Mammary Transcriptome Profile during Peak and Late Lactation Reveals Differentially Expression Genes Related to Inflammation and Immunity in Chinese Holstein
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals Selection and Samples Collection
2.3. Microbiological Study
2.4. Determination and Analysis of SCC in Milk Samples
2.5. Total RNA Extraction and cDNA Library Construction
2.6. Gene Expression Level Analysis
2.7. Functional Annotation and Pathway Analysis of DEGs
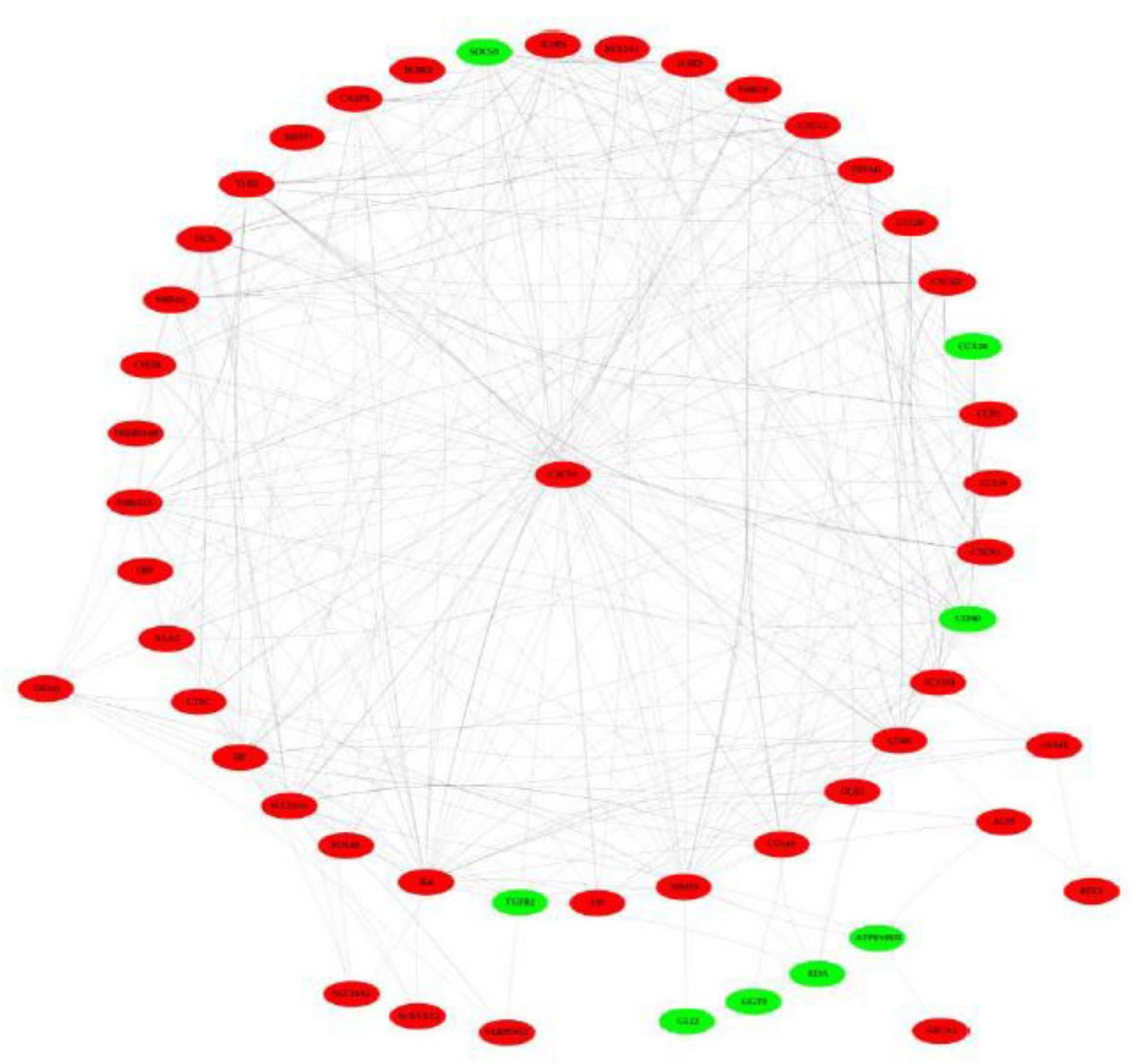
2.8. PPI Network Construction and Analysis
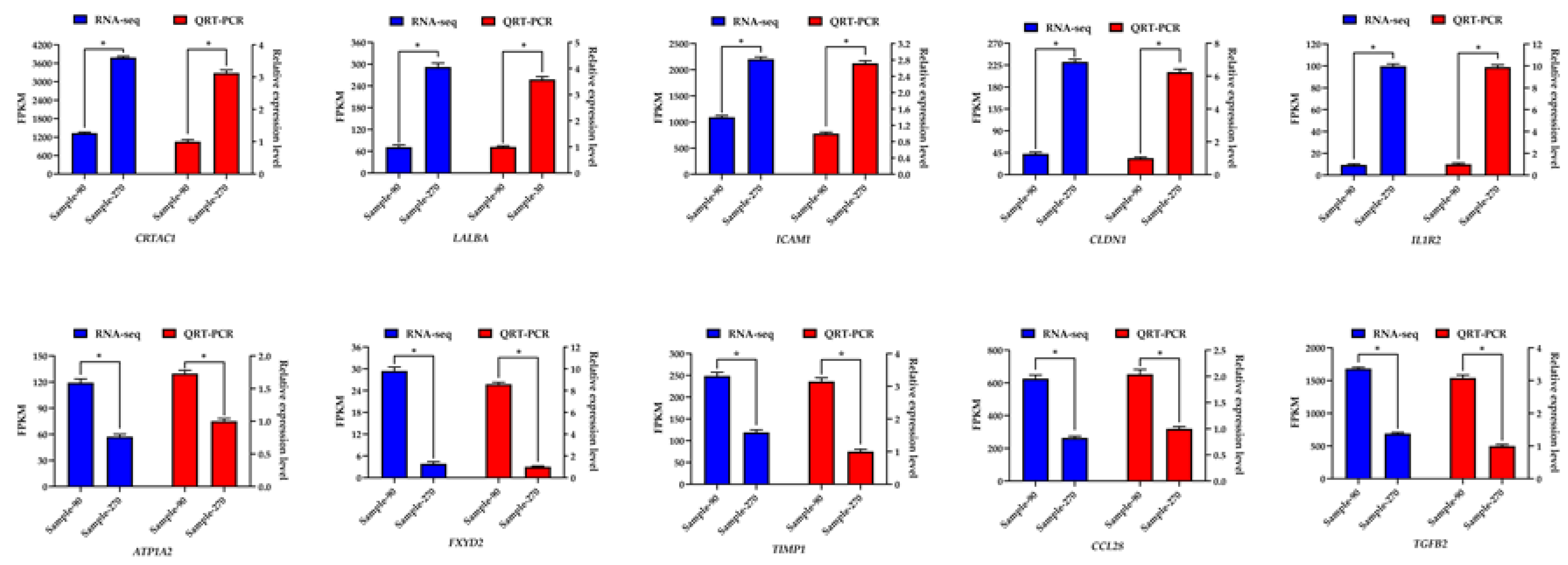
2.9. Validation of Sequencing Data by qRT-PCR
2.10. Statistical Analysis
3. Results
3.1. Microbiological Analysis
3.2. Daily Milk Yield and SCC in Milk Samples
3.3. Analysis of cDNA Libraries
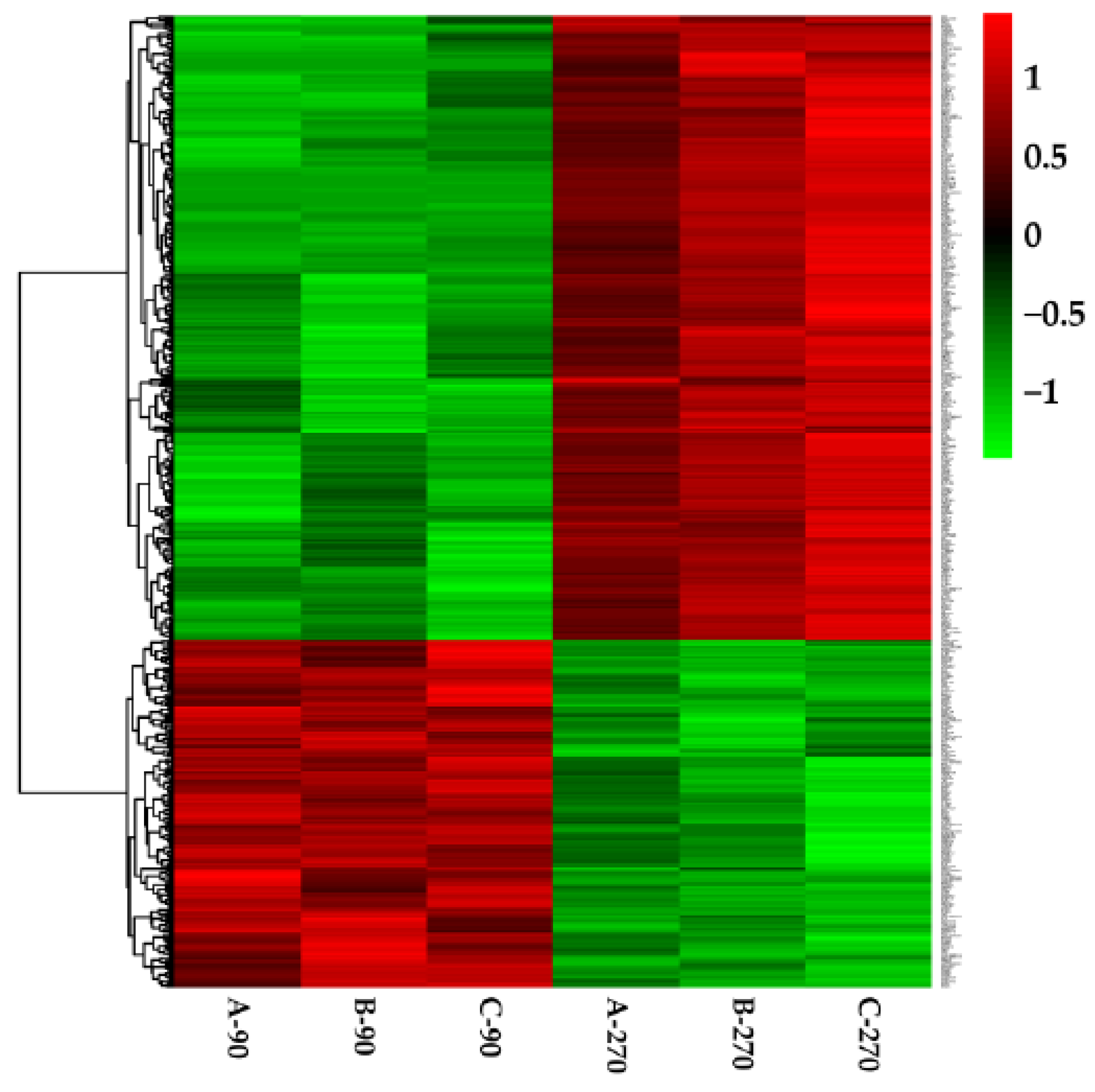
3.4. Gene Expression in Different Samples
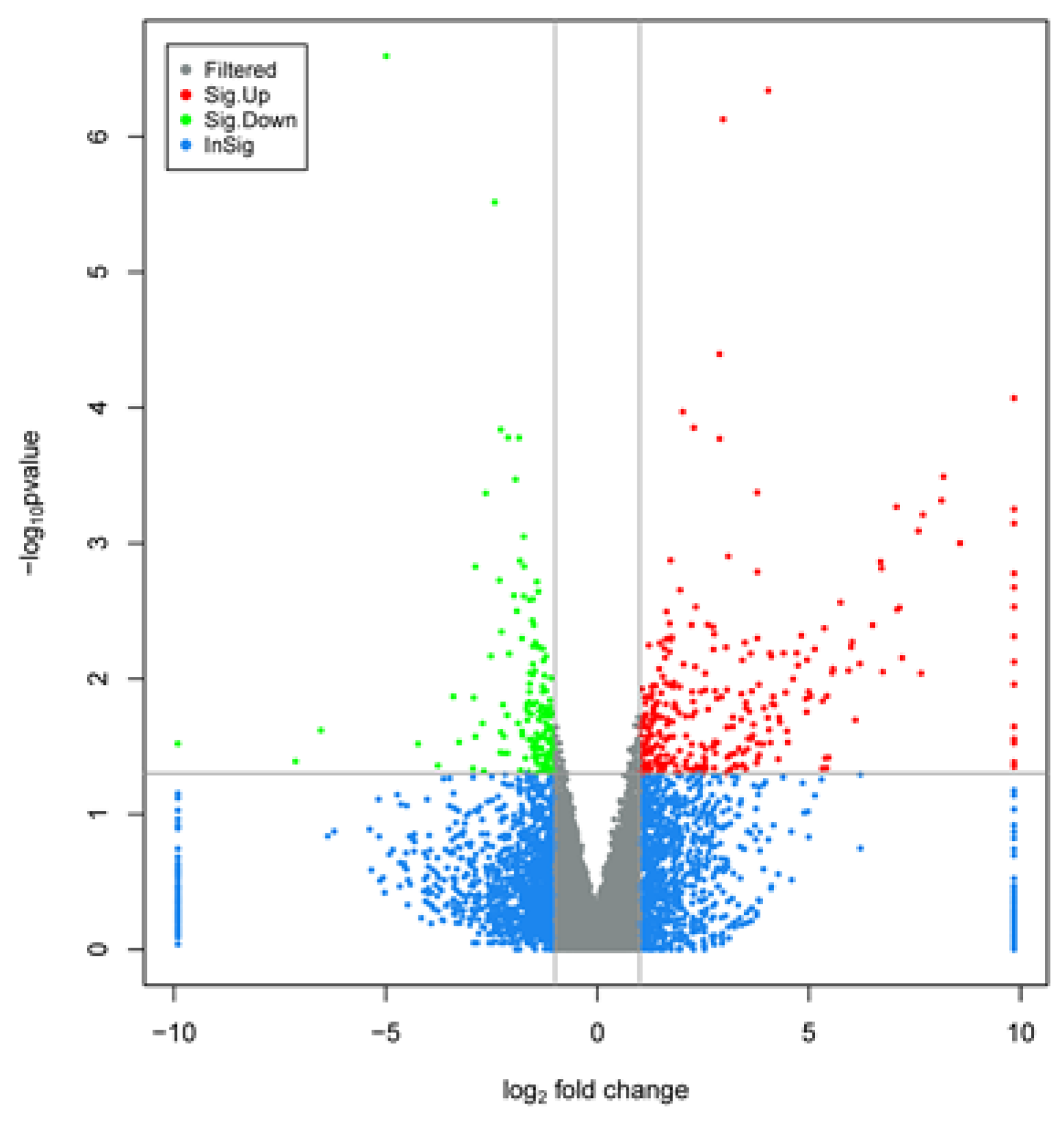
3.5. Screening of Differentially Expressed Genes
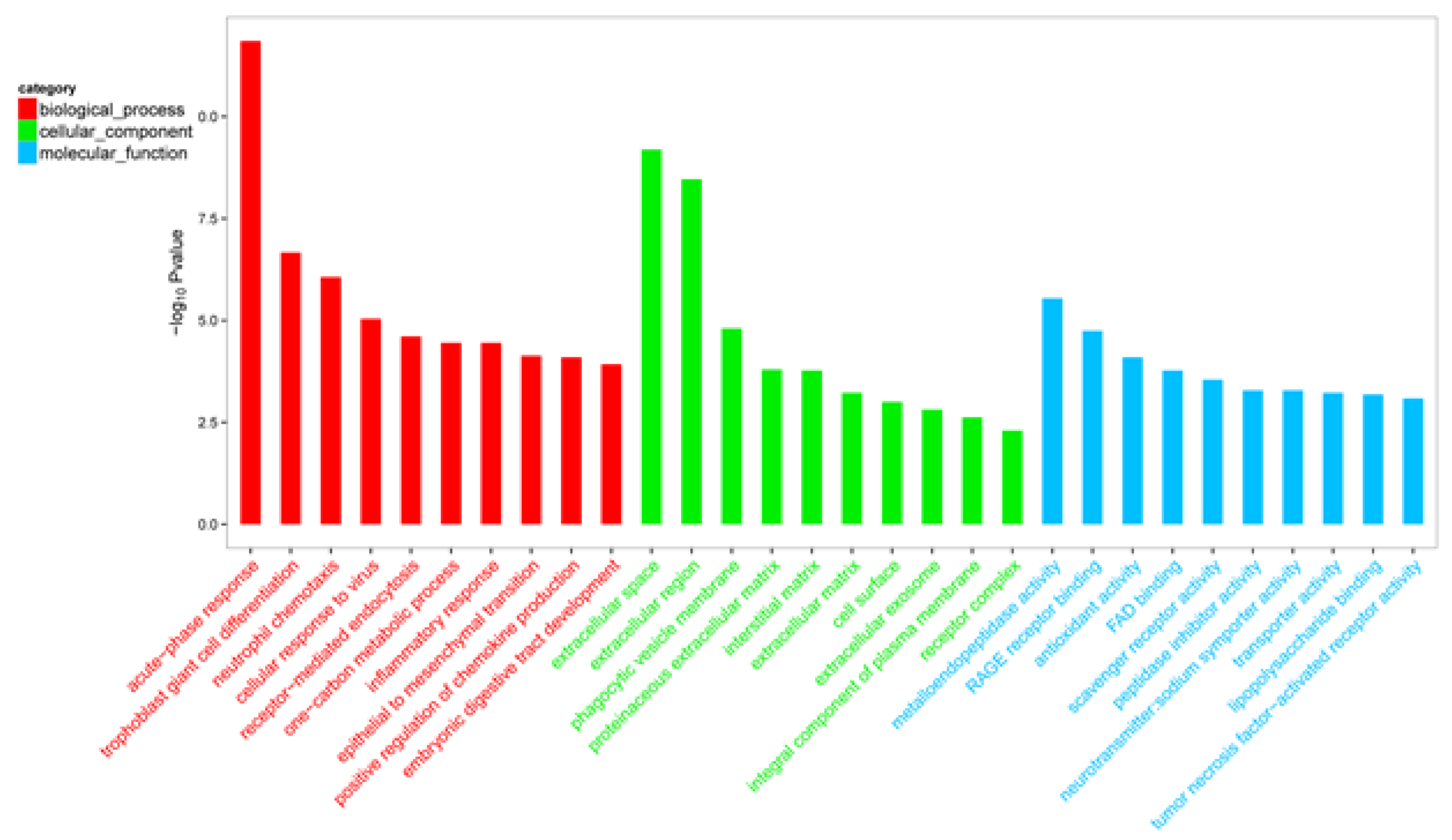
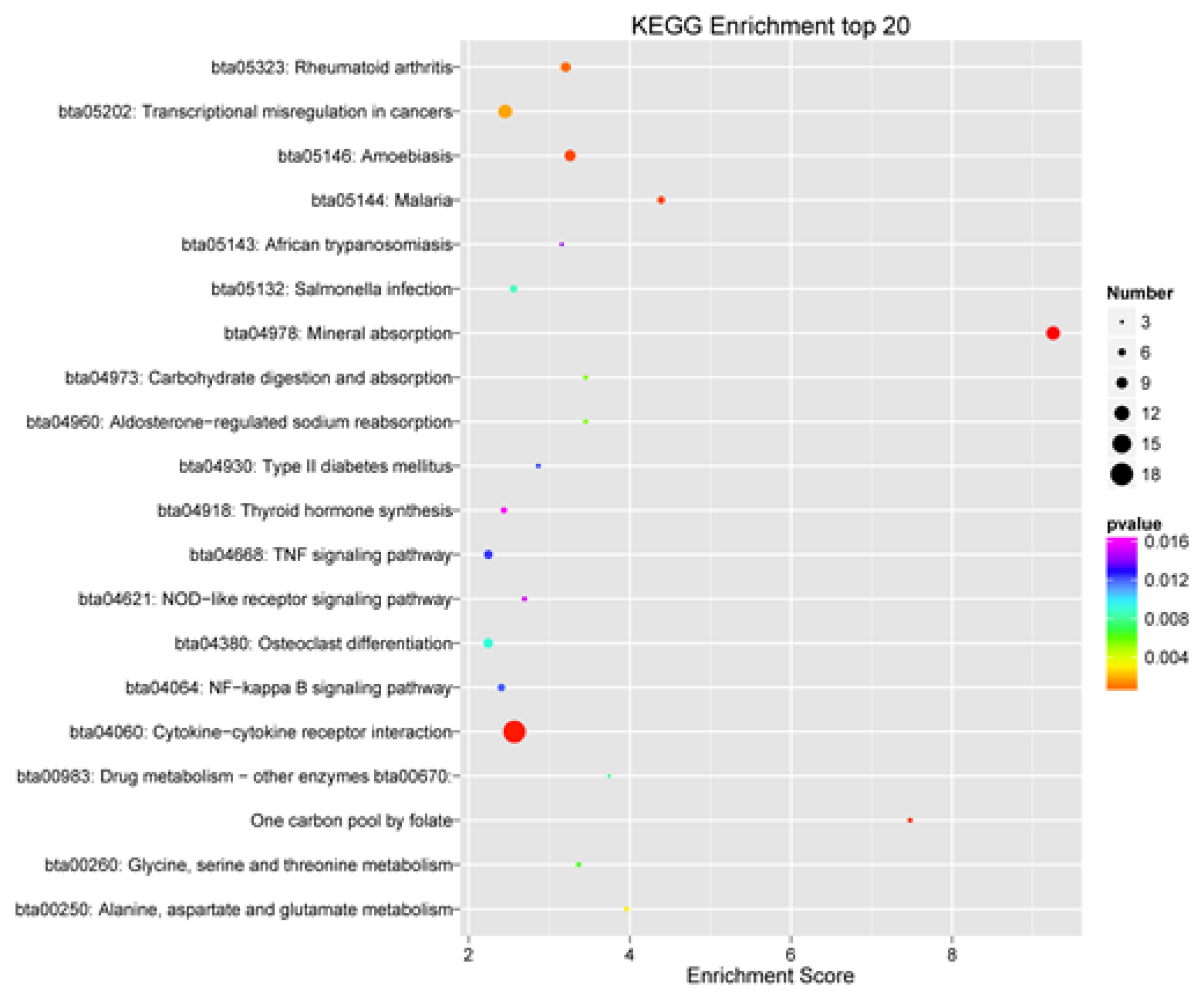
3.6. GO and KEGG Enrichment Analysis of DEGs
3.7. PPI Network Analysis
3.8. Verification Results of qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sarikaya, H.; Bruckmaier, R.M. Importance of the sampled milk fraction for the prediction of total quarter somatic cell count. J. Dairy Sci. 2006, 89, 4246–4250. [Google Scholar] [CrossRef]
- Ma, Y.; Ryan, C.; Barbano, D.M.; Galton, D.M.; Rudan, M.A.; Boor, K.J. Effects of somatic cell count on quality and shelf-life of pasteurized fluid milk. J. Dairy Sci. 2000, 83, 264–274. [Google Scholar] [CrossRef]
- Helgren, J.M.; Reinemann, D.J. Survey of milk quality on US dairy farms utilizing automatic milking systems. Trans. Asabe 2006, 49, 551–556. [Google Scholar] [CrossRef]
- Yang, H.Y.; Jian, G.; Liu, X.Q.; Su, J.L.; Bo, H. Rapid detection of milk somatic cell counts and its clinical significance. Eur. Spine J. 2010, 46. [Google Scholar] [CrossRef]
- Eltahawy, A.S.; Elfar, A.H. Influences of somatic cell count on milk composition and dairy farm profitability. Int. J. Dairy Technol. 2010, 63, 463–469. [Google Scholar] [CrossRef]
- Li, N.; Richoux, R.; Boutinaud, M.; Martin, P.; Gagnaire, V. Erratum to: Role of somatic cells on dairy processes and products: A review. Dairy Sci. Technol. 2014, 94, 517–538. [Google Scholar] [CrossRef]
- Auldist, M.J.; Coats, S.; Rogers, G.L.; Mcdowell, G.H. Changes in the composition of milk from healthy and mastitic dairy cows during the lactation cycle. Aust. J. Exp. Agric. 1995, 35, 427–436. [Google Scholar] [CrossRef]
- Hagnestam-Nielsen, C.; Emanuelson, U.; Berglund, B.; Strandberg, E. Relationship between somatic cell count and milk yield in different stages of lactation. J. Dairy Sci. 2009, 92, 3124–3133. [Google Scholar] [CrossRef]
- Ng-Kwai-Hang, K.F.; Hayes, J.F.; Moxley, J.E.; Monardes, H.G. Variability of test-day milk production and composition and relations of somatic cell counts with yield and compositional changes of bovine milk. J. Dairy Sci. 1984, 67, 361–366. [Google Scholar] [CrossRef]
- Dai, W.T.; Zou, Y.X.; White, R.R.; Liu, J.X.; Liu, H.Y. Transcriptomic profiles of the bovine mammary gland during lactation and the dry period. Funct. Integr. Genom. 2018, 18, 125–140. [Google Scholar] [CrossRef]
- Liu, D.-D.; Zhang, T.-Z.; Sun, W.; Hu, S.-X.; Yun, H.-Q.; Chi, H.-H.; Han, L.-Q.; Li, X.-H. Effects of days in milk(DIM) on production performance and correlation analysis in primiparous and older holstein cows. China Dairy Cattle 2018, 11. [Google Scholar] [CrossRef]
- Mao, Y.; Yang, Z.; Wang, X.; Hua, J.; Zhu, J.; Zhang, X. The relationship among milking traits and udder traits SCC in milk for Chinese holstein in south. China Dairy Cattle 2002, 2. [Google Scholar] [CrossRef]
- Mccoard, S.A.; Hayashi, A.A.; Sciascia, Q.; Rounce, J.; Sinclair, B.; Mcnabb, W.C.; Roy, N.C. Mammary transcriptome analysis of lactating dairy cows following administration of bovine growth hormone. Animal 2016, 10, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lv, H.; Jiang, M.; Zhou, J.; Song, S.; Hou, X. Functional analysis of the dairy cow mammary transcriptome between early lactation and mid-dry period. JDR 2019, 86, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Bauersachs, S.; Simintiras, C.A.; Sturmey, R.G.; Krebs, S.; Bick, J.; Blum, H.; Wolf, E.; Lonergan, P.; Forde, N. Effect of metabolic status on conceptus-maternal interactions on Day 19 in dairy cattle: II. Effects on the endometrial transcriptome. Biol. Reprod. 2017, 97. [Google Scholar] [CrossRef]
- Piantoni, P.; Wang, P.; Drackley, J.K.; Hurley, W.L.; Loor, J.J. Expression of metabolic, tissue remodeling, oxidative stress, and inflammatory pathways in mammary tissue during involution in lactating dairy cows. Bioinform. Biol. Insights 2010, 4, BBI.S5850. [Google Scholar] [CrossRef]
- Li, C.; Cai, W.; Zhou, C.; Yin, H.; Zhang, Z.; Loor, J.J.; Sun, D.; Zhang, Q.; Liu, J.; Zhang, S. RNA-Seq reveals 10 novel promising candidate genes affecting milk protein concentration in the Chinese Holstein population. Sci. Rep. 2016, 6, 26813. [Google Scholar] [CrossRef]
- Gao, J.; Barkema, H.W.; Zhang, L.; Liu, G.; Deng, Z.; Cai, L.; Shan, R.; Zhang, S.; Zou, J.; Kastelic, J.P. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J. Dairy Sci. 2017, 100, 4797–4806. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, M.; Hardwidge, P.R.; Cui, H.; Zhu, G. Isolation and characterization of N-acyl homoserine lactone-producing bacteria from cattle rumen and swine intestines. Front. Cell. Infect. Microbiol. 2018, 8, 155. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, W.; Mao, Y.; Yang, Z.; Lu, G.; Zhang, R.; Zhang, H.; Szeto, C.; Wang, C. Bovine leukemia virus infection in cattle of China: Association with reduced milk production and increased somatic cell score. J. Dairy Sci. 2016, 99, 3688–3697. [Google Scholar] [CrossRef]
- Patel, R.K.; Mukesh, J. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Simon, A.; Paul Theodor, P.; Wolfgang, H. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Crisà, A.; Ferrè, F.; Chillemi, G.; Moioli, B. RNA-Sequencing for profiling goat milk transcriptome in colostrum and mature milk. BMC Vet. Res. 2016, 12, 264. [Google Scholar] [CrossRef][Green Version]
- Ren, P.; Meng, Y.; Li, B.; Ma, X.; Si, E.; Lai, Y.; Wang, J.; Yao, L.; Yang, K.; Shang, X. Molecular mechanisms of acclimatization to phosphorus starvation and recovery underlying full-length transcriptome profiling in barley (Hordeum vulgare L.). Front. Plant Sci. 2018, 9, 500. [Google Scholar] [CrossRef]
- Da, W.H.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. NAR 2011, 39, 316–322. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Li, H.; Zhao, X.; Wang, J.; Zong, M.; Yang, H. Bioinformatics analysis of gene expression profile data to screen key genes involved in pulmonary sarcoidosis. Gene 2017, 596, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Identification of reference genes for quantitative real-time PCR in the bovine mammary gland during the lactation cycle. Physiol. Genom. 2007, 29, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wellnitz, O.; Bruckmaier, R.M. The innate immune response of the bovine mammary gland to bacterial infection. Vet. J. 2012, 192, 148–152. [Google Scholar] [CrossRef]
- Hospido, A.; Sonesson, U. The environmental impact of mastitis: A case study of dairy herds. Sci. Total Environ. 2005, 343, 71–82. [Google Scholar] [CrossRef]
- Hirotani, T.; Yamamoto, M.; Kumagai, Y.; Uematsu, S.; Kawase, I.; Takeuchi, O.; Akira, S. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-β. Biochem. Biophys. Res. Commun. 2005, 328, 383–392. [Google Scholar] [CrossRef]
- Zhao, Z.; Xue, Y.; Hu, Z.; Zhou, F.; Ma, B.; Long, T.; Xue, Q.; Liu, H. Toll-like receptor 2 gene polymorphisms in Chinese Holstein cattle and their associations with bovine tuberculosis. Vet. Immunol. Immunopathol. 2017, 186, 51–54. [Google Scholar] [CrossRef]
- Gilbert, F.B.; Cunha, P.; Jensen, K.; Glass, E.J.; Foucras, G.; Robert-Granié, C.; Rupp, R.; Rainard, P. Differential response of bovine mammary epithelial cells to Staphylococcus aureus or Escherichia coli agonists of the innate immune system. Vet. Res. 2013, 44, 40. [Google Scholar] [CrossRef]
- Gerard, C.; Rollins, B.J. Chemokines and disease. Nat. Immunol. 2001, 2, 108–115. [Google Scholar] [CrossRef]
- Usman, T.; Wang, Y.; Liu, C.; He, Y.; Wang, X.; Dong, Y.; Wu, H.; Liu, A.; Yu, Y. Novel SNPs in IL-17F and IL-17A genes associated with somatic cell count in Chinese Holstein and Inner-Mongolia Sanhe cattle. J. Anim. Sci. Biotechnol. 2017, 8, 5. [Google Scholar] [CrossRef]
- Wolf, F.W.; Marks, R.M.; Sarma, V.; Byers, M.G.; Katz, R.W.; Shows, T.B.; Dixit, V.M. Characterization of a novel tumor necrosis factor-alpha-induced endothelial primary response gene. J. Biol. Chem. 1992, 267, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Dixit, V.M.; Green, S.; Sarma, V.; Holzman, L.B.; Wolf, F.W.; O’Rourke, K.; Ward, P.A.; Prochownik, E.V.; Marks, R.M. Tumor necrosis factor-alpha induction of novel gene products in human endothelial cells including a macrophage-specific chemotaxin. J. Biol. Chem. 1990, 265, 2973–2978. [Google Scholar] [CrossRef] [PubMed]
- Yun-Cai, L.; Josef, P.; Michael, K. Immunity by ubiquitylation: A reversible process of modification. Nat. Rev. Immunol. 2005, 5, 941–952. [Google Scholar] [CrossRef]
- Harhaj, E.W.; Dixit, V.M. Regulation of NF-κB by deubiquitinases. Immunol. Rev. 2012, 246, 107–124. [Google Scholar] [CrossRef]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef]
- Supek, F.; Supekova, L.; Nelson, H.; Nelson, N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc. Natl. Acad. Sci. USA 1996, 93, 5105–5110. [Google Scholar] [CrossRef]
- Kumar, N.; Mitra, A.; Ganguly, I.; Singh, R.; Deb, S.M.; Srivastava, S.K.; Sharma, A. Lack of association of brucellosis resistance with (GT)(13) microsatellite allele at 3′UTR of NRAMP1 gene in Indian zebu (Bos indicus) and crossbred (Bos indicus × Bos taurus) cattle. Vet. Microbiol. 2005, 111, 139–143. [Google Scholar] [CrossRef]
- Pereira-Suárez, A.L.; Estrada-Chávez, C.; Arriaga-Díaz, C.; Espinosa-Cueto, P.; Mancilla, R. Coexpression of NRAMP1, iNOS, and nitrotyrosine in bovine tuberculosis. Vet. Pathol. 2006, 43, 709–717. [Google Scholar] [CrossRef]
- Pinedo, P.J.; Buergelt, C.D.; Donovan, G.A.; Melendez, P.; Morel, L.; Wu, R.; Langaee, T.Y.; Rae, D.O. Candidate gene polymorphisms (BoIFNG, TLR4, SLC11A1) as risk factors for paratuberculosis infection in cattle. Prev. Vet. Med. 2009, 91, 189–196. [Google Scholar] [CrossRef]
- Hébert, A.; Sayasith, K.; Sénéchal, S.; Dubreuil, P.; Lagacé, J. Demonstration of intracellular Staphylococcus aureus in bovine mastitis alveolar cells and macrophages isolated from naturally infected cow milk. FEMS Microbiol. Lett. 2000, 193, 57–62. [Google Scholar] [CrossRef]
- Joo, Y.S.; Moon, J.S.; Fox, L.K.; Suh, G.H.; Kwon, N.H.; Kim, S.H.; Park, Y.H. Comparison of natural resistance-associated macrophage protein (NRAMP)1 expression between cows with high and low milk somatic cells counts. Asian Australas. J. Anim. Sci. 2003, 16, 1830–1836. [Google Scholar] [CrossRef]
| Gene | Forward Primers (5′-3′) | Reverse Primers (5′-3′) | Length (bp) | GenBank ID |
|---|---|---|---|---|
| SLC11A2 | AGTTGACCTCCCTGGACATCT | CACGTTCGGAGGAACACTGG | 132 | NM_001101103.1 |
| CD40 | GAACAACACGTGGGGACGAA | CCGCTTCTTGGTTATGTTCCTG | 147 | NM_001105611.2 |
| ICAM1 | GGAGGTGCCGGAATATCAAT | GGCCCACTTCCTCCTTGATTA | 139 | NM_174348.2 |
| CCR1 | TCCGACTCACTCAGGACCTT | CCACGGGTCAAGGGAAATGT | 146 | NM_001077839.1 |
| IL1R2 | ACTGAAGGTGAAAGGCCTGG | CGAAGGTGGACACACCCATT | 150 | NM_001046210.2 |
| ATP1A2 | AGCTGTGGTCATCGTCACTG | TCCGCGTTGATCTGCATCTT | 138 | NM_001081524.1 |
| FXYD2 | TATGGACAGGTGGTACCTGGG | CAGCGGAATCTTTTGCTGAGG | 150 | NM_174320.4 |
| SLC30A1 | TCACGCTACCACCATTCAGC | TTTCCAGACTGGGCTTGTGG | 135 | NM_001205893.2 |
| CCL28 | AAGCAGCCAAGAAAGAGGCT | CCTCTGTGCAGCTTCATCTGT | 150 | NM_001101163.1 |
| TGFB2 | ACCCTCGGAAAATGCCATCC | GCACTCTGGCTTTTGGGTTC | 149 | NM_001113252.1 |
| RPS9 | CCTCGACCAAGAGCTGAAG | CCTCCAGACCTCACGTTTGTTC | 62 | NM_001034034.2 |
| β-actin | CATCCTGACCCTCAAGTA | CTCGTTGTAGAAGGTGTG | 91 | NM_173979.3 |
| Test Days | 90 | 150 | 210 | 270 |
|---|---|---|---|---|
| Daily milk yield (Kg) | 34.40 ± 0.05 a | 33.17 ± 0.04 b | 29.62 ± 0.04 c | 26.51 ± 0.04 d |
| Somatic cell count (SCC) (104) | 24.03 | 24.00 | 32.00 | 46.98 |
| Somatic cell score (SCS) | 4.26 ± 0.01 c | 4.26 ± 0.01 c | 4.68 ± 0.01 b | 5.23 ± 0.01 a |
| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Valid Ratio (Base) | Q30 | GC |
|---|---|---|---|---|---|---|---|
| A-90 | 61,255,240 | 7.66 Gb | 60,490,684 | 7.56 Gb | 98.72% | 97.14% | 48.50% |
| B-90 | 61,664,866 | 7.71 Gb | 60,994,408 | 7.62 Gb | 98.89% | 97.33% | 47.50% |
| C-90 | 59,050,772 | 7.38 Gb | 58,314,034 | 7.29 Gb | 98.72% | 97.11% | 49.00% |
| A-270 | 71,589,742 | 8.95 Gb | 70,550,840 | 8.82 Gb | 98.53% | 96.48% | 48.50% |
| B-270 | 77,932,606 | 9.74 Gb | 76,857,188 | 9.61 Gb | 98.60% | 96.44% | 49.00% |
| C-270 | 64,104,356 | 8.01 Gb | 62,867,970 | 7.86 Gb | 98.05% | 95.92% | 49.00% |
| Item | A-90 | B-90 | C-90 | A-270 | B-270 | C-270 |
|---|---|---|---|---|---|---|
| Total reads | 60,490,684 | 60,994,408 | 58,314,034 | 70,550,840 | 76,857,188 | 62,867,970 |
| Total mapped | 54,988,154 (90.90%) | 56,242,133 (92.21%) | 54,192,238 (92.93%) | 64,298,141 (91.14%) | 69,765,025 (90.77%) | 58,484,736 (93.03%) |
| Gene Expression | A-90 | B-90 | C-90 | A-270 | B-270 | C-270 |
|---|---|---|---|---|---|---|
| High expression genes (≥500 FPKM) | 82 | 61 | 81 | 63 | 79 | 89 |
| Medium expression genes (≥10 to 500 FPKM) | 4294 | 3207 | 5687 | 3709 | 4947 | 5641 |
| Low expression genes (<10 FPKM) | 11,585 | 12,311 | 10,692 | 11,962 | 11,404 | 10,490 |
| Nonexpressed genes | 5535 | 5917 | 5036 | 5542 | 4846 | 5056 |
| Total expressed genes | 15,961 | 15,579 | 16,460 | 15,734 | 16,430 | 16,220 |
| Term ID | Term | padj | Gene Name | Number of Genes |
|---|---|---|---|---|
| GO:0006953 | Acute-phase response | <0.001 | M-SAA3.2; ORM1; SERPINF2; SAA3; LBP; IL6; CD163; HP; IL1RN | 18 |
| GO:0030593 | Neutrophil chemotaxis | <0.001 | PDE4B; CCL19; CCL20; S100A8; S100A9; CXCL8; CSF3R; TREM1 | 8 |
| GO:0098586 | Cellular response to virus | <0.001 | IKBKE; CCL19; GLI2 | 6 |
| GO:0006954 | Inflammatory response | < 0.001 | RELT; S100A12; TNFRSF6B; OLR1; CCL19; CCL20; CCR1; MEFV; CASP4; SLC11A1; CXCL8; GGT5; CD40; TLR2 | 7 |
| GO:0032722 | Positive regulation of chemokine production | <0.001 | LBP; IL6; TLR2 | 9 |
| KEGG-Pathway | Signal Path | padj | Gene Name | Number of Genes |
|---|---|---|---|---|
| bta04060 | Cytokine–cytokine receptor interaction | <0.001 | CCL19; CCL20; CCL28; CCR1; CD40; CSF3R; CXCL2; CXCL8; CXCR1; CXCR2; EDA; IL1R2; IL6; LIF; OSMR; RELT; TGFB2; TNFRSF6B | 18 |
| bta05323 | Rheumatoid arthritis | 0.001 | ACP5; ATP6V0D2; CCL20; CXCL8; ICAM1; IL6; TGFB2; TLR2 | 8 |
| bta04064 | NF-kappa B signaling pathway | 0.012 | BCL2A1; CCL19; CD40; CXCL8; ICAM1; LBP | 6 |
| bta04668 | TNF signaling pathway | 0.013 | CCL20; CXCL2; ICAM1; IL6; LIF; MMP9; SOCS3 | 7 |
| bta04620 | Toll-like receptor signaling pathway | 0.019 | CD40; CXCL8; IKBKE; IL6; LBP; TLR2 | 6 |
| bta04142 | Lysosome | 0.024 | ABCA2; ACP5; ATP6V0D2; CD68; CTSC; SLC11A1; SLC11A2 | 7 |
| bta04062 | Chemokine signaling pathway | 0.035 | CCL19; CCL20; CCL28; CCR1; CXCL2; CXCL8; CXCR1; CXCR2; HCK | 9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Z.; Fan, Y.; Yang, Z.; Loor, J.J.; Yang, Y. Mammary Transcriptome Profile during Peak and Late Lactation Reveals Differentially Expression Genes Related to Inflammation and Immunity in Chinese Holstein. Animals 2020, 10, 510. https://doi.org/10.3390/ani10030510
Han Z, Fan Y, Yang Z, Loor JJ, Yang Y. Mammary Transcriptome Profile during Peak and Late Lactation Reveals Differentially Expression Genes Related to Inflammation and Immunity in Chinese Holstein. Animals. 2020; 10(3):510. https://doi.org/10.3390/ani10030510
Chicago/Turabian StyleHan, Ziyin, Yongliang Fan, Zhangping Yang, Juan J. Loor, and Yi Yang. 2020. "Mammary Transcriptome Profile during Peak and Late Lactation Reveals Differentially Expression Genes Related to Inflammation and Immunity in Chinese Holstein" Animals 10, no. 3: 510. https://doi.org/10.3390/ani10030510
APA StyleHan, Z., Fan, Y., Yang, Z., Loor, J. J., & Yang, Y. (2020). Mammary Transcriptome Profile during Peak and Late Lactation Reveals Differentially Expression Genes Related to Inflammation and Immunity in Chinese Holstein. Animals, 10(3), 510. https://doi.org/10.3390/ani10030510





