Copy Number Variations of Four Y-Linked Genes in Swamp Buffaloes
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
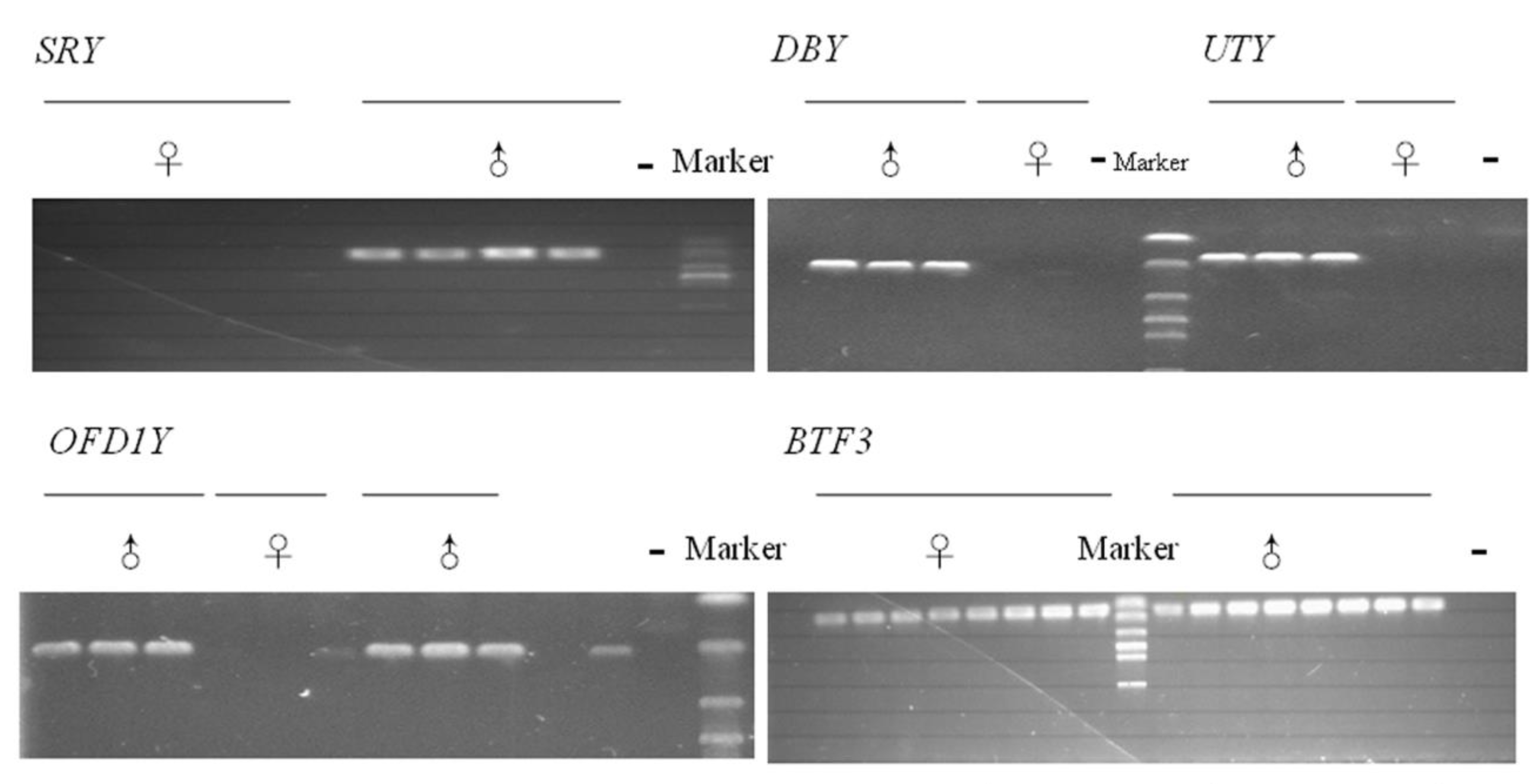
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mills, R.E.; Walter, K.; Stewart, C.; Handsaker, R.E.; Chen, K.; Alkan, C.; Abyzov, A.; Yoon, S.C.; Ye, K.; Cheetham, R.K. Mapping copy number variation by population-scale genome sequencing. Nature 2011, 470, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Beckmann, J.S.; Estivill, X.; Antonarakis, S.E. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W. Global variation in copy number in the human genome. Nature 2006, 444, 444. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Liu, G.E.; Bickhart, D.M.; Matukumalli, L.K.; Li, C.; Song, J.; Gasbarre, L.C.; Van Tassell, C.P.; Sonstegard, T.S. Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct. Integr. Genom. 2012, 12, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Prinsen, R.T.M.M.; Rossoni, A.; Gredler, B.; Bieber, A.; Bagnato, A.; Strillacci, M.G. A genome wide association study between CNVs and quantitative traits in Brown Swiss cattle. Livest. Sci. 2017, 202, 7–12. [Google Scholar] [CrossRef]
- Zhou, Y.; Connor, E.E.; Wiggans, G.R.; Lu, Y.; Tempelman, R.J.; Schroeder, S.G.; Chen, H.; Liu, G.E. Genome-wide copy number variant analysis reveals variants associated with 10 diverse production traits in Holstein cattle. BMC Genom. 2018, 19, 314. [Google Scholar] [CrossRef]
- Zhou, Y.; Utsunomiya, Y.T.; Xu, L.; Hay, E.H.A.; Bickhart, D.M.; Alexandre, P.A.; Rosen, B.D.; Schroeder, S.G.; Carvalheiro, R.; de Rezende Neves, H.H.; et al. Genome-wide CNV analysis reveals variants associated with growth traits in Bos indicus. BMC Genom. 2016, 17, 419. [Google Scholar] [CrossRef]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Gómez González, E.; Dall’Olio, S.; Davoli, R.; Russo, V.; Portolano, B. Copy Number Variation and Missense Mutations of the Agouti Signaling Protein (ASIP) Gene in Goat Breeds with Different Coat Colors. Cytogenet. Genome Res. 2009, 126, 333–347. [Google Scholar] [CrossRef]
- Rubin, C.-J.; Megens, H.-J.; Barrio, A.M.; Maqbool, K.; Sayyab, S.; Schwochow, D.; Wang, C.; Carlborg, Ö.; Jern, P.; Jørgensen, C.B.; et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 2012, 109, 19529–19536. [Google Scholar] [CrossRef]
- Sundström, E.; Imsland, F.; Mikko, S.; Wade, C.; Sigurdsson, S.; Pielberg, G.R.; Golovko, A.; Curik, I.; Seltenhammer, M.H.; Sölkner, J.; et al. Copy number expansion of the STX17 duplication in melanoma tissue from Grey horses. BMC Genom. 2012, 13, 365. [Google Scholar] [CrossRef]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Quintanamurci, L.; Fellous, M. The Human Y Chromosome: The Biological Role of a “Functional Wasteland”. J. Biomed. Biotechnol. 2001, 1, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.F.; Skaletsky, H.; Pyntikova, T.; Graves, T.A.; Daalen, S.K.M.V.; Minx, P.J.; Fulton, R.S.; Mcgrath, S.D.; Locke, D.P.; Friedman, C. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 2010, 463, 536. [Google Scholar] [CrossRef]
- Hughes, J.F.; Rozen, S. Genomics and Genetics of Human and Primate Y Chromosomes. Annu. Rev. Genom. Hum. Genet. 2012, 13, 83–108. [Google Scholar] [CrossRef]
- Chang, T.C.; Yang, Y.; Retzel, E.F.; Liu, W.S. Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development. Proc. Natl. Acad. Sci. USA 2013, 110, 12373–12378. [Google Scholar] [CrossRef]
- Jobling, M.A. Copy number variation on the human Y chromosome. Cytogenet. Genome Res. 2008, 123, 253–262. [Google Scholar] [CrossRef]
- Michelizzi, V.N.; Dodson, M.V.; Pan, Z.; Amaral, M.E.J.; Michal, J.J.; Mclean, D.J.; Womack, J.E.; Jiang, Z. Water Buffalo Genome Science Comes of Age. Int. J. Biol. Sci. 2010, 6, 333–349. [Google Scholar] [CrossRef]
- Fischer, H.; Ulbrich, F. Chromosomes of the Murrah buffalo and its crossbreds with the Asiatic swamp buffalo (Bubalus bubalis). Z. Tierz. Züchtungsbiol. 1967, 84, 110–114. [Google Scholar] [CrossRef]
- Iannuzzi, L. Standard karyotype of the river buffalo (Bubalus bubalis L., 2n = 50). Report of the committee for the standardization of banded karyotypes of the river buffalo. Cytogenet. Cell Genet. 1994, 67. [Google Scholar] [CrossRef]
- Li, W.; Bickhart, D.M.; Ramunno, L.; Iamartino, D.; Williams, J.L.; Liu, G.E. Genomic structural differences between cattle and River Buffalo identified through comparative genomic and transcriptomic analysis. Data Brief 2018, 19, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Kang, X.; Catacchio, C.R.; Liu, M.; Fang, L.; Schroeder, S.G.; Li, W.; Rosen, B.D.; Iamartino, D.; Iannuzzi, L.; et al. Computational detection and experimental validation of segmental duplications and associated copy number variations in water buffalo (Bubalus bubalis). Funct. Integr. Genom. 2019, 19, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Han, H.; Zhang, T.; Sun, T.; Xi, Y.; Chen, N.; Huang, Y.; Dang, R.; Lan, X.; Chen, H. HSFY and ZNF280BY show copy number variations within 17 water buffalo populations. Anim. Genet. 2016, 48, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Fortes, M.R.S.; Reverter, A.; Hawken, R.J.; Bolormaa, S.; Lehnert, S.A. Candidate Genes Associated with Testicular Development, Sperm Quality, and Hormone Levels of Inhibin, Luteinizing Hormone, and Insulin-Like Growth Factor 1 in Brahman Bulls1. Biol. Reprod. 2012, 87. [Google Scholar] [CrossRef]
- Paria, N.; Raudsepp, T.; Wilkerson, A.J.P.; O’Brien, P.C.M.; Fergusonsmith, M.A.; Love, C.C.; Arnold, C.; Rakestraw, P.; Murphy, W.J.; Chowdhary, B.P. A Gene Catalogue of the Euchromatic Male-Specific Region of the Horse Y Chromosome: Comparison with Human and Other Mammals. PLoS ONE 2011, 6, e21374. [Google Scholar] [CrossRef]
- Turner, M.E.; Martin, C.; Martins, A.S.; Dunmire, J.; Farkas, J.; Ely, D.L.; Milsted, A. Genomic and expression analysis of multiple Sry loci from a single Rattus norvegicus Y chromosome. BMC Genet. 2007, 8, 11. [Google Scholar] [CrossRef]
- Wilkerson, A.J.P.; Raudsepp, T.; Graves, T.; Albracht, D.; Warren, W.; Chowdhary, B.P.; Skow, L.C.; Murphy, W.J. Gene discovery and comparative analysis of X-degenerate genes from the domestic cat Y chromosome. Genomics 2008, 92, 329–338. [Google Scholar] [CrossRef]
- Geraldes, A.; Ferrand, N. A 7-bp insertion in the 3‘ untranslated region suggests the duplication and concerted evolution of the rabbit SRY gene. Genet. Sel. Evol. 2006, 38, 1–8. [Google Scholar] [CrossRef]
- Liu, Y.; Qin, X.; Song, X.-Z.H.; Jiang, H.; Shen, Y.; Durbin, K.J.; Lien, S.; Kent, M.P.; Sodeland, M.; Ren, Y.; et al. Bos taurus genome assembly. BMC Genom. 2009, 10, 180. [Google Scholar] [CrossRef]
- Liu, W.S.; Wang, A.; Yang, Y.; Chang, T.C.; Landrito, E.; Yasue, H. Molecular characterization of the DDX3Y gene and its homologs in cattle. Cytogenet. Genome Res. 2009, 126, 318–328. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, W.X.; Zuo, F.; Zhang, G.W. Comparison of mRNA expression from Y-chromosome X-degenerate region genes in taurine cattle, yaks and interspecific hybrid bulls. Anim. Genet. 2019, 50, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Skinner, B.M.; Sargent, C.A.; Churcher, C.; Hunt, T.; Herrero, J.; Loveland, J.E.; Dunn, M.; Louzada, S.; Fu, B.; Chow, W. The pig X and Y Chromosomes: Structure, sequence, and evolution. Genome Res. 2016, 26, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.-C.; Klabnik, J.L.; Liu, W.-S. Regional selection acting on the OFD1 gene family. PLoS ONE 2011, 6, e26195. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.C.; Romijn, E.P.; Zamora, I.; Yates, J.R.; Marshall, W.F. Proteomic Analysis of Isolated Chlamydomonas Centrioles Reveals Orthologs of Ciliary-Disease Genes. Curr. Biol. 2005, 15, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Xu, S. Y-SNPs Scanning and Multi-Copy Genes Identifiction in Water Buffalo; Northwest A&F University: Yangling, China, 2014. (In Chinese) [Google Scholar]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]
- Liu, G.E.; Hou, Y.; Zhu, B.; Cardone, M.F.; Jiang, L.; Cellamare, A.; Mitra, A.; Alexander, L.J.; Coutinho, L.L.; Dell’Aquila, M.E. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010, 20, 693–703. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Hou, Y.; Schroeder, S.G.; Alkan, C.; Cardone, M.F.; Matukumalli, L.K.; Song, J.; Schnabel, R.D.; Ventura, M.; Taylor, J.F. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012, 22, 778–790. [Google Scholar] [CrossRef]
- Yue, X.P.; Chang, T.C.; Dejarnette, J.M.; Marshall, C.E.; Lei, C.Z.; Liu, W.S. Copy number variation of PRAMEY across breeds and its association with male fertility in Holstein sires. J. Dairy Sci. 2013, 96, 8024–8034. [Google Scholar] [CrossRef]
- Yue, X.P.; Dechow, C.; Chang, T.C.; Dejarnette, J.M.; Marshall, C.E.; Lei, C.Z.; Liu, W.S. Copy number variations of the extensively amplified Y-linked genes, HSFY and ZNF280BY, in cattle and their association with male reproductive traits in Holstein bulls. BMC Genom. 2014, 15, 113. [Google Scholar] [CrossRef]
| Median Copy Number | ||||||
|---|---|---|---|---|---|---|
| Population | Abbreviation | Sample Size | SRY | DBY | UTY | OFD1Y |
| Dehong | DH | 20 | 1 | 2 (1–4) | 1 | 1 |
| Diandongnan | DDN | 7 | 1 | 2 (1–2) | 1 | 1 (1–2) |
| Guizhou | GZ | 4 | 1 | 2 (1–2) | 1 | 1 |
| Guangxi | GX | 11 | 1 | 1 (1–2) | 1 | 1 |
| Fuling | FL | 7 | 1 | 6 (2–8) | 1 | 1 |
| Xinyang | XY | 33 | 1 | 2 (1–3) | 1 | 1 (1–4) |
| Jianghan | JH | 14 | 1 | 3 (1–4) | 1 | 1 (1–2) |
| Guangyuan | GY | 27 | 1 | 2 (1–3) | 1 | 1 |
| Yibin | YB | 28 | 1 | 1 (1–2) | 1 | 1 (1–2) |
| Haizi | HZ | 4 | 1 | 2 (1–3) | 1 | 1 |
| Xiajiang | XJ | 25 | 1 | 2 (1–4) | 1 | 1 |
| Poyanghu | PYH | 24 | 1 | 2 (2–3) | 1 | 1 (1–2) |
| Jianghuai | JHU | 14 | 1 | 2 (1–3) | 1 | 1 (1–2) |
| Vietnamese | VN | 26 | 2 (1–4) | 2 (1–4) | 1 | 2 (1–4) |
| Laotian | LA | 12 | 2 (1–4) | 2 (1–4) | 1 | 1 (1–2) |
| Total | 254 | 1 | 2 | 1 | 1 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, T.; Hanif, Q.; Chen, H.; Lei, C.; Dang, R. Copy Number Variations of Four Y-Linked Genes in Swamp Buffaloes. Animals 2020, 10, 31. https://doi.org/10.3390/ani10010031
Sun T, Hanif Q, Chen H, Lei C, Dang R. Copy Number Variations of Four Y-Linked Genes in Swamp Buffaloes. Animals. 2020; 10(1):31. https://doi.org/10.3390/ani10010031
Chicago/Turabian StyleSun, Ting, Quratulain Hanif, Hong Chen, Chuzhao Lei, and Ruihua Dang. 2020. "Copy Number Variations of Four Y-Linked Genes in Swamp Buffaloes" Animals 10, no. 1: 31. https://doi.org/10.3390/ani10010031
APA StyleSun, T., Hanif, Q., Chen, H., Lei, C., & Dang, R. (2020). Copy Number Variations of Four Y-Linked Genes in Swamp Buffaloes. Animals, 10(1), 31. https://doi.org/10.3390/ani10010031





