Application of Meta-Analysis and Machine Learning Methods to the Prediction of Methane Production from In Vitro Mixed Ruminal Micro-Organism Fermentation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Database
2.2. Model Fitting—Meta-Analysis
2.3. Model Fitting—Machine Learning
2.3.1. Support Vector Regression
2.3.2. Artificial Neural Network—Multilayer Perceptron
2.4. Model Evaluation
3. Results
3.1. Correlation Matrix Analysis
3.2. Univariate Meta-Analysis Models
3.3. Multivariate Meta-Analysis Models
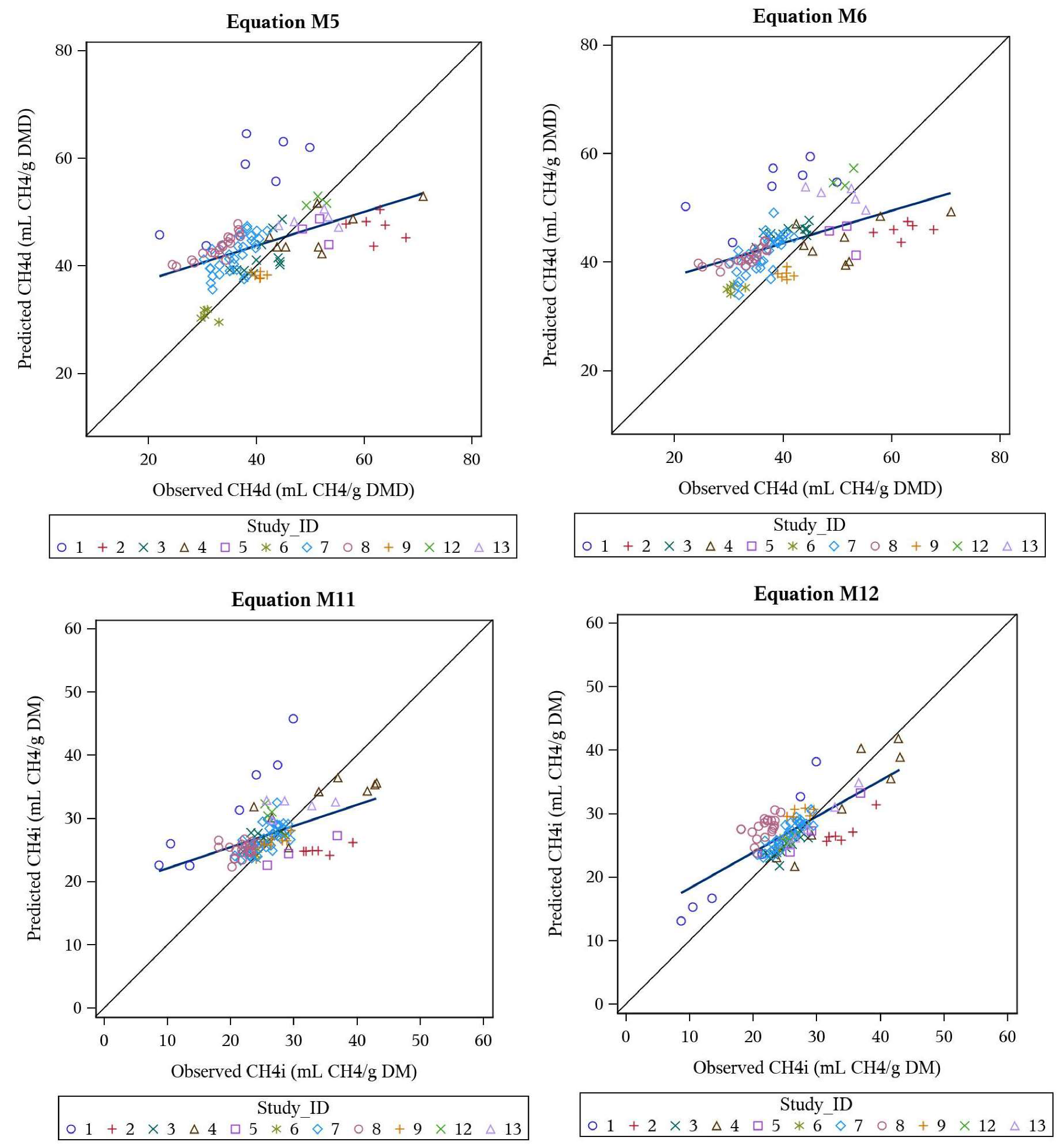
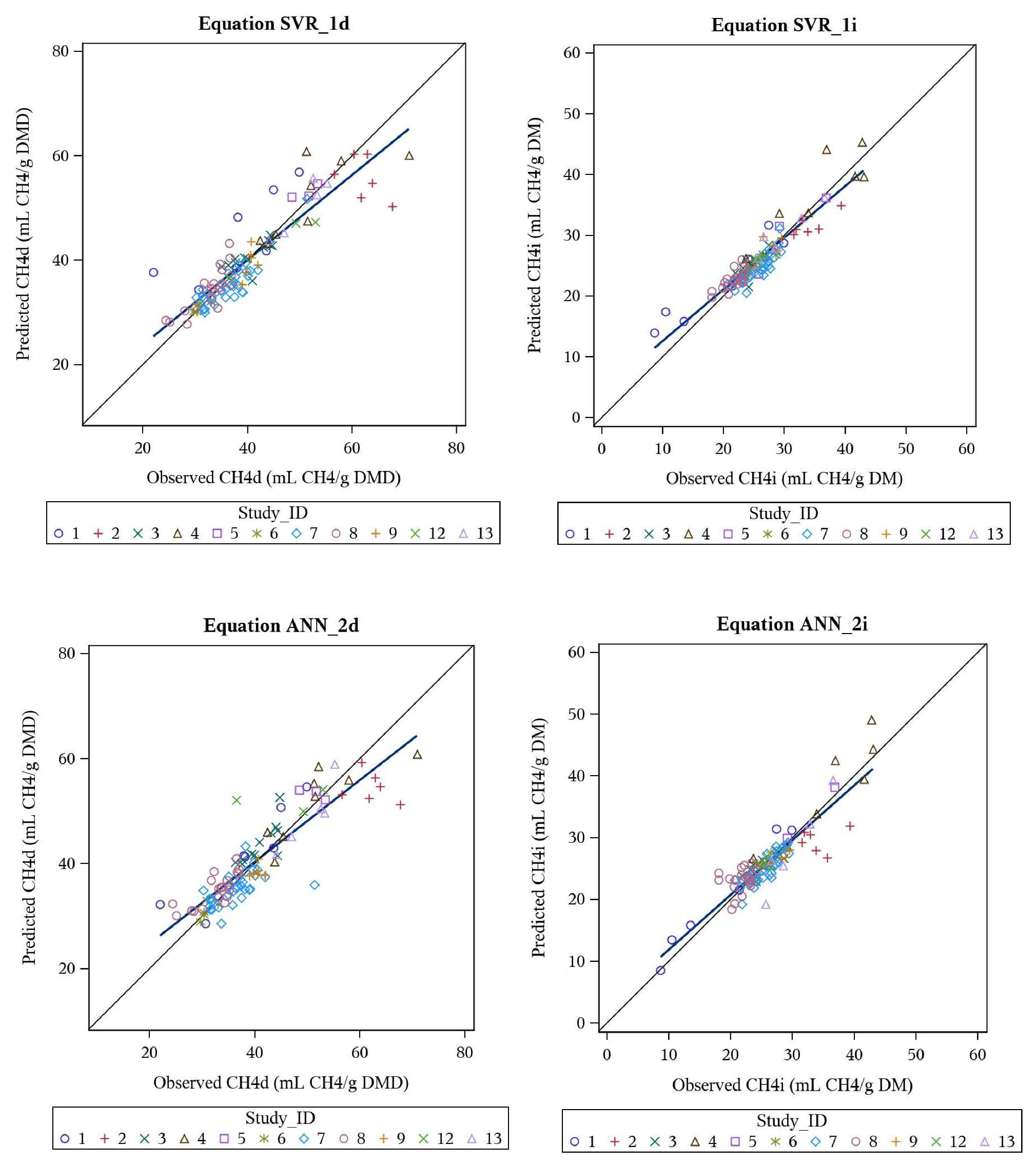
3.4. Support Vector Regression and Artificial Neural Network Models
3.5. Behaviour Analysis—Machine Learning Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Variable Abbreviation | Unit | Description |
|---|---|---|
| CH4i | mL CH4/g DM incubated | In vitro methane production |
| CH4d | mL CH4/g DM apparently digested | In vitro methane production |
| pH | - | Final pH in the incubation medium |
| DMD | g DM disappeared/g DM incubated | Apparent dry matter (DM) digestibility |
| TGP | mL gas/g DM incubated | Total gas production |
| VFA | mmol total VFA/g DM incubated | Total VFA production |
| AC | mmol AC/mol VFA | Acetic acid, proportion of total VFA |
| PR | mmol PR/mol VFA | Propionic acid, proportion of total VFA |
| BT | mmol BT/mol VFA | Butyric acid, proportion of total VFA |
| VL | mmol VL/mol VFA | Valeric acid, proportion of total VFA |
| ACp | mmol AC/g DM incubated | Acetic acid production |
| PRp | mmol PR/g DM incubated | Propionic acid production |
| BTp | mmol BT/g DM incubated | Butyric acid production |
| VLp | mmol VL/g DM incubated | Valeric acid production |
| C2C3 | AC/PR | Acetate to propionate ratio |
References
- IPCC. Climate Change 2014 Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland, 2014; pp. 1–151. [Google Scholar]
- IPCC. 2019 Refinement to the 2006 IPCC Guidelines for National Greenhouse Gas Inventories; Volume 4: Agriculture, Forestry and Other Land Use - Chapter 10: Emissions from livestock and manure management; Calvo Buendia, E., Tanabe, K., Kranjc, A., Baasansuren, J., Fukuda, M., Ngarize, S., Osako, A., Pyrozhenko, Y., Shermanau, P., Federici, S., Eds.; IPCC: Geneva, Switzerland, 2019; pp. 10.1–10.209. [Google Scholar]
- Martin, C.; Morgavi, D.P.; Doreau, M. Methane mitigation in ruminants: From microbe to the farm scale. Animal 2010, 4, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Choudhury, P.K.; Carro, M.D.; Griffith, G.W.; Dagar, S.S.; Puniya, M.; Calabro, S.; Ravella, S.R.; Dhewa, T.; Upadhyay, R.C.; et al. New aspects and strategies for methane mitigation from ruminants. Appl. Microbiol. Biotechnol. 2014, 98, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. Special Topics—Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.L.; Dijkstra, J.; Kebreab, E.; Bannink, A.; Odongo, N.E.; McBride, B.W.; France, J. Aspects of rumen microbiology central to mechanistic modelling of methane production in cattle. J. Agric. Sci. 2008, 146, 213–233. [Google Scholar] [CrossRef]
- Jayanegara, A.; Ikhsan, I.; Toharmat, T. Assessment of methane estimation from volatile fatty acid stoichiometry in the rumen in vitro. J. Ind. Trop. Anim. Agric. 2013, 38, 103–108. [Google Scholar] [CrossRef]
- Hegarty, R.S.; Nolan, J.V. Estimation of ruminal methane production from measurement of volatile fatty acid production. In Measuring Methane Production from Ruminants; Makkar, H.P.S., Vercoe, P.E., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 69–92. [Google Scholar]
- Moss, A.; Jouany, J.-P.; Newbold, J. Methane production by ruminants: Its contribution to global warming. Ann. Zootech. 2000, 49, 231–253. [Google Scholar] [CrossRef]
- McGeough, E.J.; O’Kiely, P.; O’Brien, M.; Kenny, D.A. An evaluation of the methane output associated with high-moisture grains and silages using the in vitro total gas production technique. Anim. Prod. Sci. 2011, 51, 627–634. [Google Scholar] [CrossRef]
- Navarro-Villa, A.; O’Brien, M.; López, S.; Boland, T.M.; O’Kiely, P. Modifications of a gas production technique for assessing in vitro rumen methane production from feedstuffs. Anim. Feed Sci. Technol. 2011, 166, 163–174. [Google Scholar] [CrossRef]
- Navarro-Villa, A.; O’Brien, M.; López, S.; Boland, T.M.; O’Kiely, P. In vitro rumen methane output of red clover and perennial ryegrass assayed using the gas production technique (GPT). Anim. Feed Sci. Technol. 2011, 168, 152–164. [Google Scholar] [CrossRef]
- Navarro-Villa, A.; O’Brien, M.; López, S.; Boland, T.M.; O’Kiely, P. In vitro rumen methane output of grasses and grass silages differing in fermentation characteristics using the gas-production technique (GPT). Grass Forage Sci. 2013, 68, 228–244. [Google Scholar] [CrossRef]
- Purcell, P.J.; O’Brien, M.; Boland, T.M.; O’Kiely, P. In vitro rumen methane output of perennial ryegrass samples prepared by freeze drying or thermal drying (40 °C). Anim. Feed Sci. Technol. 2011, 166, 175–182. [Google Scholar] [CrossRef]
- Purcell, P.J.; O’Brien, M.; Boland, T.M.; O’Donovan, M.; O’Kiely, P. Impacts of herbage mass and sward allowance of perennial ryegrass sampled throughout the growing season on in vitro rumen methane production. Anim. Feed Sci. Technol. 2011, 166, 405–411. [Google Scholar] [CrossRef]
- Purcell, P.J.; Boland, T.M.; O’Brien, M.; O’Kiely, P. In vitro rumen methane output of forb species sampled in spring and summer. Agric. Food Sci. 2012, 21, 83–90. [Google Scholar] [CrossRef]
- Purcell, P.J.; O’Brien, M.; Navarro-Villa, A.; Boland, T.M.; McEvoy, M.; Grogan, D.; O’Kiely, P. In vitro rumen methane output of perennial ryegrass varieties and perennial grass species harvested throughout the growing season: In vitro rumen methane output of perennial grasses. Grass Forage Sci. 2012, 67, 280–298. [Google Scholar] [CrossRef]
- Purcell, P.J.; Grant, J.; Boland, T.M.; Grogan, D.; O’Kiely, P. The in vitro rumen methane output of perennial grass species and white clover varieties, and associative effects for their binary mixtures, evaluated using a batch-culture technique. Anim. Prod. Sci. 2012, 52, 1077. [Google Scholar] [CrossRef]
- Purcell, P.J.; Boland, T.M.; O’Kiely, P. The effect of water-soluble carbohydrate concentration and type on in vitro rumen methane output of perennial ryegrass determined using a 24-hour batch-culture gas production technique. Irish J. Food Agric. Res. 2014, 53, 21–36. [Google Scholar]
- Navarro-Villa, A.; O’Brien, M.; López, S.; Boland, T.M.; O’Kiely, P. Determination of the in vitro rumen methane output of contrasting feeds using the gas production technique (GPT). (unpublished).
- SAS Institute Inc. SAS/STAT® 14.1 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2015. [Google Scholar]
- St-Pierre, N.R. Invited review: Integrating quantitative findings from multiple studies using mixed model methodology. J. Dairy Sci. 2001, 84, 741–755. [Google Scholar] [CrossRef]
- Coelho, L.P.; Richert, W.; Brucher, M. Building Machine Learning Systems with Python: Explore Machine Learning and Deep Learning Techniques for Building Intelligent Systems Using Scikit-Learn and TensorFlow; Packt Publishing: Birmingham, UK, 2018. [Google Scholar]
- Buitinck, L.; Louppe, G.; Blondel, M.; Pedregosa, F.; Mueller, A.; Grisel, O.; Niculae, V.; Prettenhofer, P.; Gramfort, A.; Grobler, J.; et al. API design for machine learning software: Experiences from the scikit-learn project. In Proceedings of the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Databases, Workshop on Languages for Data Mining and Machine Learning, Prague, Czech Republic, 23–27 September 2013; pp. 108–122. Available online: https://arxiv.org/pdf/1309.0238.pdf (accessed on 15 April 2020).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Rossum, G.V. Python Tutorial Release 3.6.4; Python Software Foundation: Wilmington, DE, USA, 2018. [Google Scholar]
- Drucker, H.; Burges, C.J.C.; Kaufman, L.; Smola, A.J.; Vapnik, V. Support vector regression machines. In Advances in Neural Information Processing Systems 9; Mozer, M.C., Jordan, M.I., Petsche, T., Eds.; MIT Press: Cambridge, MA, USA, 1997; pp. 155–161. [Google Scholar]
- Paliwal, M.; Kumar, U.A. Neural networks and statistical techniques: A review of applications. Expert Syst. Appl. 2009, 36, 2–17. [Google Scholar] [CrossRef]
- Bibby, J.; Toutenburg, T. Prediction and Improved Estimation in Linear Models; John Wiley & Sons: Chichester, UK, 1977. [Google Scholar]
- Lin, L.I. A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- Demeyer, D.; Van Nevel, C. Protein fermentation and growth by rumen microbes. Ann. Rech. Vet. 1979, 10, 277–279. [Google Scholar]
- Bannink, A.; France, J.; López, S.; Gerrits, W.J.J.; Kebreab, E.; Tamminga, S.; Dijkstra, J. Modelling the implications of feeding strategy on rumen fermentation and functioning of the rumen wall. Anim. Feed Sci. Technol. 2008, 143, 3–26. [Google Scholar] [CrossRef]
| Variable 1 | pH | DMD | TGP | CH4i | CH4d | VFA | AC | PR | BT | VL | C2C3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Training Dataset (Machine learning, n = 247) | |||||||||||
| Mean | 6.59 | 0.67 | 163 | 26.6 | 40.7 | 5.48 | 632.3 | 238.5 | 94.8 | 31.8 | 2.7 |
| Median | 6.64 | 0.68 | 160 | 25.3 | 38.0 | 5.33 | 632.2 | 237.7 | 94.4 | 31.8 | 2.7 |
| Minimum | 5.45 | 0.20 | 51 | 6.9 | 19.5 | 2.12 | 477.6 | 117.5 | 43.4 | 5.0 | 1.4 |
| Maximum | 6.78 | 0.91 | 276 | 50.8 | 71.5 | 9.79 | 812.1 | 346.9 | 181.3 | 79.3 | 7.0 |
| Training Dataset (Meta-analysis, n = 243) | |||||||||||
| Mean | 6.59 | 0.67 | 164 | 26.7 | 40.6 | 5.50 | 631.7 | 239.2 | 94.9 | 31.9 | 2.7 |
| Median | 6.64 | 0.68 | 161 | 25.3 | 38.0 | 5.35 | 632.1 | 237.9 | 94.4 | 31.8 | 2.7 |
| Minimum | 5.45 | 0.22 | 71 | 11.2 | 19.5 | 2.12 | 477.6 | 117.5 | 60.0 | 10.3 | 1.4 |
| Maximum | 6.78 | 0.91 | 276 | 50.8 | 71.5 | 9.79 | 812.1 | 346.9 | 181.3 | 79.3 | 7.0 |
| Evaluation Dataset (Machine learning/Meta-Analysis, n = 107) | |||||||||||
| Mean | 6.60 | 0.66 | 162 | 26.0 | 40.2 | 5.51 | 630.6 | 241.5 | 94.5 | 31.8 | 2.7 |
| Median | 6.64 | 0.68 | 162 | 25.4 | 37.9 | 5.25 | 629.0 | 238.5 | 96.4 | 32.5 | 2.7 |
| Minimum | 5.49 | 0.21 | 61 | 8.7 | 22.0 | 4.02 | 503.7 | 152.1 | 53.8 | 6.9 | 1.7 |
| Maximum | 6.84 | 0.87 | 246 | 43.0 | 70.9 | 9.81 | 787.5 | 333.1 | 178.9 | 60.8 | 5.1 |
| Variable | CH4d | CH4i | pH | DMD | TGP | VFA | AC | ACp | PR | PRp | BT | BTp | VL | VLp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 0.027 | 0.007 | ||||||||||||
| DMD | −0.408 | 0.399 | −0.060 | |||||||||||
| TGP | 0.146 | 0.755 | −0.206 | 0.707 | ||||||||||
| VFA | 0.223 | 0.472 | −0.437 | 0.282 | 0.648 | |||||||||
| AC | 0.405 | −0.014 | 0.208 | −0.474 | −0.341 | −0.159 | ||||||||
| ACp | 0.350 | 0.472 | −0.340 | 0.136 | 0.544 | 0.944 | 0.168 | |||||||
| PR | −0.396 | −0.045 | −0.125 | 0.426 | 0.316 | 0.094 | −0.863 | −0.184 | ||||||
| PRp | −0.050 | 0.326 | −0.431 | 0.439 | 0.663 | 0.817 | −0.622 | 0.605 | 0.640 | |||||
| BT | −0.035 | 0.045 | −0.360 | 0.065 | 0.115 | 0.250 | −0.279 | 0.136 | −0.150 | 0.143 | ||||
| BTp | 0.103 | 0.315 | −0.566 | 0.234 | 0.482 | 0.798 | −0.294 | 0.679 | −0.001 | 0.633 | 0.762 | |||
| VL | −0.113 | 0.223 | 0.273 | 0.369 | 0.096 | 0.007 | −0.246 | −0.071 | 0.023 | 0.016 | 0.145 | 0.071 | ||
| VLp | 0.059 | 0.458 | −0.021 | 0.439 | 0.435 | 0.555 | −0.279 | 0.458 | 0.069 | 0.465 | 0.236 | 0.491 | 0.820 | |
| C2C3 | 0.420 | −0.038 | 0.070 | −0.516 | −0.373 | −0.103 | 0.921 | 0.194 | −0.924 | −0.606 | −0.078 | −0.133 | −0.186 | −0.198 |
| Equation 1 | Y | X | Form | Mean 2 | SEM | RMSPE, % | EB, % | ER, % | ED, % | CCC | R | Cb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U1 | CH4d | C2C3 | Linear | 44.7 | 0.37 | 25.7 | 19 | 2 | 79 | 0.113 | 0.194 | 0.579 |
| U2 | CH4d | PR | Quad | 44.7 | 0.35 | 25.9 | 18 | 3 | 78 | 0.116 | 0.185 | 0.623 |
| U3 | CH4d | AC | Linear | 44.8 | 0.29 | 25.6 | 20 | 1 | 79 | 0.124 | 0.228 | 0.541 |
| U4 | CH4d | PR | Linear | 44.8 | 0.31 | 26.1 | 19 | 4 | 77 | 0.128 | 0.198 | 0.649 |
| U5 | CH4d | DMD | Quad | 44.3 | 0.23 | 24.0 | 18 | 2 | 81 | 0.182 | 0.391 | 0.466 |
| U6 | CH4d | DMD | Linear | 44.4 | 0.24 | 23.9 | 19 | 3 | 78 | 0.196 | 0.432 | 0.454 |
| U7 | CH4i | PRp | Quad | 27.7 | 0.29 | 21.0 | 11 | 3 | 87 | 0.303 | 0.377 | 0.803 |
| U8 | CH4i | DMD | Cubic | 28.2 | 0.19 | 21.8 | 15 | 4 | 80 | 0.305 | 0.375 | 0.813 |
| U9 | CH4i | VLp | Quad | 28.0 | 0.26 | 20.7 | 14 | 0 | 85 | 0.314 | 0.420 | 0.747 |
| U10 | CH4i | VFA | Linear | 27.4 | 0.16 | 20.9 | 7 | 7 | 86 | 0.346 | 0.390 | 0.889 |
| U11 | CH4i | TGP | Quad | 27.2 | 0.33 | 15.5 | 10 | 1 | 90 | 0.644 | 0.717 | 0.898 |
| U12 | CH4i | TGP | Linear | 27.3 | 0.31 | 15.5 | 10 | 0 | 89 | 0.650 | 0.717 | 0.906 |
| Equation 1 | Y | X | Mean 2 | SEM | RMSPE, % | EB, % | ER, % | ED, % | CCC | R | Cb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | CH4d | BTp, DMD | 44.0 | 0.36 | 22.7 | 17 | 1 | 82 | 0.306 | 0.476 | 0.643 |
| M2 | CH4d | PRp, VLp, DMD | 43.9 | 0.51 | 22.6 | 16 | 1 | 83 | 0.379 | 0.473 | 0.800 |
| M3 | CH4d | PR, VL, VFA, DMD | 43.8 | 0.55 | 22.9 | 16 | 2 | 82 | 0.383 | 0.461 | 0.830 |
| M4 | CH4d | DMD, VFA, pH, PR | 43.5 | 0.65 | 23.3 | 12 | 7 | 81 | 0.401 | 0.448 | 0.896 |
| M5 | CH4d | DMD, VFA, PR, FT, VL | 43.8 | 0.58 | 22.5 | 16 | 2 | 82 | 0.419 | 0.492 | 0.853 |
| M6 | CH4d | DMD, VFA | 43.5 | 0.53 | 21.7 | 14 | 1 | 85 | 0.425 | 0.516 | 0.823 |
| M7 | CH4i | pH, DMD, VLp, FT, BTp | 28.5 | 0.34 | 21.1 | 21 | 3 | 76 | 0.407 | 0.496 | 0.833 |
| M8 | CH4i | pH, DMD, PRp, VLp, FT | 28.4 | 0.33 | 20.8 | 21 | 2 | 78 | 0.410 | 0.520 | 0.826 |
| M9 | CH4i | pH, DMD, BTp, FT | 28.3 | 0.32 | 20.2 | 20 | 1 | 79 | 0.428 | 0.520 | 0.823 |
| M10 | CH4i | DMD, VFA, FT | 27.7 | 0.32 | 19.4 | 12 | 1 | 87 | 0.434 | 0.514 | 0.844 |
| M11 | CH4i | VFA, FT | 27.4 | 0.37 | 19.8 | 8 | 5 | 87 | 0.438 | 0.484 | 0.905 |
| M12 | CH4i | PR, VL, TGP | 27.2 | 0.40 | 14.8 | 11 | 0 | 89 | 0.703 | 0.752 | 0.936 |
| Equation ID | Y | Intercept | X1 | X2 | X3 | X4 | X5 |
|---|---|---|---|---|---|---|---|
| M5 | CH4d | 76.35 (± 4.511) | −31.03 (± 3.922) × DMD | 3.21 (± 0.352) × VFA | −0.094 (± 0.01202) × PR | −3.017 (± 1.460) (if FT = 1) | −0.133 (± 0.0380) × VL |
| M6 | CH4d | 51.35 (± 3.086) | −41.98 (± 3.429) × DMD | 3.65 (± 0.390) × VFA | |||
| M11 | CH4i | 15.8 (± 2.614) | 3.06 (± 0.241) × VFA | −5.70 (± 1.028) (if FT = 1) | |||
| M12 | CH4i | 11.58 (± 1.627) | −0.0633 (± 0.0057) × PR | 0.0947 (± 0.01728) × VL | 0.172 (± 0.0046) × TGP |
| Equation 1 | Y | X | Mean 2 | SEM | RMSPE, % | EB, % | ER, % | ED, % | CCC | R | Cb |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SVR_1d | CH4d | all, nonlinear | 40.2 | 0.82 | 9.9 | 0.5 | 0 | 99.5 | 0.899 | 0.905 | 0.994 |
| SVR_1i | CH4i | all, nonlinear | 26.1 | 0.49 | 8.3 | 0.6 | 0.1 | 99.3 | 0.917 | 0.920 | 0.997 |
| ANN_2d | CH4d | all, nonlinear | 40.5 | 0.80 | 9.5 | 0.5 | 0.8 | 98.7 | 0.907 | 0.915 | 0.991 |
| ANN_2i | CH4i | all, nonlinear | 26.0 | 0.52 | 9.1 | 0 | 2.9 | 97.1 | 0.906 | 0.906 | 1.000 |
| METd | CH4d | all, linear | 42.9 | 0.54 | 17.1 | 16 | 6 | 79 | 0.643 | 0.762 | 0.844 |
| METi | CH4i | all, linear | 27.2 | 0.40 | 14.0 | 12 | 0 | 88 | 0.734 | 0.782 | 0.939 |
| X-Variable | CH4i (on Average 25.5 and 36.3 mL CH4/g Dry Matter Incubated, for Forage and Concentrate, Respectively) | CH4d (on Average 40.0 and 46.4 mL CH4/g Dry Matter Apparently Digested for Forage and Concentrate, Respectively) | ||||||
|---|---|---|---|---|---|---|---|---|
| ANN (ANN_2i) | SVR (SVR_1i) | ANN (ANN_2d) | SVR (SVR_1d) | |||||
| Change in X-variable | +10% 2 | −10% 3 | +10% 2 | −10% 3 | +10% 2 | −10% 3 | +10% 2 | −10% 3 |
| Feed type = forage (FT = 1) | ||||||||
| pH | 14% | 36% | −6% | 5% | −14% | 7% | 9% | 35% |
| DMD | 0% | 0% | −5% | 9% | −7% | 18% | −13% | 18% |
| TGP | 12% | −10% | 20% | −16% | 15% | −8% | 20% | −16% |
| Total VFA | −1% | 1% | −1% | 0% | −1% | 1% | −1% | 0% |
| Acetate (AC) | 5% | 11% | 4% | 3% | 11% | −11% | 2% | 0% |
| Propionate (PR) | 0% | 0% | −2% | 2% | −1% | 1% | −1% | 2% |
| Butyrate (BT) | 0% | 0% | −6% | 6% | −1% | 1% | −6% | 8% |
| Valerate (VL) | −2% | 2% | 0% | −1% | −1% | 1% | 1% | −1% |
| C2C3 | −1% | 1% | 1% | 0% | −4% | 4% | 1% | 0% |
| Feed type = concentrate (FT = 2) | ||||||||
| pH | 4% | −24% | 30% | −39% | 11% | −23% | 37% | −37% |
| DMD | −2% | −1% | 5% | −2% | −8% | 8% | −3% | 5% |
| TGP | 11% | −11% | 12% | −10% | 10% | −10% | 9% | −8% |
| Total VFA | −4% | 2% | −3% | 2% | −2% | 2% | −2% | 1% |
| Acetate (AC) | −3% | 3% | 7% | −6% | 3% | −3% | 6% | −6% |
| Propionate (PR) | −3% | 3% | −4% | 4% | −3% | 0% | −3% | 3% |
| Butyrate (BT) | 2% | −2% | −1% | 1% | 2% | −3% | −2% | 2% |
| Valerate (VL) | 1% | −1% | 0% | 0% | 1% | −1% | 0% | 0% |
| C2C3 | 0% | −2% | 1% | −1% | 0% | 0% | 1% | −1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellis, J.L.; Alaiz-Moretón, H.; Navarro-Villa, A.; McGeough, E.J.; Purcell, P.; Powell, C.D.; O’Kiely, P.; France, J.; López, S. Application of Meta-Analysis and Machine Learning Methods to the Prediction of Methane Production from In Vitro Mixed Ruminal Micro-Organism Fermentation. Animals 2020, 10, 720. https://doi.org/10.3390/ani10040720
Ellis JL, Alaiz-Moretón H, Navarro-Villa A, McGeough EJ, Purcell P, Powell CD, O’Kiely P, France J, López S. Application of Meta-Analysis and Machine Learning Methods to the Prediction of Methane Production from In Vitro Mixed Ruminal Micro-Organism Fermentation. Animals. 2020; 10(4):720. https://doi.org/10.3390/ani10040720
Chicago/Turabian StyleEllis, Jennifer L., Héctor Alaiz-Moretón, Alberto Navarro-Villa, Emma J. McGeough, Peter Purcell, Christopher D. Powell, Padraig O’Kiely, James France, and Secundino López. 2020. "Application of Meta-Analysis and Machine Learning Methods to the Prediction of Methane Production from In Vitro Mixed Ruminal Micro-Organism Fermentation" Animals 10, no. 4: 720. https://doi.org/10.3390/ani10040720
APA StyleEllis, J. L., Alaiz-Moretón, H., Navarro-Villa, A., McGeough, E. J., Purcell, P., Powell, C. D., O’Kiely, P., France, J., & López, S. (2020). Application of Meta-Analysis and Machine Learning Methods to the Prediction of Methane Production from In Vitro Mixed Ruminal Micro-Organism Fermentation. Animals, 10(4), 720. https://doi.org/10.3390/ani10040720






