Simple Summary
Escherichia coli frequently causes bacterial diarrhea in piglets. Vaccine development and improved feeding and animal management strategies have reduced the incidence of bacterial diarrhea in piglets to some extent. However, current breeding strategies also have the potential to improve piglet resistance to diarrhea at a genetic level. This study sought to advance the current understanding of the functional and regulatory mechanisms whereby the candidate gene bactericidal/permeability-increasing protein (BPI) regulates piglet diarrhea at the cellular level.
Abstract
The efficacy and regulatory activity of bactericidal/permeability-increasing protein (BPI) as a mediator of Escherichia coli (E. coli) F18 resistance remains to be defined. In the present study, we evaluated lipopolysaccharide (LPS)-induced changes in BPI gene expression in porcine kidney (PK15) cells in response to E. coli F18 exposure. We additionally generated PK15 cells that overexpressed BPI to assess the impact of this gene on Toll-like receptor 4 (TLR4) signaling and glycosphingolipid biosynthesis-related genes. Through these analyses, we found that BPI expression rose significantly following LPS exposure in response to E. coli F18ac stimulation (p < 0.01). Colony count assays and qPCR analyses revealed that E. coli F18 adherence to PK15 cells was markedly suppressed following BPI overexpression (p < 0.01). BPI overexpression had no significant effect on the mRNA-level expression of genes associated with glycosphingolipid biosynthesis or TLR4 signaling. BPI overexpression suppressed the LPS-induced TLR4 signaling pathway-related expression of proinflammatory cytokines (IFN-α, IFN-β, MIP-1α, MIP-1β and IL-6). Overall, our study serves as an overview of the association between BPI and resistance to E. coli F18 at the cellular level, offering a framework for future investigations of the mechanisms whereby piglets are able to resist E. coli F18 infection.
1. Introduction
Bactericidal/permeability-increasing protein (BPI) is expressed at high levels in a range of animal cell and tissue types, with particularly pronounced expression being evident in neutrophils [1,2]. BPI exerts a range of antibacterial functions that enable it to protect against certain diseases in humans by neutralizing lipopolysaccharide (LPS) and killing Gram-negative bacteria [3]. Fan et al. [4] previously demonstrated a relationship between BPI polymorphisms and the incidence of ulcerative colitis in humans, while serum BPI levels have been utilized as a metric to analyze lipid metabolism and the functionality of the vascular endothelium in humans [5]. BPI can also prevent angiogenesis and drive the apoptotic death of endothelial cells, thereby influencing the treatment of a wide range of diseases including arthritis, atherosclerosis and cancer [6]. There is evidence that BPI promotes retinal pigment epithelial cell growth in the human eye, leading to speculation that it may be of value for the treatment of macular degeneration and related conditions [7]. The BPI family proteins BPIFA1 and BPIFB1 also exhibit antimicrobial and LPS-neutralizing activity, and there is experimental evidence for their roles in nasopharyngeal carcinoma and other respiratory diseases [8,9,10,11]. These prior studies thus highlight BPI as a broadly active antimicrobial peptide with a wide range of potential clinical applications.
Enterotoxigenic Escherichia coli (ETEC) is a primary bacterial driver of diarrhea in young animals, with F18 fimbriae-expressing ETEC being the main cause of post-weaning diarrhea in piglets [12]. Owing to their immature immune systems, piglets are unable to mount a robust adaptive immune response to ETEC infections, making it challenging to control this disease [13,14]. This has led to an increasing interest in the study of antimicrobial genes associated with porcine resistance to ETEC infections. Zhu et al. [15] compared BPI expression in Sutai piglets as a function of age and found that it was expressed at significantly higher levels in duodenal tissue samples from 35-day-old piglets relative to samples from animals in other analyzed age groups. We have also previously explored the link between BPI expression and resistance to E. coli F18 infections. Sun et al. [16] determined that the demethylation of CpG islands within the BPI gene led to its enhanced expression, thereby improving the ability of intestinal tissues to resist E. coli F18 infection. Liu et al. [17] further determined that a polymorphism in exon 10 of porcine BPI was linked to E. coli F18 resistance. To expand upon these prior findings, in this study we sought to confirm the role of BPI as a mediator of antimicrobial resistance at a cellular level. As such, we measured BPI expression in porcine kidney (PK15) cells following LPS or E. coli stimulation. We then generated BPI-overexpressing PK15 cells and evaluated the effect of such overexpression on E. coli F18 adhesion in vitro through colony counting and qPCR analyses. We have also previously utilized high-throughput data mining and functional validation approaches to identify the molecular mechanisms governing resistance to this ETEC strain, with both the Toll-like receptor signaling and glycosphingolipid biosynthesis-globo series pathways being closely associated with such resistance [18,19]. Herein, we expanded upon these prior results by assessing the impact of BPI on the expression of key genes related to the Toll-like receptor 4 (TLR4) signaling pathway (TLR4, TNF-α, IL-1β, CD14 and MyD88), TLR4 signaling-related proinflammatory cytokines (IFN-α, IFN-β, IL-6, MIP-1α and MIP-1β) and glycosphingolipid biosynthesis-related genes (FUT1 and FUT2). The overall aim of this study was to clarify the mechanisms whereby BPI facilitates porcine E. coli F18 resistance, with the goal of thereby supporting future research.
2. Materials and Methods
2.1. Experimental Materials
PK15 cells were from the American Type Culture Collection (Manassas, VA, USA). Standard porcine E. coli F18 strains with the F18ab 107/86 (O139:K12:H1) and F18ac 2134(O157:H19) fimbriae were provided by the veterinary laboratory at the Institute of Microbiology, University of Pennsylvania. LPS was purchased from Sigma-Aldrich (St. Louis, MO, USA). Dulbecco’s Modified Eagle’s Medium (DMEM), Fetal bovine serum (FBS) and trypsin-EDTA were from Gibco (Grand Island, NY, USA). Puromycin and TRIzol were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Phosphate-buffered saline (PBS) was from Beijing Labgic Technology Co., Ltd. (Beijing, China). An enzyme-linked immunosorbent assay (ELISA) kit (Porcine Interferon-α Assay Kit, Porcine Interferon-β Assay Kit, Porcine Interleukin-6 Assay Kit, Porcine Macrophage Inflammatory Protein-1α Assay Kit, Porcine Macrophage Inflammatory Protein-1β Assay Kit) was obtained from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). A DNA extraction kit (DP304) was from Tiangen Biotech Co., Ltd. (Beijing, China), while reverse transcription and real-time fluorescence quantitative kits (AceQ Universal SYBR qPCR Master Mix) were obtained from Vazyme Biotech Co., Ltd. (Nanjing, China).
2.2. BPI Overexpression in PK15 Cells
A BPI overexpression lentiviral vector (pGLV5-BPI) and a corresponding negative control (pGLV5-NC) were prepared by GenePharma (Suzhou, China). Prior to lentiviral transduction, PK15 cells were plated at 5.0 × 105 cells/well in 12-well plates in DMEM containing 10% FBS and were grown at 37 °C in a 5% CO2 incubator until 80% confluent, at which time four replicate samples were each transduced with the pGLV5-BPI or pGLV5-NC lentiviral vectors. Cells were incubated for 48 h following transduction, at which time positive cells were identified via fluorescence microscopy. Puromycin (10 µg/mL every 24 h) was then used to select for pGLV5-BPI-positive cells, and qPCR was used to confirm successful BPI overexpression in these cells.
2.3. LPS and E. coli F18 Stimulation
Cells of the blank, pGLV5-NC-positive and pGLV5-BPI-positive cells were plated in 12-well plates (5.0 × 105 per well) until 80% confluent, they were induced with 0.1 µg/mL LPS for 0, 2, 4, 6, 8, 12, 24 and 36 h, and then three replicates were performed per group. Total cellular RNA was extracted to detect Changes in BPI gene expression, and cell culture supernatants were collected for ELISA analysis.
Standard porcine E. coli strains carrying F18ab and F18ac fimbriae were inoculated into Luria-Bertani (LB) medium for 12 h in a 37 °C with constant agitation Bacteria were then collected via centrifugation at 3000 rpm for 10 min, washed three times with PBS and diluted to 1.0 × 109 colony-forming units (CFU/mL) in cell culture medium.
2.4. Analysis of E. coli F18 Adhesion to PK15 Cells
2.4.1. Colony Counting
Cells of the pGLV5-BPI and pGLV5-NC groups were inoculated to the wells of 12-well cell culture plates at a density of 5.0 × 105 cells/well and cultured until the cells reached approximately 80%. Diluents were made from the precipitation residues of the two E. coli strains, and 1 mL of the diluent was added to each well, with three replicates per group. The plates were incubated in a 5% CO2 incubator at 37 °C for 2 h. After discarding the culture medium, the cells were washed thrice with PBS and immediately treated for 20 min with 0.5% Triton X-100 (prepared with ultrapure water). After serially diluting the culture ten times, LB agar plates were coated with the culture and incubated overnight at 37 °C. Finally, bacterial count was determined by counting the number of colonies on the plate coated with 1000× the bacterial diluent using ImageJ software. The final number of bacteria adhering to the plate (CFU/mL) was equal to the number of colonies on the plate × 103.
2.4.2. Fluorescence Quantitative Polymerase Chain Reaction
Cells of the pGLV5-NC-positive and pGLV5-BPI-positive groups were seeded at 5.0 × 105 cells/well in 12-well plates until 80% confluent, at which time 1 mL of either of the two experimental E. coli strains was added to each well. Cells were then incubated for 1 h at 37 °C, after which supernatants were discarded, and cells were washed three times with PBS. Total DNA was then isolated using a DNA extraction kit. After extraction, this DNA was used as the amplification template, and qPCR primers were designed based on the PILIN gene of E. coli F18ab and F18ac, and the porcine β-ACTIN gene, which were detected by fluorescence quantitative polymerase chain reaction (PCR). [20]. All analyses were conducted in triplicate.
2.5. Primer Design
qPCR primers for BPI, TLR4, MyD88, CD14, TNF-α, IL-1β, FUT1, FUT2 and PILIN were designed with Primer Premier 5.0 based upon sequences in GenBank. GAPDH and β-ACTIN served as reference controls. All primer synthesis was conducted by Sangon Biotechnology (Shanghai, China), and the corresponding sequences are shown in Table 1.

Table 1.
qPCR primer sequences.
2.6. RNA Extraction and Preparation
TRIzol was used to extract total RNA based upon provided protocols, after which formaldehyde denaturing gel electrophoresis was conducted to gauge RNA integrity, and a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) was employed to assess RNA concentration and purity.
Isolated RNA was then reverse transcribed to produce cDNA in reactions containing 2 µL of 5× qRT SuperMix II, 500 ng of total RNA and up to 10 µL of RNase-free H2O. Thermocycler settings were as follows: 25 °C for 10 min, 50 °C for 30 min and 85 °C for 5 min. After preparation, cDNA was stored at 4 °C.
2.7. qPCR
All qPCR reactions were conducted in a 20-µL volume composed of 2 µL of cDNA, 0.4 µL of each primer (10 µmol/L), 10 µL of 2 × AceQ Universal SYBR qPCR Master Mix and 7.2 µL of ddH2O. Thermocycler settings were as follows: 95 °C for 5 min; 40 cycles of 95 °C for 5 s, 60 °C for 30 s. Melting curves were then used to confirm amplified product specificity. Three independent experimental replicates were conducted for all analyses.
2.8. Cytokine ELISAs
We obtained culture supernatants of pGLV5-NC-positive and pGLV5-BPI-positive groups at appropriate time points after LPS stimulation and measured the levels of proinflammatory cytokines (IFN-α, IFN-β, IL-6, MIP-1α and MIP-1β) via ELISA based on the provided kit instructions.
2.9. Statistical Analysis
The 2–ΔΔCt method was used to quantify relative gene expression [21], which was normalized to appropriate internal control genes. SPSS 25.0 (SPSS, Inc., Chicago, IL, USA) was used to compare data via ANOVAs with LSD tests. * p < 0.05, ** p < 0.01 and *** p < 0.001.
3. Results
3.1. LPS and E. coli F18 Induce BPI Upregulation in PK15 Cells
We began by treating PK15 cells with 0.1 µg/mL LPS for 0, 2, 4, 6, 8, 12, 24 and 36 h in order to evaluate the impact of such treatment of BPI expression, revealing that this gene was rapidly upregulated following stimulation for 4 h (Figure 1a). When cells were instead stimulated with E. coli F18ab or F18ac (1.0 × 109 CFU/mL), we found that the F18ac strain markedly enhanced BPI expression (p < 0.001), whereas F18ab strain stimulation had no impact on the expression of this gene (p > 0.05) (Figure 1b).

Figure 1.
The impact of LPS and E. coli F18 stimulation on BPI gene expression. (a) LPS was used to stimulate porcine kidney (PK15) cells for 0, 2, 4, 6, 8, 12, 24 and 36 h, after which bactericidal/permeability-increasing protein (BPI) expression was analyzed. Different lowercase letters indicate significant differences (p < 0.05), whereas identical letters indicate a lack of any difference between groups (p > 0.05). (b) BPI expression was assessed in PK15 cells following stimulation with E. coli F18ab or F18ac. Blank: untreated control group. Samples were analyzed in triplicate. Data are means ± standard deviation. *** p < 0.001.
3.2. Preparation of BPI-Overexpressing PK15 Cells
Next, we confirmed that we were able to successfully transduce PK15 cells with the pGLV5-BPI and pGLV5-NC lentiviral vectors, as confirmed based on the presence of detectable green fluorescent protein within these cells (Figure 2a). Subsequent qPCR analyses confirmed that cells transduced with the pGLV5-BPI plasmid exhibited significant BPI overexpression (5556-fold higher than in control cells; Figure 2b). These findings thus indicated that we had successfully prepared BPI-overexpressing PK15 cells, which were then used for a series of experiments.
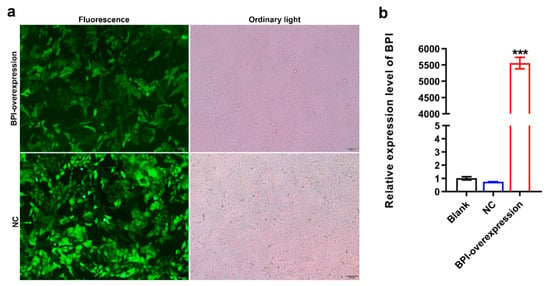
Figure 2.
Preparation of BPI-overexpressing PK15 cells. (a) Green fluorescent protein (GFP) expression in the indicated PK15 cells was assessed at 96 h following lentiviral transduction. (b) BPI expression was measured via qPCR. Blank: untreated control group, NC: PK15 cells transfected with the negative control lentivirus (pGLV5-NC), BPI-overexpression: PK15 cells transfected with the BPI gene overexpression lentivirus (pGLV5-BPI). Samples were analyzed in triplicate. Data are means ± standard deviation. *** p < 0.001.
3.3. The Impact of BPI Overexpression on E. coli F18 Adhesion to PK15 Cells
A colony counting assay revealed that the overexpression of BPI was sufficient to markedly suppress the adhesion of E. coli F18ab (Figure 3a) and E. coli F18ac (Figure 3b) to PK15 cells (p < 0.01 and p < 0.001, respectively). In line with this, qPCR analyses of PILIN gene confirmed that BPI overexpression significantly impaired E. coli F18ab (Figure 3c) and E. coli F18ac (Figure 3d) adhesion to these PK15 cells (p < 0.01 and p < 0.001, respectively). As such, these findings indicate that BPI overexpression can interfere with E. coli F18 adherence to PK15 cells.
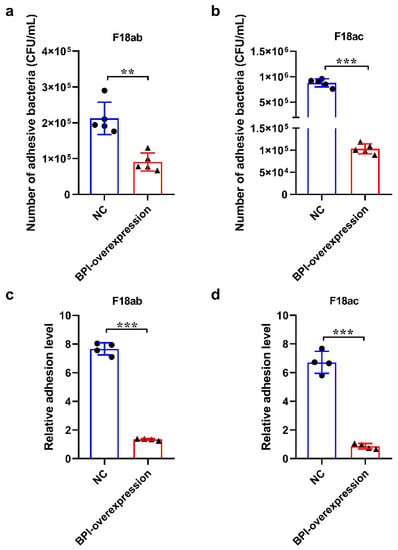
Figure 3.
The impact BPI overexpression on E. coli F18 adhesion to PK15 cells in vitro. (a,b) E. coli F18ab and F18ac adhesion to PK15 cells was assessed via a colony counting approach (n = 5). (c,d) E. coli F18ab and F18ac adhesion to PK15 cells was assessed via qPCR. NC: PK15 cells transfected with the negative control lentivirus (pGLV5-NC), BPI-overexpression: PK15 cells transfected with the BPI gene overexpression lentivirus (pGLV5-BPI). Data are means ± standard deviation. (n = 4). ** p < 0.01, *** p < 0.001.
3.4. The Impact of BPI Overexpression on TLR4 and Glycosphingolipid Biosynthesis-Globo Series Pathway-Related Gene Expression
Next, we assessed the expression of the glycosphingolipid biosynthesis-globo series pathway genes FUT1 and FUT2 in control or BPI-overexpressing PK15 cells, in which we also evaluated the expression of TLR4 signaling pathway-related genes (TLR4, TNF-α, IL-1β, CD14 and MyD88). We found that BPI overexpression did not impact TLR4, CD14, TNF-α, IL-1β, MyD88, FUT1 or FUT2 expression (p > 0.05). We also found that MyD88, FUT1 and FUT2 were highly expressed in PK15 cells (Figure 4).
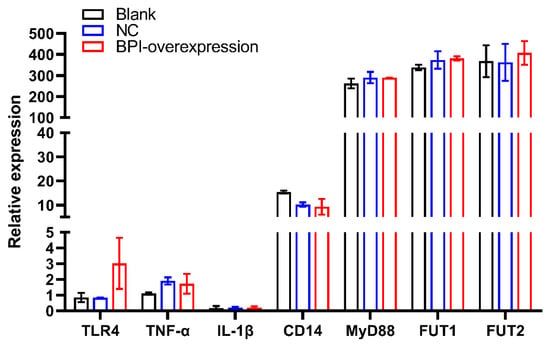
Figure 4.
The impact of BPI overexpression on Toll-like receptor 4 (TLR4) signaling pathway-related gene expression and on α-(1,2) fucosyltransferase (FUT1, FUT2) gene expression. Blank: untreated control group, NC: PK15 cells transfected with the negative control lentivirus (pGLV5-NC), BPI-overexpression: PK15 cells transfected with the BPI gene overexpression lentivirus (pGLV5-BPI). Samples were analyzed in triplicate. Data are means ± standard deviation.
3.5. The Impact of BPI Overexpression on the Upregulation of TLR4-Related Proinflammatory Cytokines
Lastly, we evaluated the production of BPI-overexpressing or control PK15 cells in response to LPS treatment (0.1 µg/mL for 0–36 h). ELISAs revealed that supernatant levels of proinflammatory cytokines such as IFN-α, IFN-β, IL-6, MIP-1α and MIP-1β initially rose and then declines over time. Furthermore, we found that BPI overexpression significantly reduced the LPS-induced upregulation of the TLR4-related proinflammatory cytokines IFN-α, IFN-β, IL-6, MIP-1α and MIP-1β in these cells (Figure 5).

Figure 5.
The impact of LPS stimulation on TLR4-related inflammatory cytokine production. NC: PK15 cells transfected with the negative control lentivirus (pGLV5-NC), BPI-overexpression: PK15 cells transfected with the BPI gene overexpression lentivirus (pGLV5-BPI). Samples were analyzed in triplicate. Data are means ± standard deviation. * p < 0.05.
4. Discussion
E. coli F18 can infect porcine cells and is characterized in part by the presence of high levels of cell wall-associated LPS [22]. Prior genetic analyses have demonstrated that the ability of piglets to resist E. coli F18 infection is tied to both innate immunity and the expression of the E. coli F18 receptor by intestinal epithelial cells [23,24,25]. PK15 cells have commonly been utilized as a model cell line for analyses of pathogenic E. coli adhesion and associated immune responses [26,27]. As such, we stimulated PK15 cells with LPS or with porcine E. coli F18, revealing that both of these treatments resulted in significant BPI upregulation. Glycosphingolipid biosynthesis-globo series pathway genes (FUT1, FUT2) are involved in the formation of the E. coli F18 receptor, and its expression level is closely related to resistance to E. coli F18 in piglets [28,29]. TLRs recognize different microbial components, sense microbial populations in the intestinal tract, initiate proinflammatory signaling pathways to resist the invasion of pathogenic microorganisms and play an important role in immune regulation in the process of resisting E. coli F18 infection [30,31]. To further assess the mechanistic role of BPI in the context of E. coli F18 infection, we next generated BPI-overexpressing PK15 cells. While such overexpression did not alter the mRNA level expression of FUT1, FUT2 or TLR4 signaling pathway-related genes (TLR4, CD14, TNF-α, IL-1β, IFN-α and MyD88), it did markedly impair E. coli F18 adhesion to these cells in vitro. We also found that BPI overexpression suppressed LPS-induced IFN-γ, IFN-α, IFN-β, IL-6, MIP-1α and MIP-1β expression. Balakrishnan et al. [32] have previously shown that LPS can increase BPI protein levels within cells. At baseline, proinflammatory cytokine production is minimal, whereas it is rapidly induced at high levels in response to LPS stimulation. BPI can bind to the conserved LPS lipid A/inner core, and it can thereby inhibit its ability to interact with TLR4 and to thereby activate proinflammatory signaling [33,34]. We therefore speculate that BPI inhibits the LPS-mediated induction of the host cellular immune response by forming a complex with LPS, thereby enabling cells to resist E. coli F18 infection.
We confirmed the link between BPI expression and E. coli F18 infection at the cellular level in porcine PK15 cells in vitro, underscoring the role of BPI as an inhibitor of inflammatory responses at least in part owing to its ability to neutralize LPS, thereby enabling these cells to better resist infection by this ETEC strain. Our data further demonstrate that BPI can markedly reduce E. coli adherence to PK15 cells in vitro. Future GST pull-down, co-immunoprecipitation (Co-IP) and gene knockout experiments will be necessary in order to fully understand the mechanisms whereby BPI influences E. coli F18 receptor molecules in this experimental context. It is also important to note that IFN-α/β are key antiviral cytokines that can induce an antiviral state in both infected and uninfected adjacent cells [35,36] by upregulating a range of IFN-stimulated genes with diverse antiviral activities [37,38]. As we found that BPI regulates IFN-α/β production by PK15 cells, this suggests that it may additionally serve as a potential regulator of antiviral immunity in these porcine cells. However, future experimental work will be needed to test this possibility.
5. Conclusions
In conclusion, we found that the overexpression of porcine BPI significantly reduced the adhesion of E. coli F18 to porcine kidney cells in vitro, although it had no impact on the expression of TLR4 or glycosphingolipid biological signaling pathway-related genes in these cells. In addition, BPI overexpression was sufficient to markedly suppress the LPS-induced upregulation of TLR4 signaling-related proinflammatory cytokines including IFN-α, IFN-β, MIP-1α, MIP-1β and IL-6.
Author Contributions
Conceptualization, W.B.; data curation, J.J. and Z.W.; formal analysis, J.J. and Y.H.; methodology, J.J.; software, J.J.; supervision, W.B. and S.W.; validation, Y.H. and S.S.; writing—original draft, J.J.; writing—review and editing, Z.Y., Z.W. and W.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31772560), Key Research and Development Project (Modern Agriculture) of Jiangsu Province (BE2019344, BE2019341), Natural Science Foundation of Jiangsu province, China (BK20180899), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Conflicts of Interest
The author declares that there is no potential conflict of interest.
References
- Weiss, J.; Elsbach, P.; Olsson, I.; Odeberg, H. Purification and characterization of a potent bactericidal and membrane active protein from the granules of human polymorphonuclear leukocytes. J. Biol. Chem. 1978, 253, 2664–2672. [Google Scholar]
- Nishimura, H.; Gogami, A.; Miyagawa, Y.; Nanbo, A.; Murakami, Y.; Baba, T.; Nagasawa, S. Bactericidal/permeability-increasing protein promotes complement activation for neutrophil-mediated phagocytosis on bacterial surface. Immunology 2001, 103, 519–525. [Google Scholar] [CrossRef]
- Wilde, C.G.; Seilhamer, J.J.; McGrogan, M.; Ashton, N.; Snable, J.L.; Lane, J.C.; Leong, S.R.; Thornton, M.B.; Miller, K.L.; Scott, R.W.; et al. Bactericidal/permeability-increasing protein and lipopolysaccharide (LPS)-binding protein. LPS binding properties and effects on LPS-mediated cell activation. J. Biol. Chem. 1994, 269, 17411–17416. [Google Scholar]
- Fan, L.; Fu, G.; Ding, Y.Y.; Lv, P.; Li, H. Bactericidal/permeability increasing protein gene polymorphism and inflammatory bowel diseases: Meta-analysis of five case–control studies. Int. J. Colorectal. Dis. 2017, 32, 433–435. [Google Scholar] [CrossRef]
- Esteve, E.; Castro, A.; Moreno, J.M.; Vendrell, J.; Ricart, W.; Fernández-Real, J.M. Circulating bactericidal/permeability-increasing protein (BPI) is associated with serum lipids and endothelial function. Thromb. Haemost. 2010, 103, 780–787. [Google Scholar] [CrossRef] [PubMed]
- Der Schaft, D.W.; Wagstaff, J.; Mayo, K.H.; Griffioen, A.W. The antiangiogenic properties of bactericidal/permeability-increasing protein (BPI). Ann. Med. 2002, 34, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, M.; Rook, S.L.; Sassa, Y.; Ma, R.C.; Geraldes, P.; Goddard, L.; Clermont, A.; Gao, B.; Salti, H.; Gundel, R.; et al. Bactericidal/permeability-increasing protein’s signaling pathways and its retinal trophic and anti-angiogenic effects. FASEB J. 2006, 20, 2058–2067. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Wu, Y.F.; Tang, L.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.J.; Guo, C.; Li, X.Y.; Liao, Q.J.; et al. BPIFB1 (LPLUNC1) inhibits migration and invasion of nasopharyngeal carcinoma by interacting with VTN and VIM. Br. J. Cancer 2018, 118, 233–247. [Google Scholar] [CrossRef] [PubMed]
- De Smet, E.G.; Seys, L.J.; Verhamme, F.M.; Vanaudenaerde, B.M.; Brusselle, G.G.; Bingle, C.D.; Bracke, K.R. Association of innate defense proteins bpifa1 and bpifb1 with disease severity in copd. Int. J. Chronic Obstruct. Pulm. Dis. 2017, 13, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Little, M.S.; Redinbo, M.R. Crystal structure of the mouse innate immunity factor bacterial permeability-increasing family member a1. Acta Crystallogr. F Struct. Biol. Commun. 2018, 74, 268–276. [Google Scholar] [CrossRef]
- Leclair, E.E. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem. Soc. Trans. 2003, 31, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Seo, H.; Moxley, R.A.; Zhang, W. Mapping the neutralizing epitopes of F18 fimbrial adhesin subunit FedF of enterotoxigenic Escherichia coli (ETEC). Vet. Microbiol. 2019, 230, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Basha, S.; Surendran, N.; Pichichero, M. Immune responses in neonates. Expert Rev. Clin. Immunol. 2014, 10, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Fairbrother, J.M.; Nadeau, É.; Gyles, C.L. Escherichia coli in postweaning diarrhea in pigs: An update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 2005, 6, 17–39. [Google Scholar] [CrossRef]
- Zhu, J.; Zi, C.; Wu, Z.C.; Liu, L.; Zheng, X.R.; Su, X.M.; Zhu, G.Q.; Huang, X.G.; Wu, S.L.; Bao, W.B. Age-dependent expression of the BPI gene in Sutai piglets. Genet. Mol. Res. 2013, 12, 2120–2126. [Google Scholar] [CrossRef]
- Sun, L.; Wang, J.; Yin, X.M.; Sun, S.Y.; Zi, C.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Identification of a 5-Methylcytosine Site that may Regulate C/EBPβ Binding and Determine Tissue-Specific Expression of the BPI Gene in Piglets. Sci. Rep. 2016, 6, 28506. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Zhao, Q.H.; Zi, C.; Wu, Z.C.; Su, X.M.; Huo, Y.J.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Genetic variation in exon 10 of the BPI gene is associated with Escherichia coli F18 susceptibility in Sutai piglets. Gene 2013, 523, 70–75. [Google Scholar] [CrossRef]
- Liu, Y.; Gan, L.N.; Qin, W.Y.; Sun, S.Y.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Differential expression of Toll-like receptor 4 signaling pathway genes in Escherichia coli F18-resistant and-sensitive Meishan piglets. Pol. J. Vet. Sci. 2016, 19, 303–308. [Google Scholar] [CrossRef]
- Dong, W.H.; Dai, C.H.; Sun, L.; Wang, J.; Sun, S.Y.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Expression of key glycosphingolipid biosynthesis-globo series pathway genes in Escherichia coli F18-resistant and Escherichia coli F18-sensitive piglets. Anim. Genet. 2016, 47, 428–435. [Google Scholar] [CrossRef]
- Dai, C.H.; Gan, L.N.; Qin, W.U.; Zi, C.; Zhu, G.Q.; Wu, S.L.; Bao, W.B. Use of Fluorescence Quantitative Polymerase Chain Reaction (PCR) for the Detection of Escherichia coli Adhesion to Pig Intestinal Epithelial Cells. Pol. J. Vet. Sci. 2016, 19, 619–625. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Preston, A.; Mandrell, R.E.; Gibson, B.W.; Apicella, M.A. The lipooligosaccharides of pathogenic gram-negative bacteria. Crit. Rev. Microbiol. 1996, 22, 139–180. [Google Scholar] [CrossRef] [PubMed]
- Benin, A.M.; Ducher-Suchaux, M.F. Relationship between virulence and adherence of various enterotoxigenic Escherichia coli: Strains to isolated intestinal epithelial cells from Chinese Meishan and European large white pigs. Am. J. Vet. Res. 1991, 52, 45–49. [Google Scholar]
- Boldin, B. Persistence and Spread of Gastro-Intestinal Infections: The case of enterotoxigenic Escherichia coli in piglets. Bull. Math Biol. 2008, 70, 2077–2101. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Hilliard, G.M.; Hamilton, D.J.; Luo, J.; Ostmann, M.M.; Fleckenstein, J.M. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 2009, 457, 594–598. [Google Scholar] [CrossRef]
- Jiang, S.; Li, F.Q.; Li, X.L.; Wang, L.I.; Zhang, L.; Lu, C.; Zheng, L.; Yan, M.H. Transcriptome analysis of PK-15 cells in innate immune response to porcine deltacoronavirus infection. PLoS ONE 2019, 14, e0223177. [Google Scholar] [CrossRef]
- Frommel, U.; Bohm, A.; Nitschke, J.; Weinreich, J.; Groß, J.; Rödiger, S.; Wex, T.; Ansorge, H.; Zinke, O.; Schröder, C.; et al. Adhesion patterns of commensal and pathogenic Escherichia coli from humans and wild animals on human and porcine epithelial cell lines. Gut. Pathog. 2013, 5, 31. [Google Scholar] [CrossRef][Green Version]
- Meijerink, E.; Neuenschwander, S.; Fries, R.; Dinter, A.; Bertschinger, H.U.; Stranzinger, G.; Vögeli, P. A DNA polymorphism influencing α(1,2) fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics 2000, 52, 129–136. [Google Scholar] [CrossRef]
- Wu, Z.; Feng, H.; Cao, Y.; Huang, Y.; Dai, C.; Wu, S.; Bao, W. New Insight into the Molecular Mechanism of the FUT2 Regulating Escherichia coli F18 Resistance in Weaned Piglets. Int. J. Mol. Sci. 2018, 19, 3301. [Google Scholar] [CrossRef]
- De Nardo, D. Toll-like receptors: Activation, signalling and transcriptional modulation. Cytokine 2015, 74, 181–189. [Google Scholar] [CrossRef]
- Barton, G.M.; Medzhitov, R. Toll-like receptor signaling pathways. Science 2003, 300, 1524–1525. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, A.; Schnare, M.; Chakravortty, D. Of men not mice: Bactericidal/permeability-increasing protein expressed in human macrophages acts as a phagocytic receptor and modulates entry and replication of Gram-negative bacteria. Front. Immunol. 2016, 7, 455. [Google Scholar] [CrossRef] [PubMed]
- Canny, G.; Cario, E.; Lennartsson, A.; Gullberg, U.; Brennan, C.; Levy, O.; Colgan, S.P. Functional and biochemical characterization of epithelial bactericidal/permeability-increasing protein. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G557–G567. [Google Scholar] [CrossRef] [PubMed]
- Schultz, H.; Weiss, J.; Carroll, S.F.; Gross, W.L. The endotoxin-binding bactericidal/permeability-increasing protein (BPI): A target antigen of autoantibodies. J. Leukoc. Biol. 2001, 69, 505–512. [Google Scholar] [PubMed]
- Xie, S.; Chen, X.; Qiao, S.L.; Li, R.; Sun, Y.G.; Xia, S.F.; Wang, L.J.; Luo, X.G.; Deng, R.G.; Zhou, E.M.; et al. Identification of the RNA pseudoknot within the 3′ end of the porcine reproductive and respiratory syndrome virus genome as a pathogen-associated molecular pattern to activate antiviral signaling via RIG-I and toll-like receptor 3. J. Virol. 2018, 92, e00097-18. [Google Scholar] [CrossRef]
- Sato, M.; Suemori, H.; Hata, N.; Asagiri, M.; Ogasawara, K.; Nakao, K.; Nakaya, T.; Katsuki, M.; Noguchi, S.; Tanaka, N.; et al. Distinct and Essential Roles of Transcription Factors IRF-3 and IRF-7 in Response to Viruses for IFN-α/β Gene Induction. Immunity 2000, 13, 539–548. [Google Scholar] [CrossRef]
- Schoggins, J.W.; Rice, C.M. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2011, 1, 519–525. [Google Scholar] [CrossRef]
- Lenschow, D.J.; Lai, C.; Frias-Staheli, N.; Giannakopoulos, N.V.; Lutz, A.; Wolff, T.; Osiak, A.; Levine, B.; Schmidt, R.E.; García-Sastre, A.; et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 2007, 104, 1371–1376. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).