Epidermal Growth Factor Stimulates Fatty Acid Synthesis Mainly via PLC-γ1/Akt Signaling Pathway in Dairy Goat Mammary Epithelial Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Collection of Goat Mammary Gland Tissue
2.3. Phosphorylation Site Analysis of Goat EGFR
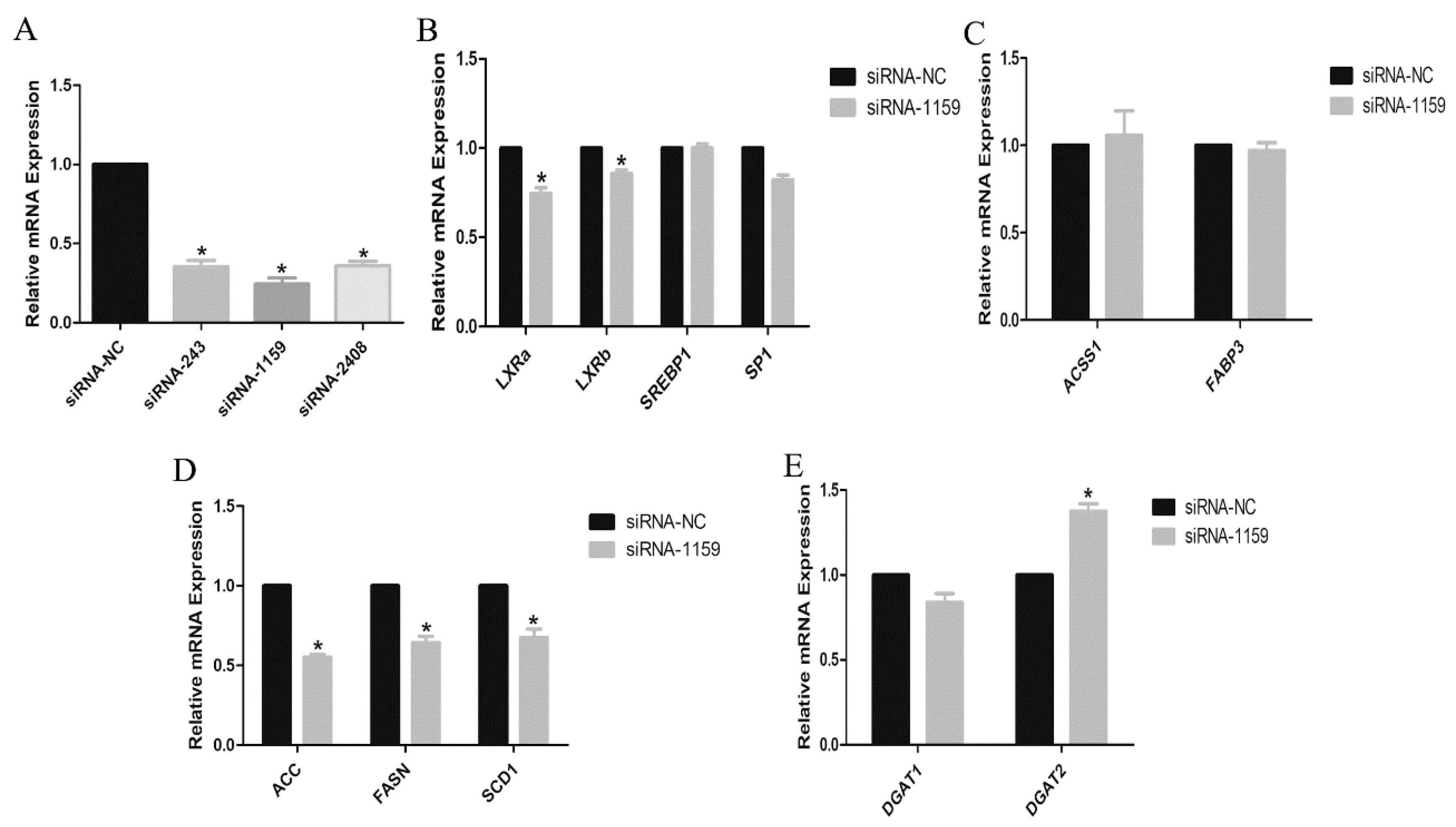
2.4. Small Interference RNA Design
2.5. Cell Culture and Transfection
2.6. Cell Treatment
2.7. Total RNA Extraction and Real-Time Quantitative PCR
2.8. Western Blot Analysis
2.9. Cellular TG Analysis
2.10. Statistical Analysis
3. Results
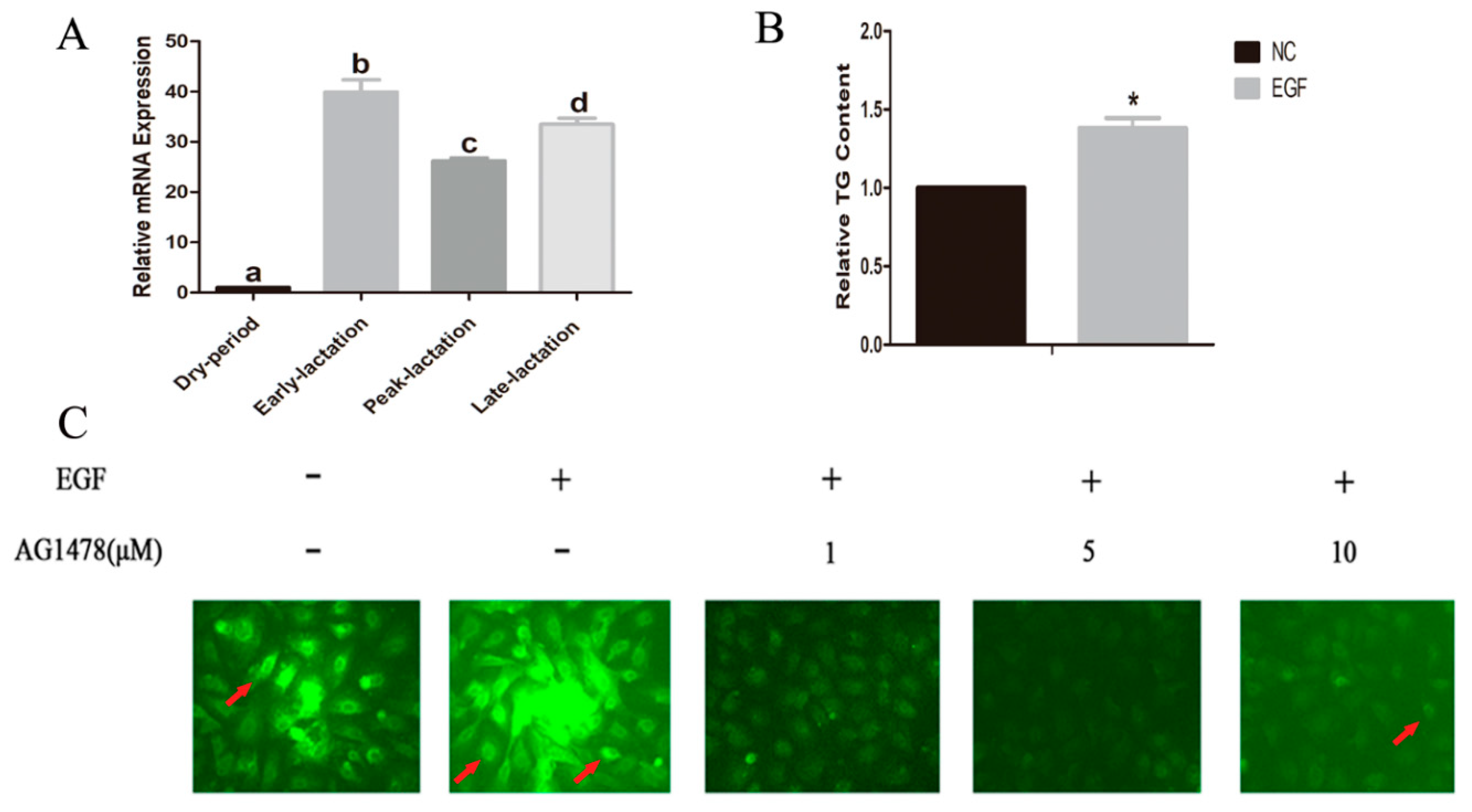
3.1. mRNA Abundance of EGFR in Goat Mammary Gland Tissues and Lipid Content in GMECs Treated with EGF
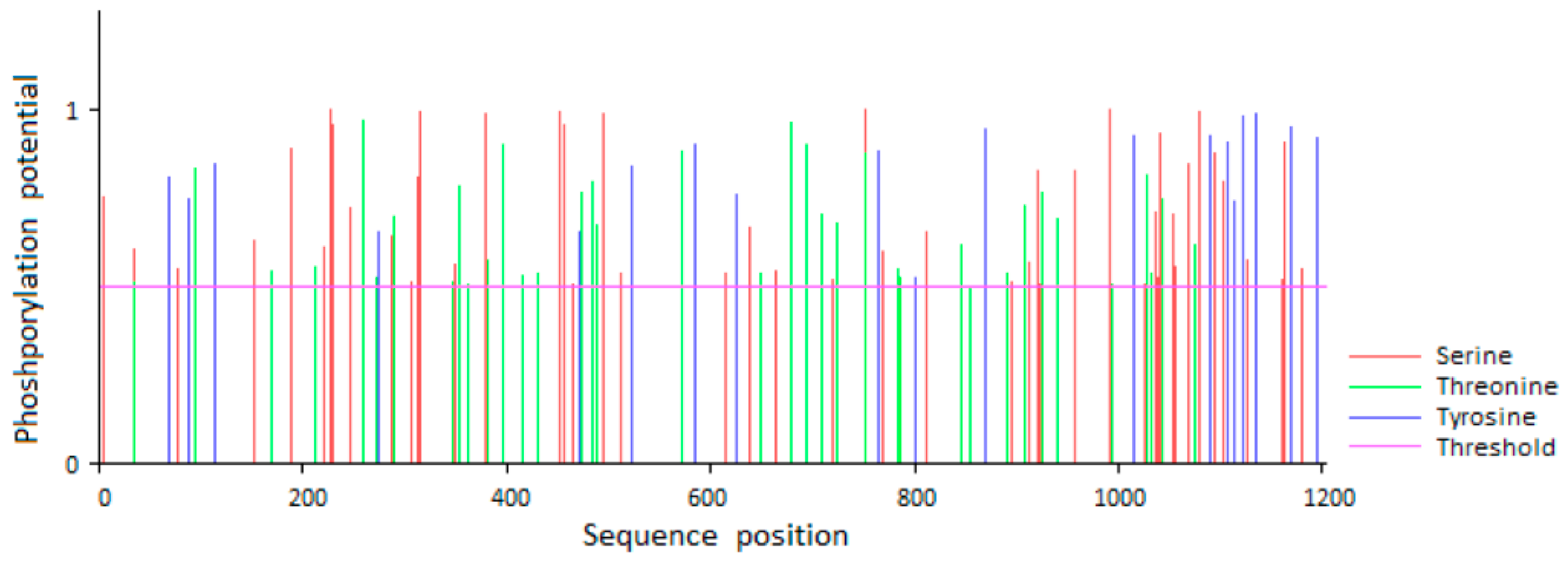
3.2. Prediction of Phosphorylation Sites of Goat EGFR Protein
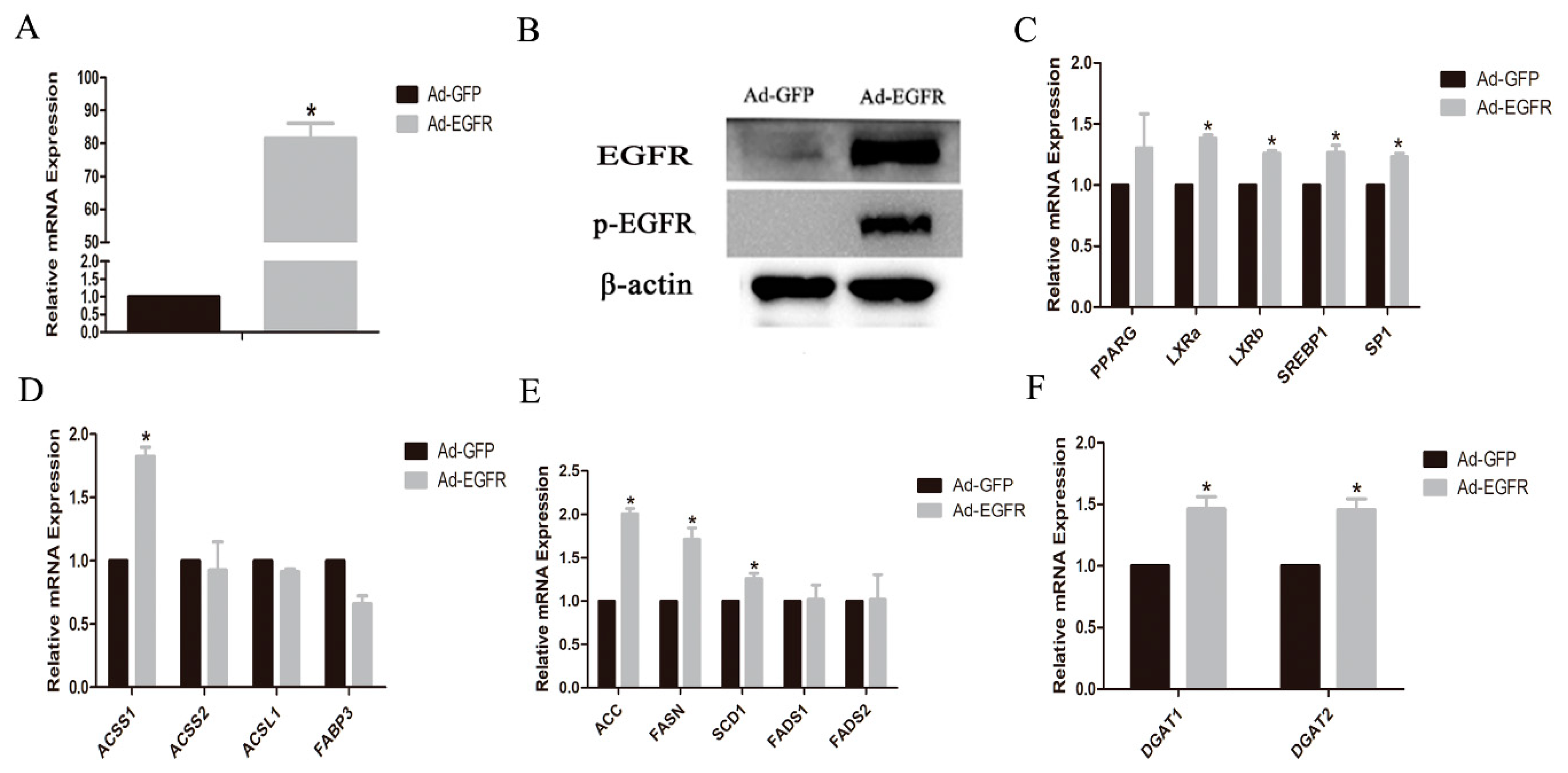
3.3. Effects of EGFR Gene on the Lipogenic Genes in GMECs
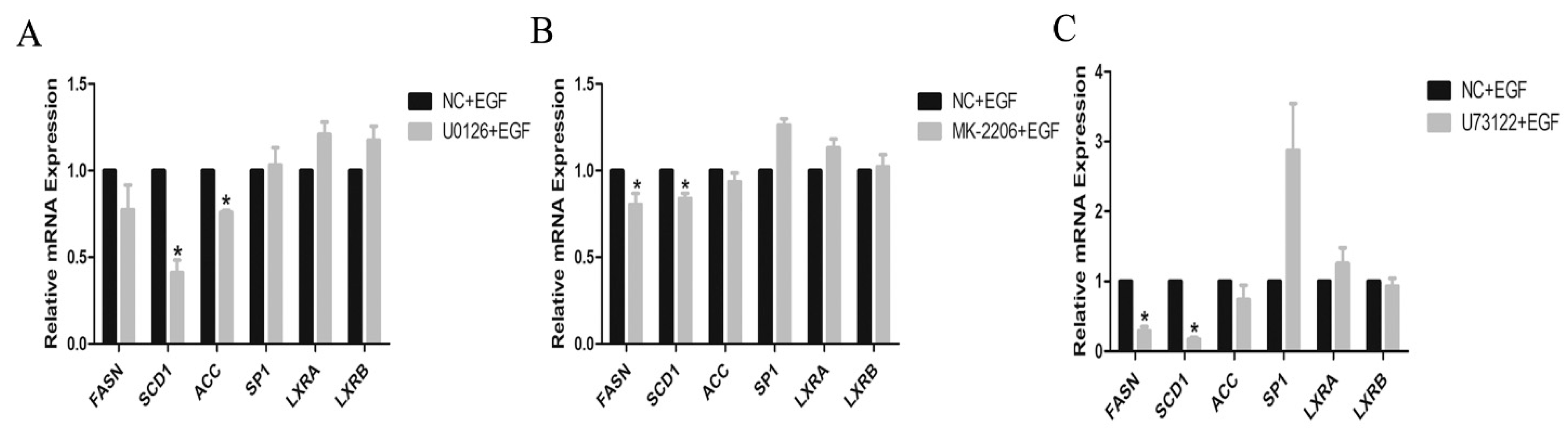
3.4. Effects of EGF or Inhibitor on the Signal Protein Activity in GMECs
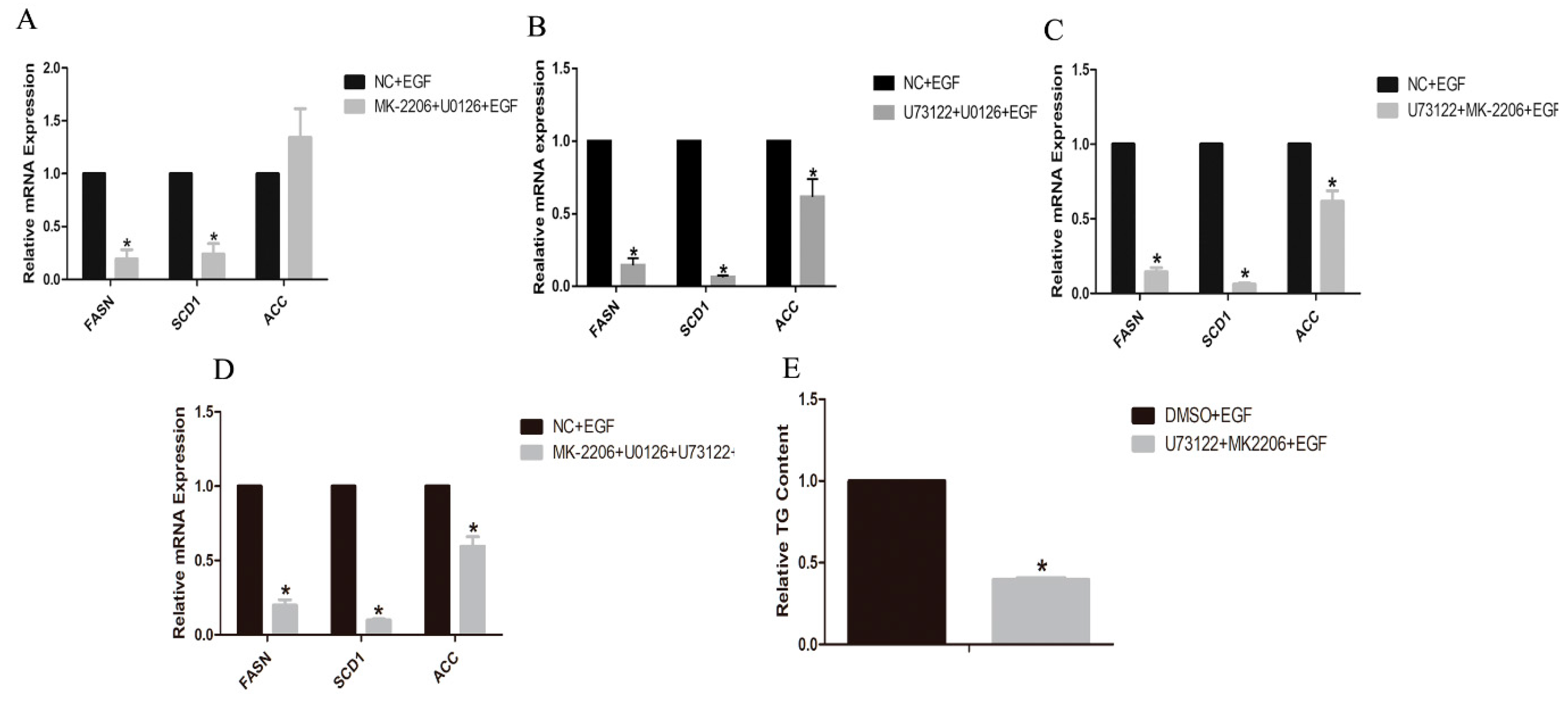
3.5. Expressions of Lipogenic Genes and TG Content in Inhibitor-Treated GMECs
4. Discussion
4.1. EGF and Lipid Metabolism
4.2. EGFR Activation in Regulating Lipid Metabolism in GMECs
4.3. PLC-γ1 and PI3K/Akt Signaling Is Involved in the Lipid Metabolism in GMECs
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MEK | MAPK/ERK kinase |
| LXRa | Liver X receptor a |
| LXRb | Liver X receptor b |
| Sp1 | Specificity protein 1 |
| RTK | Receptor tyrosine kinase |
| PPARG | Peroxisome proliferator-activated receptor gamma |
| ACSS1 | Acetyl coenzyme synthetase 1 |
| DGAT1 | Diacylglycerol transferase 1 |
| DGAT2 | Diacylglycerol transferase 2 |
| FABP3 | Fatty acid binding protein 3 |
| FADS1 | Fatty acid desaturase 1 |
| FADS2 | Fatty acid desaturase 2 |
| Ad-GFP | adenovirus-green fluorescent protein |
Appendix A
| Site | Sense Strand | Antisense Strand |
|---|---|---|
| SiRNA-EGFR-342 | CCUUGGGAAUUUGGAAAUUTT | AAUUUCCAAAUUCCCAAGGTT |
| SiRNA-EGFR-1159 | GCCAAAGUUUGUAACGGAATT | UUCCGUUACAAACUUUGGCTT |
| SiRNA-EGFR-2409 | CCACAUCUCCCAAAGCCAATT | UUGGUUUGGGAGAUGUGGTT |
| SiRNA-NC | UUCUCCGAACGUGUCAGUTT | ACGUGACACGUUCGGAGAATT |
| NCBI Accession ID | Gene | Primer Sequence (5′ to 3′) | Size (bp) | Reference | |
|---|---|---|---|---|---|
| JN236219.1 | ACC | F | CTCCAACCTCAACCACTACGG | 171 | [47] |
| R | GGGGAATCACAGAAGCAGCC | ||||
| NM_00107608.1 | ACSL1 | F | GTGGGCTCCTTTGAAGAACTGT | 120 | [48] |
| R | ATAGATGCCTTTGACCTGTTCAAAT | ||||
| XM_018057282.1 | ACSS1 | F | TCCTTGGCTGGGAGGATCAA | 163 | This manuscript |
| R | TGTTGTCTGTCCTGTGAGCCA | ||||
| BC134532 | ACSS2 | F | GGCGAATGCCTCTACTGCTT | 100 | [49] |
| R | GGCCAATCTTTTCTCTAATCTGCTT | ||||
| BC134532 | ACSS2 | F | GGCGAATGCCTCTACTGCTT | 100 | [49] |
| R | GGCCAATCTTTTCTCTAATCTGCTT | ||||
| XM_005688895.1 | DGAT1 | F | CCACTGGGACCTGAGGTGTC | 101 | [49] |
| R | GCATCACCACACACCAATTCA | ||||
| HM566448.1 | DGAT2 | F | CATGTACACATTCTGCACCGATT | 100 | [48] |
| R | TGACCTCCTGCCACCTTTCT | ||||
| NM_001285701.1 | FABP3 | F | GATGAGACCACGGCAGATG | 120 | [47] |
| R | GTCAACTATTTCCCGCACAAG | ||||
| XM_018043053.1 | FADS1 | F | GGTGGACTTGGCCTGGATG | 101 | [50] |
| R | TGACCATGAAGACAAGCCCC | ||||
| HM443643.1 | FADS2 | F | GCCTGTAGGCTCAGATGTTTGTTC | 101 | [50] |
| R | TGCCTGGCAGTAACAGAGCAC | ||||
| DQ915966.3 | FASN | F | GGGCTCCACCACCGTGTTCCA | 226 | [47] |
| R | GCTCTGCTGGGCCTGCAGCTG | ||||
| XM_01290735.2 | LXRb | F | CAGCCTCCGTTGTGGTCAT | 150 | This manuscript |
| R | CTTCACAGCTGAGCACGTTG | ||||
| HQ589347.1 | PPARG | F | CCTTCACCACCGTTGACTTCT | 145 | [47] |
| R | GATACAGGCTCCACTTTGATTGC | ||||
| GU947654 | SCD1 | F | CCATCGCCTGTGGAGTCAC | 256 | [47] |
| R | GTCGGATAAATCTAGCGTAGCA | ||||
| HM_236311 | SP1 | F | TAGTGAAGGAAGGGGCTCTGG | 108 | [50] |
| R | AAGTGTGCCCGTAGGTGTGAG | ||||
| HM443643.1 | SREBP1 | F | ACGCCATCGAGAAACGCTAC | 181 | [51] |
| R | GTGCGCAGACTCAGGTTCTC | ||||
| XM_005700842.1 | UXT | F | TGTGGCCCTTGGATATGGTT | 101 | [52] |
| R | GGTTGTCGCTGAGCTCTGTG | ||||
References
- Haenlein, G.F.W. Goat milk in human nutrition. Small Rumin. Res. 2004, 51, 155–163. [Google Scholar] [CrossRef]
- Xu, H.F.; Luo, J.; Ma, G.Z.; Zhang, X.Y.; Yao, D.W.; Li, M.; Loor, J.J. Acyl-CoA synthetase short-chain family member 2 (ACSS2) is regulated by SREBP-1 and plays a role in fatty acid synthesis in caprine mammary epithelial cells. J. Cell. Physiol. 2018, 233, 1005–1016. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Huang, J.T.; Tian, H.B.; Ma, Y.; Chen, Z.; Wang, J.J.; Shi, H.P.; Luo, J.J. Trans-10, cis-12 conjugated linoleic acid alters lipid metabolism of goat mammary epithelial cells by regulation of de novo synthesis and the AMPK signaling pathway. J. Dairy Sci. 2018, 101, 5571–5581. [Google Scholar] [CrossRef]
- Yao, D.; Luo, J.; He, Q.; Shi, H.; Li, J.; Wang, H.; Xu, H.; Chen, Z.; Yi, Y.; Loor, J.J. SCD1 Alters Long-Chain Fatty Acid (LCFA) Composition and its Expression is Directly Regulated by SREBP-1 and PPARγ 1 in Dairy Goat Mammary Cells. J. Cell. Physiol. 2017, 232, 635–649. [Google Scholar] [CrossRef]
- Ichinose, J.; Murata, M.; Yanagida, T.; Sako, Y. EGF signalling amplification induced by dynamic clustering of EGFR. Biochem. Biophys. Res. Commun. 2004, 324, 1143–1149. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, J.; Chen, Z.; Wang, L.; Wu, X.; Ou, M. Epidermal growth factor (EGF) triggers the malignancy of hemangioma cells via activation of NF-κB signals. Biomed. Pharmacother. 2016, 82, 133–140. [Google Scholar] [CrossRef]
- Honegger, A.M.; Schmidt, A.; Ullrich, A.; Schlessinger, J. Evidence for epidermal growth factor (EGF)-induced intermolecular autophosphorylation of the EGF receptors in living cells. Mol. Cell. Biol. 1990, 10, 4035–4044. [Google Scholar] [CrossRef]
- Wu, D.; Peng, F.; Zhang, B.; Ingram, A.J.; Krepinsky, J.C. EGFR-PLC 1 signaling mediates high glucose-induced PKC 1-Akt activation and collagen I upregulation in mesangial cells. Am. J. Physiol. Ren. Physiol. 2009, 297, F822–F834. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.Y.; Zhang, M.M.; Jin, M.L.; Bai, C.X.; Wang, X.D. Potential Mechanism of Interleukin-8 Production From Lung Cancer Cells: An Involvement of EGF-EGFR-PI3K-Akt-Erk Pathway. J. Cell. Physiol. 2012, 227, 35–43. [Google Scholar] [CrossRef]
- Papaiahgari, S.; Zhang, Q.; Kleeberger, S.R.; Cho, H.Y.; Reddy, S.P. Hyperoxia Stimulates an Nrf2-ARE Transcriptional Response via ROS-EGFR-PI3K-Akt/ERK MAP Kinase Signaling in Pulmonary Epithelial Cells. Antioxid. Redox Signal. 2006, 8, 43–52. [Google Scholar] [CrossRef]
- Berrahmoune, H.; Lamont, J.V.; Herbeth, B.; Lambert, D.; Masson, C.; Mcphillips, M.; Fitzgerald, P.S.; Visvikis-Siest, S. Association between EGF and lipid concentrations: A benefit role in the atherosclerotic process? Clin. Chim. Acta 2009, 402, 196–198. [Google Scholar] [CrossRef]
- Gibson, N.J.; Tolbert, L.P.; Oland, L.A. Roles of Specific Membrane Lipid Domains in EGF Receptor Activation and Cell Adhesion Molecule Stabilization in a Developing Olfactory System. PLoS ONE 2009, 4, e7222. [Google Scholar] [CrossRef]
- Thomas, P.; Beatrice, G.; Yuen-Li, C.; Oona, D.; Griffiths, J.R.; Julian, D.; Almut, S. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene 2005, 24, 6465–6481. [Google Scholar]
- Bian, Y.; Yu, Y.; Wang, S.; Li, L. Up-regulation of fatty acid synthase induced by EGFR/ERK activation promotes tumor growth in pancreatic cancer. Biochem. Biophys. Res. Commun. 2015, 463, 612–617. [Google Scholar] [CrossRef]
- Wang, H.; Yang, C.; Leskow, F.C.; Sun, J.; Canagarajah, B.; Hurley, J.H.; Kazanietz, M.G. Phospholipase Cgamma/diacylglycerol-dependent activation of beta2-chimaerin restricts EGF-induced Rac signaling. Embo J. 2006, 25, 2062–2074. [Google Scholar] [CrossRef]
- Jeanne-Marie, K.; Yenya, H.; Guillermo, G.C.A.; Gimbrone, M.A., Jr. Lipid raft localization of cell surface E-selectin is required for ligation-induced activation of phospholipase C gamma. J. Immunol. 2003, 171, 3216–3224. [Google Scholar]
- Tian, H.; Luo, J.; Zhang, Z.; Wu, J.; Zhang, T.; Busato, S.; Huang, L.; Song, N.; Bionaz, M. CRISPR/Cas9-mediated Stearoyl-CoA Desaturase 1 (SCD1) Deficiency Affects Fatty Acid Metabolism in Goat Mammary Epithelial Cells. J. Agric. Food Chem. 2018, 66, 10041–10052. [Google Scholar] [CrossRef]
- Farr, V.C.; Stelwagen, K.; Cate, L.R.; Molenaar, A.J.; Mcfadden, T.B.; Davis, S.R.J. An Improved Method for the Routine Biopsy of Bovine Mammary Tissue. J. Dairy Sci. 1996, 79, 543–549. [Google Scholar] [CrossRef]
- Zhang, T.; Huang, J.; Yi, Y.; Zhang, X.; Loor, J.J.; Cao, Y.; Shi, H.; Luo, J. Akt Serine/Threonine Kinase 1 Regulates de Novo Fatty Acid Synthesis through the Mammalian Target of Rapamycin/Sterol Regulatory Element Binding Protein 1 Axis in Dairy Goat Mammary Epithelial Cells. J. Agric. Food Chem. 2018, 66, 1197–1205. [Google Scholar] [CrossRef]
- Yang, X.; Dunning, K.R.; Wu, L.L.-Y.; Hickey, T.E.; Norman, R.J.; Russell, D.L.; Liang, X.; Robker, R.L.J. Development. Identification of Perilipin-2 as a lipid droplet protein regulated in oocytes during maturation. Reprod. Fertil. Dev. 2010, 22, 1262. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef]
- Gou, D.; Mishra, A.; Weng, T.; Su, L.; Chintagari, N.R.; Wang, Z.; Zhang, H.; Gao, L.; Wang, P.; Stricker, H.M. Annexin A2 Interactions with Rab14 in Alveolar Type II Cells. J. Biol. Chem. 2008, 283, 13156–13164. [Google Scholar] [CrossRef]
- Xu, H.F.; Luo, J.; Zhao, W.S.; Yang, Y.C.; Bionaz, M.J. Overexpression of SREBP1 (sterol regulatory element binding protein 1) promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells. J. Dairy Sci. 2016, 99, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Tyner, J.W.; Kim, E.Y.; Kyotaro, I.; Pelletier, M.R.; Roswit, W.T.; Morton, J.D.; Battaile, J.T.; Patel, A.C.; Patterson, G.A.; Alexander, P.; et al. Blocking airway mucous cell metaplasia by inhibiting EGFR antiapoptosis and IL-13 transdifferentiation signals. J. Clin. Investig. 2006, 116, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Edgar, B. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development 2009, 136, 483–493. [Google Scholar] [CrossRef]
- Hynes, N.E.; Horsch, K.; Olayioye, M.A.; Badache, A. The ErbB receptor tyrosine family as signal integrators. Endocr. Relat. Cancer 2001, 8, 151. [Google Scholar] [CrossRef]
- Wong, R.W.C. Transgenic and knock-out mice for deciphering the roles of EGFR ligands. Cell. Mol. Life Sci. CMLS 2003, 60, 113–118. [Google Scholar] [CrossRef]
- Luetteke, N.C.; Qiu, T.H.; Fenton, S.E.; Troyer, K.L.; Riedel, R.F.; Chang, A.; Lee, D.C. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development 1999, 126, 2739–2750. [Google Scholar]
- Sobolewska, A.; Gajewska, M.J.; Gajkowska, B.; Motyl, T.J. IGF-I, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. Eur. J. Cell Biol. 2009, 88, 117–130. [Google Scholar] [CrossRef]
- Chen, Y.; Henson, E.S.; Xiao, W.; Huang, D.; Mcmillan-Ward, E.M.; Israels, S.J.; Gibson, S.B. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy 2016, 12, 1029–1046. [Google Scholar] [CrossRef]
- Shuping, X.; Zhang, W. Loss of EGFR induced autophagy sensitizes hormone refractory prostate cancer cells to adriamycin. Prostate 2011, 71, 1216–1224. [Google Scholar]
- Suburu, J.; Shi, L.; Wu, J.; Wang, S.; Samuel, M.; Thomas, M.J.; Kock, N.D.; Yang, G.; Kridel, S.; Chen, Y.Q. Fatty acid synthase is required for mammary gland development and milk production during lactation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1132. [Google Scholar] [CrossRef]
- Yarden, Y. The EGFR family and its ligands in human cancer. signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 2001, 37, 3–8. [Google Scholar] [CrossRef]
- Jena, M.K.; Jaswal, S.; Kumar, S.; Mohanty, A.K. Molecular Mechanism of Mammary Gland Involution: An Update. Dev. Biol. 2018, 445, 145–155. [Google Scholar] [CrossRef]
- Penrose, H.; Heller, S.; Cable, C.; Makboul, R.; Chadalawada, G.; Chen, Y.; Crawford, S.E.; Savkovic, S.D. Epidermal growth factor receptor mediated proliferation depends on increased lipid droplet density regulated via a negative regulatory loop with FOXO3/Sirtuin6. Biochem. Biophys. Res. Commun. 2016, 469, 370–376. [Google Scholar] [CrossRef]
- Guo, D.; Prins, R.; Hildebrandt, I.; Dang, J.; Kuga, D.; Lin, K.; Huang, T.; Kofman, A.; Soto, H.; Mazzotta, M.J.C.R. Abstract #1985: EGFR Signaling through the PI3K Pathway Promotes Tumor Survival via SREBP1-Mediated Lipogenesis in Glioblastoma Patients; Amer Assoc Cancer Research: Philadelphia, PA, USA, 2009; Volume 69, p. 615. [Google Scholar]
- Iwasa, H.; Yu, S.; Xue, J.; Driscoll, M. Novel EGF pathway regulators modulate C. Elegans healthspan and lifespan via EGF receptor, PLC-γ, and IP3R activation. Aging Cell 2010, 9, 490–505. [Google Scholar] [CrossRef]
- Worster, D.T.; Schmelzle, T.; Solimini, N.L.; Lightcap, E.S.; Millard, B.; Mills, G.B.; Brugge, J.S.; Albeck, J.G. Akt and ERK Control the Proliferative Response of Mammary Epithelial Cells to the Growth Factors IGF-1 and EGF Through the Cell Cycle Inhibitor p57Kip2. Sci. Signal. 2012, 5, ra19. [Google Scholar] [CrossRef]
- Díaz, M.E.; González, L.; Miquet, J.G.; Martínez, C.S.; Sotelo, A.I.; Bartke, A.; Turyn, D. Growth hormone modulation of EGF-induced PI3K-Akt pathway in mice liver. Cell. Signal. 2012, 24, 514–523. [Google Scholar] [CrossRef]
- Zhang, C.; Hu, J.; Sheng, L.; Yuan, M.; Qiu, Z. Ellagic acid ameliorates AKT-driven hepatic steatosis in mice by suppressing de novo lipogenesis via the AKT/SREBP-1/FASN pathway. Food Funct. 2019, 10, 3410–3420. [Google Scholar] [CrossRef]
- Hao, J.; Zhu, L.; Zhao, S.; Liu, S.; Liu, Q.; Duan, H. PTEN ameliorates high glucose-induced lipid deposits through regulating SREBP-1/FASN/ACC pathway in renal proximal tubular cells. Exp. Cell Res. 2011, 317, 1629–1639. [Google Scholar] [CrossRef]
- Vasco, V.R.L.; Leopizzi, M.; Maio, V.D.; Rocca, C.D. U-73122 reduces the cell growth in cultured MG-63 ostesarcoma cell line involving Phosphoinositide-specific Phospholipases C. Springerplus 2016, 5, 156. [Google Scholar] [CrossRef]
- Jiang, Q.; Zhou, C.; Bi, Z.; Wan, Y. EGF-induced cell migration is mediated by ERK and PI3K/AKT pathways in cultured human lens epithelial cells. J. Ocul. Pharmacol. Ther. 2006, 22, 93. [Google Scholar] [CrossRef]
- Yun, S.; Byun, H.Y.; Oh, Y.S.; Yong, R.Y.; Ryu, S.H.; Suh, P.G. Protein kinase C-α negatively regulates EGF-induced PLC-? Activity through direct phosphorylation. Adv. Enzym. Regul. 2010, 50, 178–189. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Wang, Z. Akt binds to and phosphorylates phospholipase C-gamma 1 in response to epidermal growth factor. Mol. Biol. Cell 2006, 17, 2267–2277. [Google Scholar] [CrossRef]
- Carpenter, G.; Ji, Q.S. Phospholipase C-γ as a Signal-Transducing Element. Exp. Cell Res. 1999, 253, 15–24. [Google Scholar] [CrossRef]
- Shi, H.; Luo, J.; Zhu, J.; Li, J.; Sun, Y.; Lin, X.; Zhang, L.; Yao, D.; Shi, H.J.P.R. PPARγ Regulates Genes Involved in Triacylglycerol Synthesis and Secretion in Mammary Gland Epithelial Cells of Dairy Goats. PPAR Res. 2013, 2013, 1–10. [Google Scholar] [CrossRef]
- Piantoni, P.; Bionaz, M.; Graugnard, D.E.; Daniels, K.M.; Akers, R.M.; Loor, J.J.J.J.o.N. Gene Expression Ratio Stability Evaluation in Prepubertal Bovine Mammary Tissue from Calves Fed Different Milk Replacers Reveals Novel Internal Controls for Quantitative Polymerase Chain Reaction. J. Nuri. 2008, 138, 1158–1164. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.; Bioinformatics, J.J.; Insights, B. Gene Networks Driving Bovine Mammary Protein Synthesis During the Lactation Cycle. BBI 2011, 5, 83–98. [Google Scholar]
- Zhu, J.; Sun, Y.; Luo, J.; Wu, M.; Li, J.; Cao, Y. Specificity Protein 1 Regulates Gene Expression Related to Fatty Acid Metabolism in Goat Mammary Epithelial Cells. Int. J. Mol. Sci. 2015, 16, 1806–1820. [Google Scholar] [CrossRef]
- Zhu, J.J.; Luo, J.; Wang, W.; Yu, K.; Li, J.J.A. Inhibition of FASN reduces the synthesis of medium-chain fatty acids in goat mammary gland. Animal 2014, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks regulating lipid synthesis in bovine mammary gland during the lactation cycle. FASEB J. 2007, 21, A1106. [Google Scholar]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Dai, B.; Qu, H.; Zhong, Y.; Ma, Y.; Luo, J.; Hadsell, D.; Shi, H. Epidermal Growth Factor Stimulates Fatty Acid Synthesis Mainly via PLC-γ1/Akt Signaling Pathway in Dairy Goat Mammary Epithelial Cells. Animals 2020, 10, 930. https://doi.org/10.3390/ani10060930
Huang J, Dai B, Qu H, Zhong Y, Ma Y, Luo J, Hadsell D, Shi H. Epidermal Growth Factor Stimulates Fatty Acid Synthesis Mainly via PLC-γ1/Akt Signaling Pathway in Dairy Goat Mammary Epithelial Cells. Animals. 2020; 10(6):930. https://doi.org/10.3390/ani10060930
Chicago/Turabian StyleHuang, Jiangtao, Bangguo Dai, Hexuan Qu, Yuling Zhong, Yue Ma, Jun Luo, Darryl Hadsell, and Huaiping Shi. 2020. "Epidermal Growth Factor Stimulates Fatty Acid Synthesis Mainly via PLC-γ1/Akt Signaling Pathway in Dairy Goat Mammary Epithelial Cells" Animals 10, no. 6: 930. https://doi.org/10.3390/ani10060930
APA StyleHuang, J., Dai, B., Qu, H., Zhong, Y., Ma, Y., Luo, J., Hadsell, D., & Shi, H. (2020). Epidermal Growth Factor Stimulates Fatty Acid Synthesis Mainly via PLC-γ1/Akt Signaling Pathway in Dairy Goat Mammary Epithelial Cells. Animals, 10(6), 930. https://doi.org/10.3390/ani10060930





