Use of the HRM Method in Quick Identification of FecXO Mutation in Highly Prolific Olkuska Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Methods
2.2.1. DNA Extraction
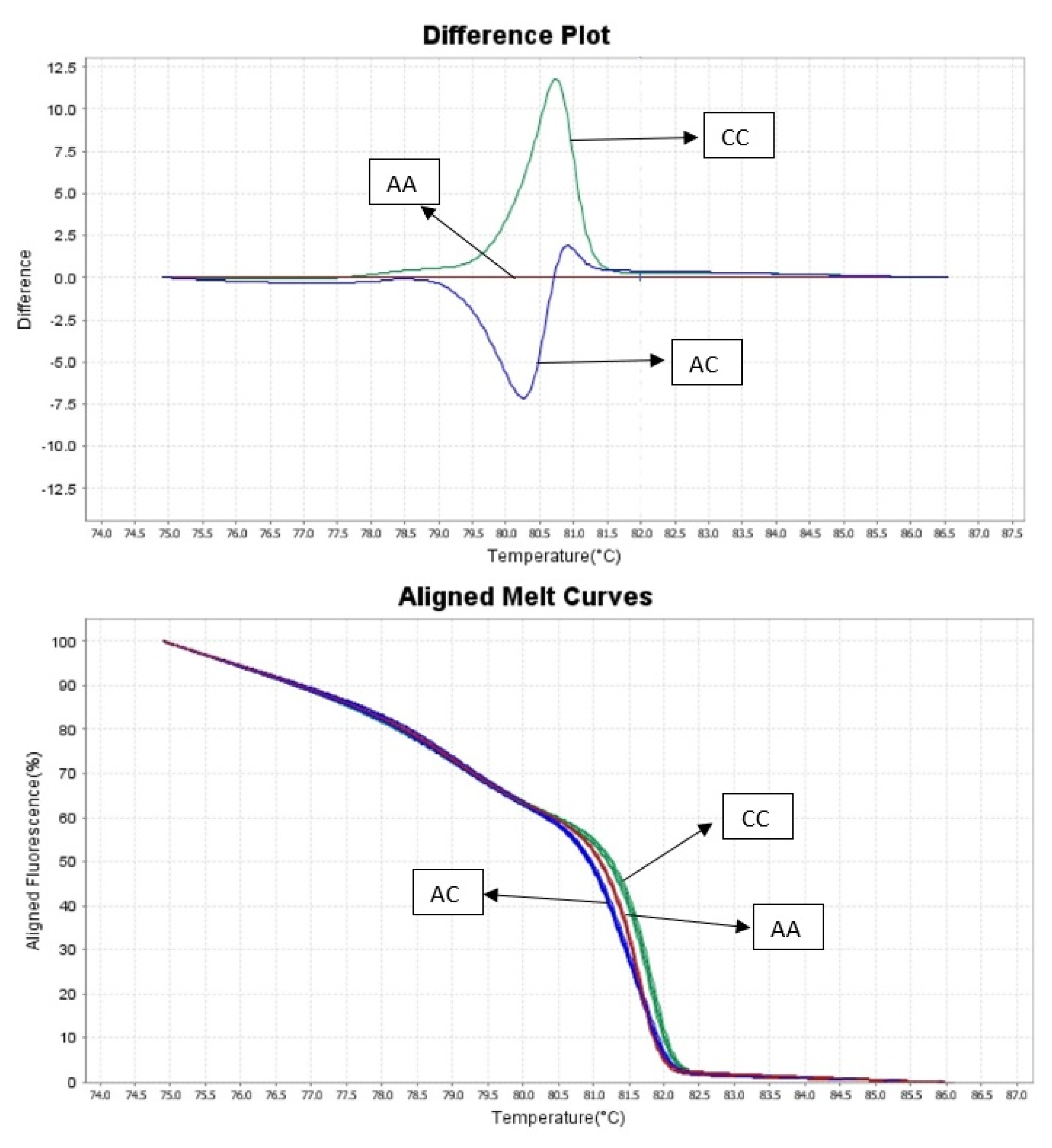
2.2.2. HRM Analysis
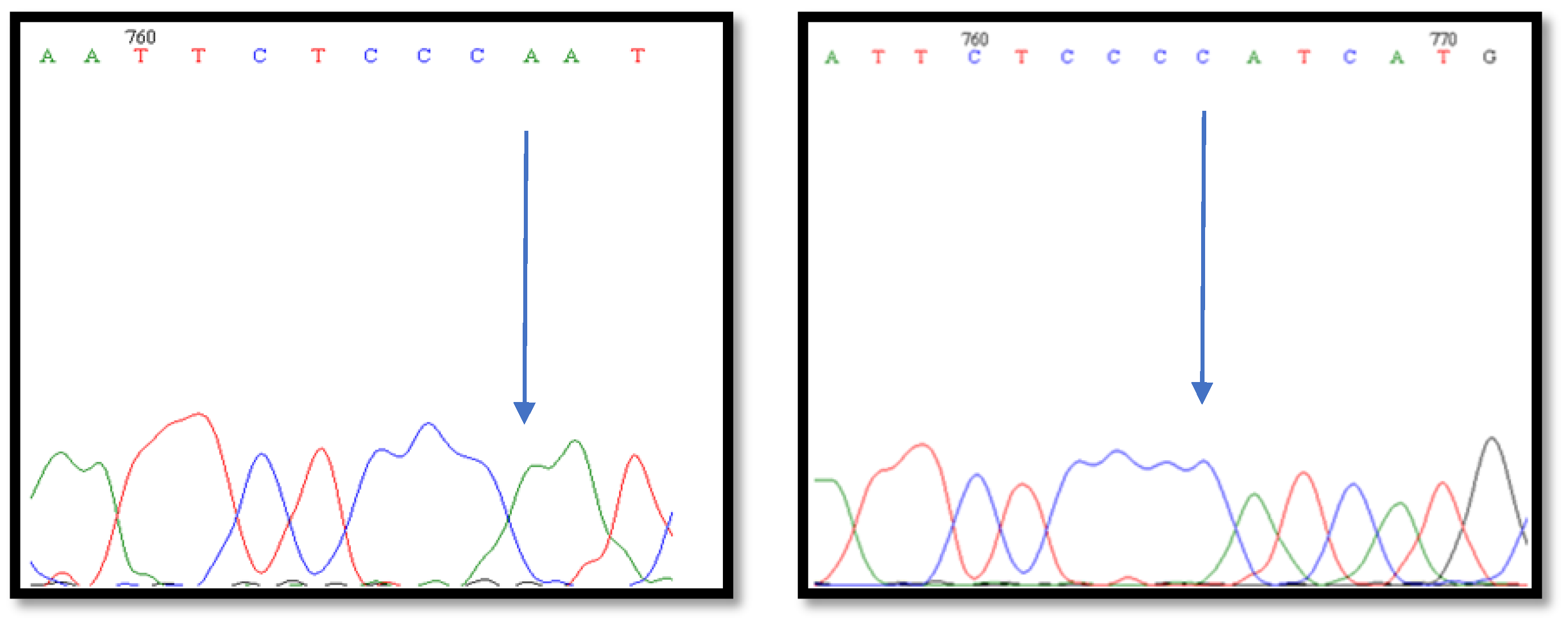
2.2.3. BMP-15 Fragment Gene Sequencing
2.2.4. Statistical Analysis
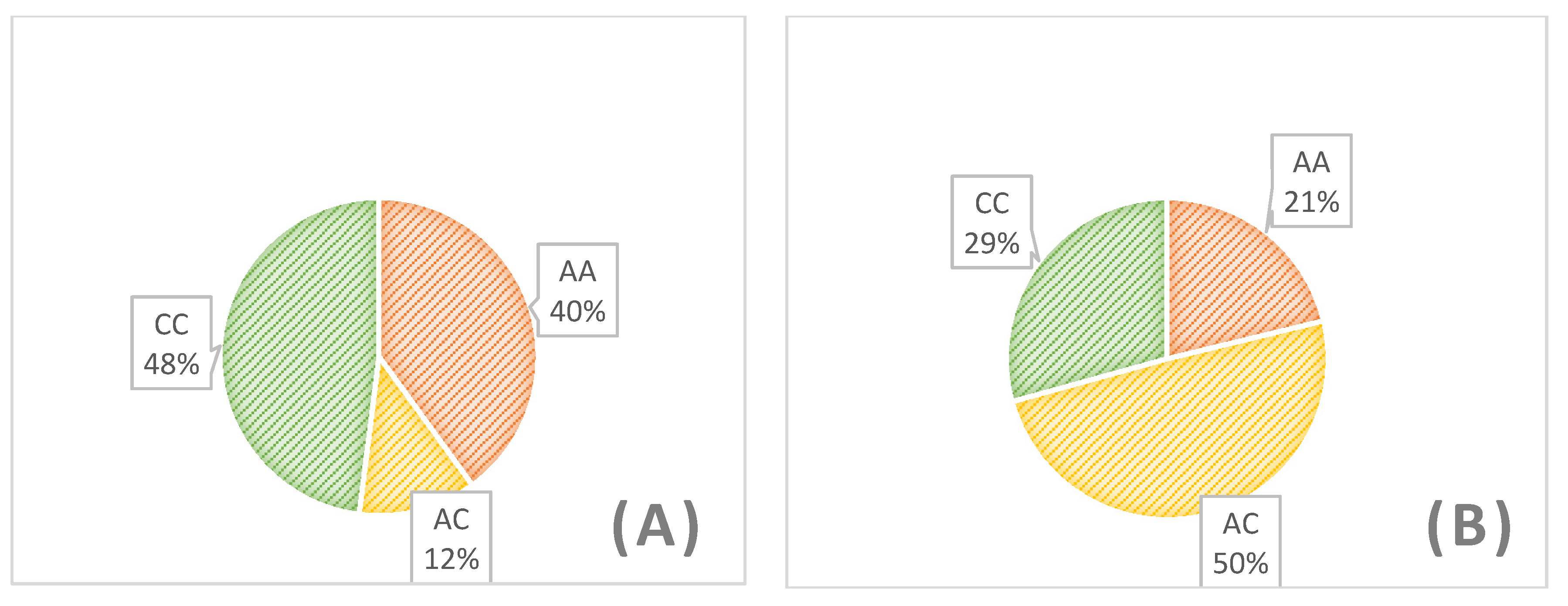
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Davis, G.H. Major genes affecting ovulation rate in sheep. Genet. Sel. Evol. GSE 2005, 37 (Suppl. 1), S11–S23. [Google Scholar] [CrossRef]
- Mandon-Pépin, B.; Oustry-Vaiman, A.; Vigier, B.; Piumi, F.; Cribiu, E.; Cotinot, C. Expression Profiles and Chromosomal Localization of Genes Controlling Meiosis and Follicular Development in the Sheep Ovary. Biol. Reprod. 2003, 68, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Safari, E.; Fogarty, N.M.; Gilmour, A.R. A review of genetic parameter estimates for wool, growth, meat and reproduction traits in sheep. Livest. Prod. Sci. 2005, 92, 271–289. [Google Scholar] [CrossRef]
- Gebreselassie, G.; Berihulay, H.; Jiang, L.; Ma, Y. Review on Genomic Regions and Candidate Genes Associated with Economically Important Production and Reproduction Traits in Sheep (Ovies aries). Animals 2020, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Snowder, G.D. Genetic Improvement of Overall Reproductive Successin Sheep: A Review. 2009. Available online: https://tspace.library.utoronto.ca/handle/1807/53119 (accessed on 26 January 2009).
- Abdoli, R.; Zamani, P.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Nadri, S. A review on prolificacy genes in sheep. Reprod. Domest. Anim. Zuchthyg. 2016, 51, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Demars, J.; Fabre, S.; Sarry, J.; Rossetti, R.; Gilbert, H.; Persani, L.; Tosser-Klopp, G.; Mulsant, P.; Nowak, Z.; Drobik, W.; et al. Genome-Wide Association Studies Identify Two Novel BMP15 Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013, 9, e1003482. [Google Scholar] [CrossRef] [PubMed]
- Chantepie, L.; Bodin, L.; Sarry, J.; Woloszyn, F.; Plisson-Petit, F.; Ruesche, J.; Drouilhet, L.; Fabre, S. Genome-Wide identification of a regulatory mutation in BMP15 controlling prolificacy in sheep. bioRxiv 2019. [Google Scholar] [CrossRef]
- Juengel, J.L.; Davis, G.H.; McNatty, K.P. Using sheep lines with mutations in single genes to better understand ovarian function. Reproduction 2013, 146, R111–R123. [Google Scholar] [CrossRef] [PubMed]
- Knight, P.G.; Glister, C. TGF-β superfamily members and ovarian follicle development. Reproduction 2006, 132, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Davis, G.H. Fecundity genes in sheep. Anim. Reprod. Sci. 2004, 82–83, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Pan, Z.; Wang, X.; Hu, W.; Di, R.; Yao, Y.; Chu, M. Progress on major genes for high fecundity in ewes. Front. Agric. Sci. Eng. 2014, 1, 282. [Google Scholar] [CrossRef]
- Kaczor, U. Genes Involved Litter Size in Olkuska Sheep. Genet. Polymorph. 2017. [Google Scholar] [CrossRef]
- Kucharski, M.; Kaczor, U.; Kaczor, A. Identification of FecXO mutation in the BMP15 gene in prolific Olkuska sheep. Gene 2015, 14, 93–100. [Google Scholar]
- Vossen, R.H.; Aten, E.; Roos, A.; den Dunnen, J.T. High-Resolution Melting Analysis (HRMA)—More than just sequence variant screening. Human Mutation 2009, 30, 860–866. [Google Scholar] [CrossRef] [PubMed]
- Notter, D.R. Genetic improvement of reproductive efficiency of sheep and goats. Anim. Reprod. Sci. 2012, 130, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Słomka, M.; Sobalska-Kwapis, M.; Wachulec, M.; Bartosz, G.; Strapagiel, D. High Resolution Melting (HRM) for High-Throughput Genotyping—Limitations and Caveats in Practical Case Studies. Int. J. Mol. Sci. 2017, 18, 316. [Google Scholar] [CrossRef] [PubMed]
- Tucker, E.J.; Huynh, B.L. Genotyping by High-Resolution Melting Analysis. In Crop Breeding; Fleury, D., Whitford, R., Eds.; Springer: New York, NY, USA, 2014; Volume 1145, pp. 59–66. ISBN 978-1-4939-0445-7. [Google Scholar]
- Montgomery, J.L.; Sanford, L.N.; Wittwer, C.T. High-Resolution DNA melting analysis in clinical research and diagnostics. Expert Rev. Mol. Diagn. 2010, 10, 219–240. [Google Scholar] [CrossRef] [PubMed]
- Galloway, S.M.; Gregan, S.M.; Wilson, T.; McNatty, K.P.; Juengel, J.L.; Ritvos, O.; Davis, G.H. Bmp15 mutations and ovarian function. Mol. Cell. Endocrinol. 2002, 191, 15–18. [Google Scholar] [CrossRef]

| Name | Sequence [5′–3′] | Analysis Type | PCR Product Length (bp) |
|---|---|---|---|
| BMP-15F | CAGAAGACCAAACCTCTCCCTA | Sanger sequencing | 498 |
| BMP-15R | CTGATTACGCCAGTTTGCAC | ||
| FecXO-F | TCCACCCTTTTCAAGTCAGC | HRM-PCR | 248 |
| FecXO-R | ACTCCCATTTGCCTCAATCA |
| Olkuska Breed | Genotype | Allele | HWE p-Value | |||
|---|---|---|---|---|---|---|
| AA | AC | CC | A | C | ||
| Flock 1 (n = 20) | 0.2 | 0.3 | 0.5 | 0.35 | 0.65 | 0.12764 |
| Flock 2 (n = 20) | 0.5 | 0.05 | 0.45 | 0.525 | 0.475 | 0.000057 |
| Flock 3 (n = 20) | 0.3 | 0.1 | 0.6 | 0.35 | 0.65 | 0.000484 |
| Flock 4(n = 20) | 0.3 | 0.05 | 0.65 | 0.325 | 0.675 | 0.000074 |
| Flock 5 (n = 20) | 0.7 | 0.1 | 0.2 | 0.75 | 0.25 | 0.001040 |
| Total (n = 100) | 0.4 | 0.12 | 0.48 | 0.46 | 0.54 | 0.0000001 |
| Flocks | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | * | 0.04 | ns | ns | 0.006 |
| 2 | * | ns | ns | ns | |
| 3 | * | ns | 0.01 | ||
| 4 | * | 0.01 | |||
| 5 | * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smołucha, G.; Piórkowska, K.; Ropka-Molik, K.; Sikora, J. Use of the HRM Method in Quick Identification of FecXO Mutation in Highly Prolific Olkuska Sheep. Animals 2020, 10, 844. https://doi.org/10.3390/ani10050844
Smołucha G, Piórkowska K, Ropka-Molik K, Sikora J. Use of the HRM Method in Quick Identification of FecXO Mutation in Highly Prolific Olkuska Sheep. Animals. 2020; 10(5):844. https://doi.org/10.3390/ani10050844
Chicago/Turabian StyleSmołucha, Grzegorz, Katarzyna Piórkowska, Katarzyna Ropka-Molik, and Jacek Sikora. 2020. "Use of the HRM Method in Quick Identification of FecXO Mutation in Highly Prolific Olkuska Sheep" Animals 10, no. 5: 844. https://doi.org/10.3390/ani10050844
APA StyleSmołucha, G., Piórkowska, K., Ropka-Molik, K., & Sikora, J. (2020). Use of the HRM Method in Quick Identification of FecXO Mutation in Highly Prolific Olkuska Sheep. Animals, 10(5), 844. https://doi.org/10.3390/ani10050844





