3_Old | −0.453041 | 0.412831 | −0.304223 |
| LW_4_Old | −0,410942 | −0.075118 | −0.388568 |
| LW_5_Old | 0.009080 | 0.883065 | 0.229365 |
| LW_6_Old | 0.593214 | −0.192177 | 0.485617 |
| LW_1_Imp | −0.687686 | 0.641848 | −0.457983 |
| York_1_Imp | 0.458763 | −0.774221 | 0.218848 |
| LW_2_Imp | −0.852489 | 0.334876 | −0.683160 |
| York_2_Imp | 1.216740 | 1.077880 | 1.364776 |
| York_3_Imp | 0.085042 | −0.801193 | −0.124170 |
| York_4_Imp | 0.866407 | 1.090529 | 1.052726 |
| York_5_Imp | 0.330318 | 0.614465 | 0.451124 |
| York_6_Imp | −0.525864 | 0.103496 | −0.447231 |
| LW_1_New | −0.115446 | −0.590256 | −0.251726 |
| York_1_New | −0.157773 | −0.466608 | −0.258837 |
| LW_2_New | −0.651816 | −0.072000 | −0.604517 |
| LW_3_New | −0.418594 | 0.283185 | −0.305703 |
| York_3_New | −0.345033 | −0.566545 | −0.452361 |
| LW_4_New | 0.548433 | −1.142167 | 0.207365 |
| York_4_New | 0.453650 | −0.404940 | 0.306747 |
| LW_5_New | 0.341198 | −0.315651 | 0.227932 |
| LW_6_New | −0.470192 | 0.287218 | −0.351119 |
| LW_7_New | 0.801264 | 0.068671 | 0.738151 |
| LW_8_New | 0.569170 | −0.241109 | 0.451726 |
| LW_9_New | −0.489366 | 0.028054 | −0.433288 |
| LW_10_New | −0.767598 | 0.166413 | −0.648975 |
| LW_11_New | 0.758969 | 0.044248 | 0.693978 |
| Clusters | Two-Tailed Fisher’s Exact Test | ||
|---|---|---|---|
| Haplotype | Haplogroup | Asia/Europe | |
| Old-Imp | 0.0005 *** | 0.0225 * | 0.3843 *** |
| Old-New | 0.0005 *** | 0.0005 *** | 9.521 × 10−5 *** |
| Imp-New | 0.0005 *** | 0.0005 *** | 9.446 ×10−9 *** |
| Clusters | Two-Tailed Fisher’s Exact Test | ||
|---|---|---|---|
| Haplotype | Haplogroup | Asia/Europe | |
| Old_Center—Old_Siberia | 0.0010 *** | 0.0270 * | 0.0247 * |
| Old_Center—Old_South | 0.0005 *** | 0.3348 | 0.5837 |
| Old_Siberia—Old_South | 0.0001 *** | 0.1439 | 0.1615 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Getmantseva, L.; Bakoev, S.; Bakoev, N.; Karpushkina, T.; Kostyunina, O. Mitochondrial DNA Diversity in Large White Pigs in Russia. Animals 2020, 10, 1365. https://doi.org/10.3390/ani10081365
Getmantseva L, Bakoev S, Bakoev N, Karpushkina T, Kostyunina O. Mitochondrial DNA Diversity in Large White Pigs in Russia. Animals. 2020; 10(8):1365. https://doi.org/10.3390/ani10081365
Chicago/Turabian StyleGetmantseva, Lyubov, Siroj Bakoev, Nekruz Bakoev, Tatyana Karpushkina, and Olga Kostyunina. 2020. "Mitochondrial DNA Diversity in Large White Pigs in Russia" Animals 10, no. 8: 1365. https://doi.org/10.3390/ani10081365
APA StyleGetmantseva, L., Bakoev, S., Bakoev, N., Karpushkina, T., & Kostyunina, O. (2020). Mitochondrial DNA Diversity in Large White Pigs in Russia. Animals, 10(8), 1365. https://doi.org/10.3390/ani10081365





