Respiratory and Neurological Disease across Different Ethnic Groups Is Influenced by the Microbiome
Abstract
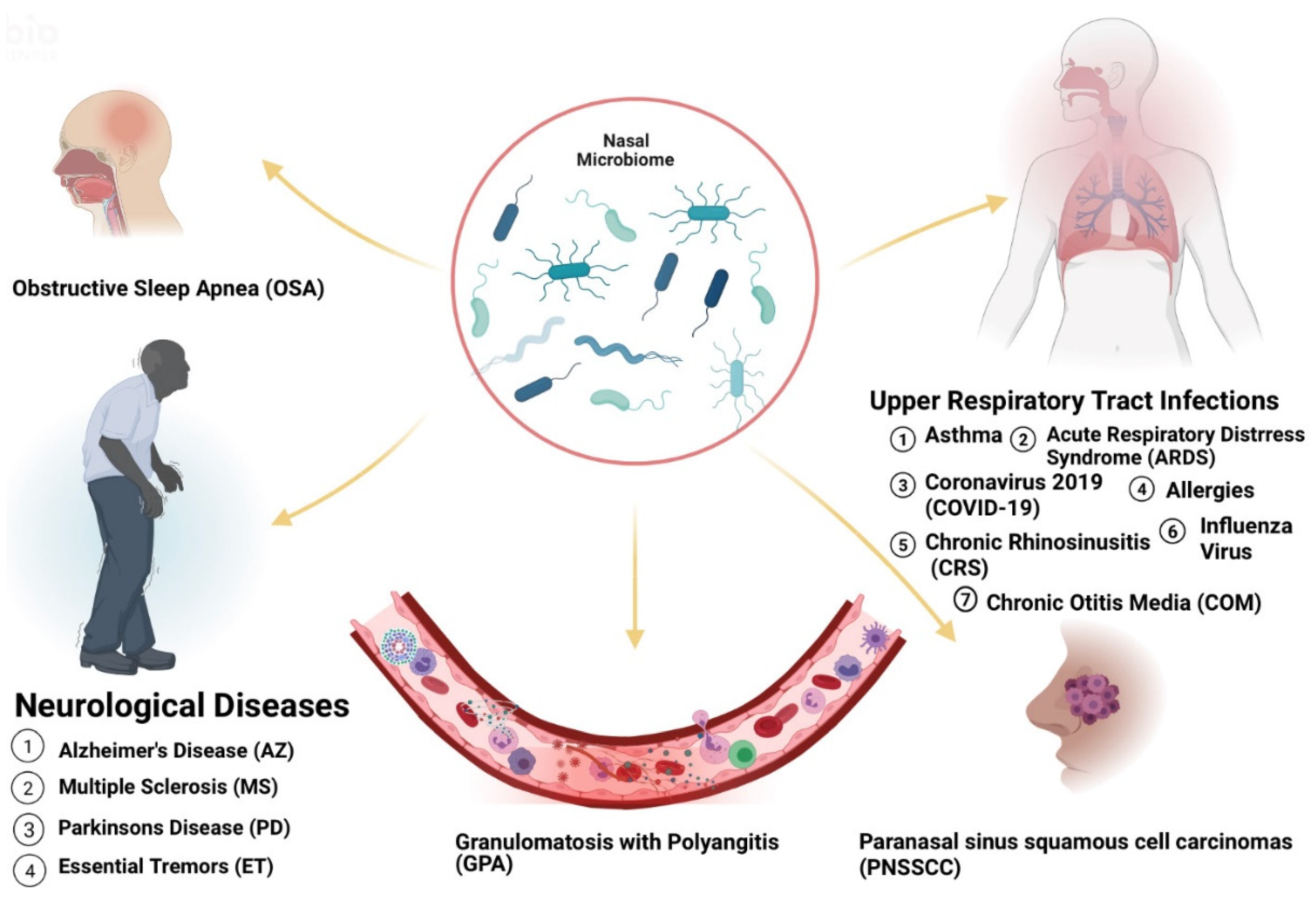
:1. Introduction
2. Microbiome Populations in the Nose and Impacts of Dysbiosis in Disease States
2.1. Asthma
2.2. Acute Respiratory Illnesses
2.3. Rhinitis and Chronic Rhinosinusitis
2.4. Otitis Media
2.5. Granulomatosis with Polyangiitis
2.6. Atopic Dermatitis and Psoriasis
2.7. Neurological Illness: Alzheimer’s, Parkinson’s and Multiple Sclerosis
3. Antibiotic Properties of Nasal Microbiome and Impacts on Local Immune Response
3.1. Antibiotic Microorganisms
3.2. Impact of Nasal Microbiome on the Immune System
4. Diet, Behavior and Environmental Changes and Impacts on Respiratory Disease
4.1. Diet
4.2. Tobacco/Smoking
4.3. Environment
5. More on Health Disparities and Limitations
6. Potential Pitfalls
7. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Findley, K.; Williams, D.R.; Grice, E.A.; Bonham, V.L. Health Disparities and the Microbiome. Trends Microbiol. 2016, 24, 847–850. [Google Scholar] [CrossRef] [Green Version]
- Braveman, P. What are health disparities and health equity? We need to be clear. Public Health Rep. 2014, 129 (Suppl. 2), 5–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, D.R.; Mohammed, S.A.; Leavell, J.; Collins, C. Race, socioeconomic status, and health: Complexities, ongoing challenges, and research opportunities. Ann. N. Y. Acad. Sci. 2010, 1186, 69–101. [Google Scholar] [CrossRef]
- Solovieff, N.; Hartley, S.W.; Baldwin, C.T.; Klings, E.S.; Gladwin, M.T.; Taylor, J.G.; Kato, G.J.; Farrer, L.A.; Steinberg, M.H.; Sebastiani, P. Ancestry of African Americans with sickle cell disease. Blood Cells Mol. Dis. 2011, 47, 41–45. [Google Scholar] [CrossRef] [Green Version]
- El-Hazmi, M.A.; Al-Hazmi, A.M.; Warsy, A.S. Sickle cell disease in Middle East Arab countries. Indian J. Med. Res.. 2011, 134, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Purohit, P.; Dehury, S.; Patel, S.; Patel, D.K. Prevalence of deletional alpha thalassemia and sickle gene in a tribal dominated malaria endemic area of eastern India. ISRN Hematol. 2014, 2014, 745245. [Google Scholar] [CrossRef]
- Lim, S.H.; Morris, A.; Li, K.; Fitch, A.C.; Fast, L.; Goldberg, L.; Quesenberry, M.; Sprinz, P.; Methé, B. Intestinal microbiome analysis revealed dysbiosis in sickle cell disease. Am. J. Hematol. 2018, 93, E91–E93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fettweis, J.M.; Brooks, J.P.; Serrano, M.G.; Sheth, N.U.; Girerd, P.H.; Edwards, D.J.; Strauss, J.F., III; Jefferson, K.K.; Buck, G.A. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology 2014, 160, 2272–2282. [Google Scholar] [CrossRef] [Green Version]
- Greenberg, J.D.; Spruill, T.M.; Shan, Y.; Reed, G.; Kremer, J.M.; Potter, J.; Yazici, Y.; Ogedegbe, G.; Harrold, L.R. Racial and ethnic disparities in disease activity in patients with rheumatoid arthritis. Am. J. Med. 2013, 126, 1089–1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espina, M.d.T.; Gabarrini, G.; Harmsen, H.J.M.; Westra, J.; jan van Winkelhoff, A.; van Dijl, J. Talk to your gut: The oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol. Rev. 2019, 43, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Gotschlich, E.C.; Colbert, R.A.; Gill, T. Methods in microbiome research: Past, present, and future. Best Pract. Res. Clin. Rheumatol. 2019, 33, 101498. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E.; et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 15, R66. [Google Scholar] [CrossRef]
- Toivonen, L.; Hasegawa, K.; Waris, M.; Ajami, N.J.; Petrosino, J.F.; Camargo, C.A., Jr.; Peltola, V. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 2019, 74, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Frank, D.N.; Ramakrishnan, V. Microbiome of the paranasal sinuses: Update and literature review. Am. J. Rhinol. Allergy 2016, 30, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Swidsinski, A.; Göktas, O.; Bessler, C.; Loening-Baucke, V.; Hale, L.P.; Andree, H.; Weizenegger, M.; Hölzl, M.; Scherer, H.; Lochs, H. Spatial organisation of microbiota in quiescent adenoiditis and tonsillitis. J. Clin. Pathol. 2007, 60, 253–260. [Google Scholar] [CrossRef]
- Walker, R.E.; Walker, C.G.; Camargo, C.A.; Bartley, J.; Flint, D.; Thompson, J.M.D.; Mitchell, E.A. Nasal microbial composition and chronic otitis media with effusion: A case-control study. PLoS ONE. 2019, 14, e0212473. [Google Scholar] [CrossRef] [PubMed]
- Lasisi, A.O.; Olaniyan, F.A.; Muibi, S.A.; Azeez, I.A.; Abdulwasiu, K.G.; Lasisi, T.J.; Imam, Z.O.; Yekinni, T.O.; Olayemi, O. Clinical and demographic risk factors associated with chronic suppurative otitis media. Int. J. Pediatr. Otorhinolaryngol. 2007, 71, 1549–1554. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, M.; Zhang, J.; Zeng, L.; Wang, Y.; Zheng, Q.Y. Risk factors for chronic and recurrent otitis media-a meta-analysis. PLoS ONE 2014, 9, e86397. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Jackson, D.; Bacharier, L.B.; Mauger, D.; Boushey, H.; Castro, M.; Durack, J.; Huang, Y.; Lemanske, R.F.; Storch, G.A.; et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat. Commun. 2019, 10, 5714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forno, E.; Celedón, J.C. Health disparities in asthma. Am. J. Respir. Crit Care Med. 2012, 15, 1033–1035. [Google Scholar] [CrossRef] [PubMed]
- Castro-Rodriguez, J.A.; Forno, E.; Rodriguez-Martinez, C.E.; Celedón, J.C. Risk and Protective Factors for Childhood Asthma: What Is the Evidence? J. Allergy Clin. Immunol. Pract. 2016, 4, 1111–1122. [Google Scholar] [CrossRef] [Green Version]
- Aligne, C.A.; Auinger, P.; Byrd, R.S.; Weitzman, M. Risk factors for pediatric asthma. Contributions of poverty, race, and urban residence. Am. J. Respir. Crit. Care Med. 2000, 162, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Wagner Mackenzie, B.; Waite, D.W.; Hoggard, M.; Taylor, M.W.; Biswas, K.; Douglas, R.G. Moving beyond descriptions of diversity: Clinical and research implications of bacterial imbalance in chronic rhinosinusitis. Rhinology 2017, 55, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Soto, P.F.; Padhye, L.; Moore, D.; Codispoti, C.; Tobin, M.; Batra, P.; Mahdavinia, M. Latino Ethnicity Is Associated with Variations in the Nasal Microbiome in Patients With CRS. J. Allergy Clin. Immunol. 2020, 145, AB64. [Google Scholar] [CrossRef]
- Min, J.Y.; Tan, B.K. Risk factors for chronic rhinosinusitis. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rueca, M.; Fontana, A.; Bartolini, B.; Piselli, P.; Mazzarelli, A.; Copetti, M.; Binda, E.; Perri, F.; Gruber, C.E.M.; Nicastri, E.; et al. Investigation of Nasal/Oropharyngeal Microbial Community of COVID-19 Patients by 16S rDNA Sequencing. Int. J. Environ. Res. Public Health 2021, 18, 2174. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Salazar, C.; Kimura, K.S.; Shilts, M.H.; Strickland, B.A.; Freeman, M.H.; Wessinger, B.C.; Gupta, V.; Brown, H.M.; Rajagopala, S.V.; Turner, J.H.; et al. SARS-CoV-2 infection and viral load are associated with the upper respiratory tract microbiome. J. Allergy Clin. Immunol. 2021, 147, 1226–1233. [Google Scholar] [CrossRef]
- Ko, J.Y.; Danielson, M.L.; Town, M.; Derado, G.; Greenlund, K.J.; Kirley, P.D.; Alden, N.B.; Yousey-Hindes, K.; Anderson, E.J.; Ryan, P.A.; et al. COVID-NET Surveillance Team. Risk Factors for Coronavirus Disease 2019 (COVID-19)-Associated Hospitalization: COVID-19-Associated Hospitalization Surveillance Network and Behavioral Risk Factor Surveillance System. Clin Infect Dis. 2021, 72, e695–e703. [Google Scholar] [CrossRef]
- Sze, S.; Pan, D.; Nevill, C.; Gray, L.; Martin, C.; Nazareth, J.; Minhas, J.; Divall, P.; Khunti, K.; Abrams, K.; et al. Ethnicity and clinical outcomes in COVID-19: A systematic review and meta-analysis. EclinicalMedicine 2021, 100630. [Google Scholar] [CrossRef]
- Rhee, R.L.; Sreih, A.G.; Najem, C.E.; Grayson, P.C.; Zhao, C.; Bittinger, K.; Collman, R.G.; Merkel, P.A. Characterisation of the nasal microbiota in granulomatosis with polyangiitis. Ann. Rheum Dis. 2018, 77, 1448–1453. [Google Scholar] [CrossRef]
- Fiona, A.; Pearce, M.J.; Lanyon, P.C.; Watts, R.A.; Hubbard, R.B. The incidence, prevalence and mortality of granulomatosis with polyangiitis in the UK Clinical Practice Research Datalink. Rheumatology 2017, 56, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Lindberg, H.; Colliander, C.; Nise, L.; Dahlqvist, J.; Knight, A. Are Farming and Animal Exposure Risk Factors for the Development of Granulomatosis With Polyangiitis? Environmental Risk Factors Revisited: A Case-Control Study. J. Rheumatol. 2021, 48, 894–897. [Google Scholar] [CrossRef]
- Onuora, S. New insights into risk factors for GPA. Nat. Rev. Rheumatol. 2018, 14, 248. [Google Scholar] [CrossRef] [PubMed]
- Vernacchio, L.; Lesko, S.M.; Vezina, R.M.; Corwin, M.J.; Hunt, C.E.; Hoffman, H.J.; Mitchell, A.A. Racial/ethnic disparities in the diagnosis of otitis media in infancy. Int. J. Pediatr. Otorhinolaryngol. 2004, 68, 795–804. [Google Scholar] [CrossRef] [PubMed]
- Tsang, T.K.; Lee, K.H.; Foxman, B.; Balmaseda, A.; Gresh, L.; Sanchez, N.; Ojeda, S.; Lopez, R.; Yang, Y.; Kuan, G.; et al. Association Between the Respiratory Microbiome and Susceptibility to Influenza Virus Infection. Clin. Infect. Dis. 2020, 71, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Ezzine, H.; Cherkaoui, I.; Rguig, A.; Oumzil, H.; Mrabet, M.; Bimouhen, A.; Falaki, F.E.; Regragui, Z.; Tarhda, Z.; Youbi, M.; et al. Influenza epidemiology and risk factors for severe acute respiratory infection in Morocco during the 2016/2017 and 2017/2018 seasons. Pan. Afr. Med. J. 2020, 36, 159. [Google Scholar] [CrossRef]
- Planet, P.J.; Parker, D.; Cohen, T.S.; Smith, H.; Leon, J.D.; Ryan, C.; Hammer, T.J.; Fierer, N.; Chen, E.I.; Prince, A.S. Lambda Interferon Restructures the Nasal Microbiome and Increases Susceptibility to Staphylococcus aureus Superinfection. mBio 2016, 7, e01939-15. [Google Scholar] [CrossRef] [Green Version]
- Ryb, G.E.; Cooper, C. Race/ethnicity and acute respiratory distress syndrome: A National Trauma Data Bank study. J. Natl. Med. Assoc. 2010, 102, 865–869. [Google Scholar] [CrossRef]
- Odeyemi, Y.; Moraes, A.G.D.; Gajic, O. What factors predispose patients to acute respiratory distress syndrome? Evid.-Based Pract. Crit. Care 2020, 103–108.e1. [Google Scholar] [CrossRef]
- Wu, B.G.; Sulaiman, I.; Wang, J.; Shen, N.; Clemente, J.C.; Li, Y.; Laumbach, R.J.; Lu, S.E.; Udasin, I.; Le-Hoang, O.; et al. Severe Obstructive Sleep Apnea Is Associated with Alterations in the Nasal Microbiome and an Increase in Inflammation. Am. J. Respir. Crit. Care Med. 2019, 199, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Gulotta, G.; Iannella, G.; Vicini, C.; Polimeni, A.; Greco, A.; de Vincentiis, M.; Visconti, I.C.; Meccariello, G.; Cammaroto, G.; De Vito, A.; et al. Risk Factors for Obstructive Sleep Apnea Syndrome in Children: State of the Art. Int. J. Environ. Res. Public Health 2019, 16, 3235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, J.S.; Spencer, J.I.; Yates, R.L.; Yee, S.A.; Jacobs, B.M.; DeLuca, G.C. Invited Review: From nose to gut—The role of the microbiome in neurological disease. Neuropathol. Appl. Neurobiol. 2019, 45, 195–215. [Google Scholar] [CrossRef] [Green Version]
- Berstad, K.; Berstad, J.E.R. Parkinson’s disease; the hibernating spore hypothesis. Med Hypotheses. 2017, 104, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.A.; Saint-Hilaire, M.H.; Cupples, L.A.; Thomas, C.A.; Burchard, A.E.; Feldman, R.G.; Myers, R.H. Environmental, medical, and family history risk factors for Parkinson’s disease: A New England-based case control study. Am. J. Med. Genet. 1999, 88, 742–749. [Google Scholar] [CrossRef]
- Armstrong, R. Risk factors for Alzheimer’s disease. Folia Neuropathol. 2019, 57, 87–105. [Google Scholar] [CrossRef] [Green Version]
- Louis, E.D.; Barnes, L.F.; Ford, B.; Pullman, S.L.; Yu, Q. Ethnic differences in essential tremor. Arch. Neurol. 2000, 57, 723–727. [Google Scholar] [CrossRef] [Green Version]
- Ong, Y.L.; Deng, X.; Tan, E.K. Etiologic links between environmental and lifestyle factors and Essential tremor. Ann. Clin. Transl. Neurol. 2019, 6, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Jiménez, F.J.; de Toledo-Heras, M.; Alonso-Navarro, H.; Ayuso-Peralta, L.; Arévalo-Serrano, J.; Ballesteros-Barranco, A.; Puertas, I.; Jabbour-Wadih, T.; Barcenilla, B. Environmental risk factors for essential tremor. Eur. Neurol. 2007, 58, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Totté, J.E.E.; Pardo, L.M.; Fieten, K.B.; Vos, M.C.; van den Broek, T.J.; Schuren, F.H.J.; Pasmans, S.G. Nasal and skin microbiomes are associated with disease severity in paediatric atopic dermatitis. Br. J. Dermatol. 2019, 181, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Wollina, U. Microbiome in atopic dermatitis. Clin. Cosmet Investig. Dermatol. 2017, 10, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.W.; Yan, D.; Singh, R.; Liu, J.; Lu, X.; Ucmak, D.; Lee, K.; Afifi, L.; Fadrosh, D.; Leech, J.; et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome 2018, 6, 154. [Google Scholar] [CrossRef]
- Alexis, A.F.; Blackcloud, P. Psoriasis in skin of color: Epidemiology, genetics, clinical presentation, and treatment nuances. J. Clin. Aesthet Dermatol. 2014, 7, 16–24. [Google Scholar]
- Kamiya, K.; Kishimoto, M.; Sugai, J.; Komine, M.; Ohtsuki, M. Risk Factors for the Development of Psoriasis. Int. J. Mol. Sci. 2019, 20, 4347. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Li, J.; Zhu, W.; Kuang, Y.; Liu, T.; Zhang, W.; Chen, X.; Peng, C. Skin and Gut Microbiome in Psoriasis: Gaining Insight Into the Pathophysiology of It and Finding Novel Therapeutic Strategies. Front. Microbiol. 2020, 11, 589726. [Google Scholar] [CrossRef]
- Mason, M.R.; Nagaraja, H.N.; Camerlengo, T.; Joshi, V.; Kumar, P.S. Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS ONE. 2013, 8, e77287. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Guo, L.; Gu, H.; Huo, Y.; Lin, H. Alterations in Oral-Nasal-Pharyngeal Microbiota and Salivary Proteins in Mouth-Breathing Children. Front. Microbiol. 2020, 11, 575550. [Google Scholar] [CrossRef]
- Lemon, K.P.; Klepac-Ceraj, V.; Schiffer, H.K.; Brodie, E.L.; Lynch, S.V.; Kolter, R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. mBio 2010, 1, e00129-10. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Li, Y.; Zhao, J. The gut microbiome of healthy long-living people. Aging 2019, 11, 289–290. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Authelet, K.J.; Hoptay, C.E.; Kwak, C.; Crandall, K.A.; Freishtat, R.J. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome 2018, 6, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teräsjärvi, J.T.; Toivonen, L.; Vuononvirta, J.; Mertsola, J.; Peltola, V.; He, Q. Polymorphism, Nasopharyngeal Bacterial Colonization, and the Development of Childhood Asthma: A Prospective Birth-Cohort Study in Finnish Children. Genes 2020, 11, 768. [Google Scholar] [CrossRef] [PubMed]
- Toivonen, L.; Karppinen, S.; Schuez-Havupalo, L.; Waris, M.; He, Q.; Hoffman, K.L.; Petrosino, J.F.; Dumas, O.; Camargo, C.A.; Hasegawa, K.; et al. Longitudinal Changes in Early Nasal Microbiota and the Risk of Childhood Asthma. Pediatrics 2020, 146. [Google Scholar] [CrossRef]
- Leong, A.B.; Ramsey, C.D.; Celedón, J.C. The challenge of asthma in minority populations. Clin. Rev. Allergy Immunol. 2012, 43, 156–183. [Google Scholar] [CrossRef]
- Espuela-Ortiz, A.; Lorenzo-Diaz, F.; Baez-Ortega, A.; Eng, C.; Hernandez-Pacheco, N.; Oh, S.S.; Lenoir, M.; Burchard, E.G.; Flores, C.; Pino-Yanes, M. Bacterial salivary microbiome associates with asthma among african american children and young adults. Pediatr. Pulmonol. 2019, 54, 1948–1956. [Google Scholar] [CrossRef]
- Bime, C.; Poongkunran, C.; Borgstrom, M.; Natt, B.; Desai, H.; Parthasarathy, S.; Garcia, J.G. Racial Differences in Mortality from Severe Acute Respiratory Failure in the United States, 2008–2012. Ann. Am. Thorac. Soc. 2016, 13, 2184–2189. [Google Scholar] [CrossRef] [PubMed]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ta, L.D.H.; Yap, G.C.; Tay, C.J.X.; Lim, A.S.M.; Huang, C.H.; Chu, C.W.; De Sessions, P.F.; Shek, L.P.; Goh, A.; Van Bever, H.P.; et al. Establishment of the nasal microbiota in the first 18 months of life: Correlation with early-onset rhinitis and wheezing. J. Allergy Clin. Immunol. 2018, 142, 86–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, V.R.; Hauser, L.J.; Feazel, L.M.; Ir, D.; Robertson, C.E.; Frank, D.N. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J. Allergy Clin. Immunol. 2015, 136, 334–342. [Google Scholar] [CrossRef] [PubMed]
- van Mierlo, M.M.F.; Pasmans, S.G.M.A.; Totté, J.E.E.; de Wit, J.; Herpers, B.L.; Vos, M.C.; Klaassen, C.H.W.; Pardo, L.M. Temporal Variation in Staphylococcus aureus Protein A Genotypes from Nose and Skin in Atopic Dermatitis Patients. Dermatology 2021, 237, 506–512. [Google Scholar] [CrossRef]
- Chiesa Fuxench, Z.C. Atopic Dermatitis: Disease Background and Risk Factors. Adv. Exp. Med. Biol. 2017, 1027, 11–19. [Google Scholar] [CrossRef]
- Brunner, P.M.; Guttman-Yassky, E. Racial differences in atopic dermatitis. Ann. Allergy Asthma Immunol. 2019, 122, 449–455. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.M.; Jin, H.Z. Skin Microbiome: An Actor in the Pathogenesis of Psoriasis. Chin. Med. J. 2018, 131, 95–98. [Google Scholar] [CrossRef]
- Krahel, J.A.; Baran, A.; Kamiński, T.W.; Flisiak, I. Proprotein Convertase Subtilisin/Kexin Type 9, Angiopoietin-Like Protein 8, Sortilin, and Cholesteryl Ester Transfer Protein-Friends of Foes for Psoriatic Patients at the Risk of Developing Cardiometabolic Syndrome? Int. J. Mol. Sci. 2020, 21, 3682. [Google Scholar] [CrossRef]
- Yin, G.; Li, J.F.; Sun, Y.F.; Ding, X.; Zeng, J.Q.; Zhang, T.; Peng, L.H.; Yang, Y.S.; Zhao, H. Fecal microbiota transplantation as a novel therapy for severe psoriasis. Zhonghua Nei Ke Za Zhi 2019, 58, 782–785. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Erny, D.; Hrabě de Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Debelius, J.W.; Thron, T.; Janssen, S.; Shastri, G.G.; Ilhan, Z.E.; Challis, C.; Schretter, C.E.; Rocha, S.; Gradinaru, V.; et al. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell 2016, 167, 1469–1480. [Google Scholar] [CrossRef] [Green Version]
- Ferini-Strambi, L.; Lombardi, G.E.; Marelli, S.; Galbiati, A. Neurological Deficits in Obstructive Sleep Apnea. Curr. Treat. Options Neurol. 2017, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Zhang, N.; Van Crombruggen, K.; Gevaert, E.; Bachert, C. Barrier function of the nasal mucosa in health and type-2 biased airway diseases. Allergy 2016, 71, 295–307. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; May, D.S.; Chevrette, M.G.; Temkin, M.I.; Wendt-Pienkowski, E.; Cagnazzo, J.; Carlson, C.M.; Gern, J.E.; Currie, C.R. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Appl. Environ. Microbiol. 2019, 85, e02406-18. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; Jo, A.; Jeon, Y.J.; An, S.; Lee, K.M.; Yoon, S.S.; Choi, J.Y. Nasal commensal Staphylococcus epidermidis enhances interferon-λ-dependent immunity against influenza virus. Microbiome 2019, 7, 80. [Google Scholar] [CrossRef] [Green Version]
- Salzano, F.A.; Marino, L.; Salzano, G.; Botta, R.M.; Cascone, G.; D’Agostino Fiorenza, U.; Selleri, C.; Casolaro, V. Microbiota Composition and the Integration of Exogenous and Endogenous Signals in Reactive Nasal Inflammation. J. Immunol. Res. 2018, 2018, 2724951. [Google Scholar] [CrossRef] [PubMed]
- Zrelli, S.; Amairia, S.; Zrelli, M. Respiratory syndrome coronavirus-2 response: Microbiota as lactobacilli could make the difference. J. Med. Virol. 2021, 93, 3288–3293. [Google Scholar] [CrossRef]
- Blander, J.M.; Longman, R.S.; Iliev, I.D.; Sonnenberg, G.F.; Artis, D. Regulation of inflammation by microbiota interactions with the host. Nat. Immunol. 2017, 18, 851–860. [Google Scholar] [CrossRef]
- Calzada, D.; Baos, S.; Cremades-Jimeno, L.; Cárdaba, B. Immunological Mechanisms in Allergic Diseases and Allergen Tolerance: The Role of Treg Cells. J. Immunol. Res. 2018, 2018, 6012053. [Google Scholar] [CrossRef] [PubMed]
- Quinn, G.A.; Cole, A.M. Suppression of Innate Immunity by a Nasal Carriage Strain of Staphylococcus Aureus Increases Its Colonization on Nasal Epithelium. Immunology 2007, 122, 80–89. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.; Kumamoto, Y.; Peaper, D.; Ho, J.; Murray, T.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Nat. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Wisnivesky, J.P. Racial and ethnic differences in the use of environmental control practices among children with asthma. J. Asthma. 2010, 47, 507–512. [Google Scholar] [CrossRef]
- Noverr, M.C.; Huffnagle, G.B. The ‘microflora hypothesis’ of allergic diseases. Clin. Exp. Allergy 2005, 35, 1511–1520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, L.; Pang, W.; Liu, W.; Li, J.; Wang, H.; Shi, G. Dietary Fiber Intake Regulates Intestinal Microflora and Inhibits Ovalbumin-Induced Allergic Airway Inflammation in a Mouse Model. PLoS ONE 2016, 11, e0147778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, P.; Whelan, F.J.; Schenck, L.P.; McGrath, J.J.C.; Vanderstocken, G.; Bowdish, D.M.; Surette, M.G.; Stämpfli, M.R. Streptococcus pneumoniae Colonization Is Required To Alter the Nasal Microbiota in Cigarette Smoke-Exposed Mice. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [Green Version]
- Bugova, G.; Janickova, M.; Uhliarova, B.; Babela, R.; Jesenak, M. The effect of passive smoking on bacterial colonisation of the upper airways and selected laboratory parameters in children. Acta Otorhinolaryngol. Ital. 2018, 38, 431–438. [Google Scholar] [CrossRef]
- Hickman, E.; Zorn, B.; Rebuli, M.; Robinette, C.; Wolfgang, M.; Jaspers, I. The Effects of E-cigarette and Cigarette Use on the Nasal Microbiome. 2021. Available online: https://www.atsjournals.org/doi/pdf/10.1164/ajrccm-conference.2020.201.1_MeetingAbstracts.A6033 (accessed on 13 August 2021).
- Wagner Mackenzie, B.; Chang, K.; Zoing, M.; Jain, R.; Hoggard, M.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci Rep. 2019, 9, 17416. [Google Scholar] [CrossRef] [Green Version]
- Voĭtovich, A.V. Microbiocenosis of the human nasal mucous membrane in the conditions of industrial city. Georgian Med. News 2013, 225, 49–53. [Google Scholar]
- Shukla, S.K.; Ye, Z.; Sandberg, S.; Reyes, I.; Fritsche, T.R.; Keifer, M. The nasal microbiota of dairy farmers is more complex than oral microbiota, reflects occupational exposure, and provides competition for staphylococci. PLoS ONE 2017, 12, e0183898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roghmann, M.C.; Lydecker, A.D.; Hittle, L.; DeBoy, R.T.; Nowak, R.G.; Johnson, J.K.; Mongodin, E.F. Comparison of the Microbiota of Older Adults Living in Nursing Homes and the Community. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, N.; Mahmoud, N.F.; Solyman, S.; Hanora, A. Human Nasal Microbiome as Characterized by Metagenomics Differs Markedly Between Rural and Industrial Communities in Egypt. OMICS 2019, 23, 573–582. [Google Scholar] [CrossRef]
- Chen, C.H.; Liou, M.L.; Lee, C.Y.; Chang, M.C.; Kuo, H.Y.; Chang, T.H. Diversity of nasal microbiota and its interaction with surface microbiota among residents in healthcare institutes. Sci. Rep. 2019, 9, 6175. [Google Scholar] [CrossRef] [PubMed]
- Aarab, R.; Vijverberg, S.J.H.; Prins, M.; Snijder, M.B.; van Ree, R.; Fokkens, W.J.; Zwinderman, A.H.; Bel, E.H.; van der Zee, A.M. Prevalence of and factors associated with adult-onset asthma in different ethnic groups: The HELIUS study. Respir Med. 2019, 150, 113–119. [Google Scholar] [CrossRef]
- Ruokolainen, L.; Paalanen, L.; Karkman, A.; Laatikainen, T.; von Hertzen, L.; Vlasoff, T.; Markelova, O.; Masyuk, V.; Auvinen, P.; Paulin, L.; et al. Significant disparities in allergy prevalence and microbiota between the young people in Finnish and Russian Karelia. Clin. Exp. Allergy. 2017, 47, 665–674. [Google Scholar] [CrossRef]
- Smith, S.P.; Russell, J.L.; Chen, N.W.; Kuo, Y.F.; Resto, V.A. Sinonasal Carcinoma: Racial and Ethnic Disparities in Survival—A Review of 4714 Patients. Otolaryngol. Head Neck Surg. 2015, 153, 551–560. [Google Scholar] [CrossRef]
- Ansa, B.; Goodman, M.; Ward, K.; Kono, S.A.; Owonikoko, T.K.; Higgins, K.; Beitler, J.J.; Grist, W.; Wadsworth, T.; El-Deiry, M.; et al. Paranasal sinus squamous cell carcinoma incidence and survival based on Surveillance, Epidemiology, and End Results data, 1973 to 2009. Cancer 2013, 119, 2602–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wegienka, G.; Joseph, C.L.; Havstad, S.; Zoratti, E.; Ownby, D.; Johnson, C.C. Sensitization and allergic histories differ between black and white pregnant women. J. Allergy Clin. Immunol. 2012, 130, 657–662.e2. [Google Scholar] [CrossRef] [Green Version]
- Pecha, P.P.; Hamberis, A.; Patel, T.A.; Melvin, C.L.; Ford, M.E.; Andrews, A.L.; White , D.R.; Schlosser, R.J. Racial Disparities in Pediatric Endoscopic Sinus Surgery. Laryngoscope 2021, 131, E1369–E1374. [Google Scholar] [CrossRef] [PubMed]
- Schwager, M.; Song, Y.; Saiganesh, A.; Guo, J.; Laing, I.; Le Souëf, P.; Zhang, G. Increased nasal Streptococcus pneumoniae presence in Western environment associated with atopic eczema in Chinese immigrants. World Allergy Organ. J. 2020, 13, 100165. [Google Scholar] [CrossRef]
- Sharma, M.; Li, Y.; Stoll, M.L.; Tollefsbol, T.O. The Epigenetic Connection Between the Gut Microbiome in Obesity and Diabetes. Front. Genet. 2019, 10, 1329. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.; Forno, E.; Herrera-Luis, E.; Pino-Yanes, M.; Qi, C.; Rios, R.; Han, Y.Y.; Kim, S.; Oh, S.; Acosta-Pérez, E.; et al. A genome-wide association study of severe asthma exacerbations in Latino children and adolescents. Eur. Respir J. 2021, 57. [Google Scholar] [CrossRef]
| Disease | Greater Abundance Linked to Disease | Affected Group | Known Risk Factors |
|---|---|---|---|
| Chronic otitis media with effusion (COME) | Corynebacterium, Streptococcus, Moraxella [16] | Caucasian Americans (CAs) [17] | allergy or atopy, upper respiratory tract infection, snoring, acute otitis media, passive smoke exposure and low social status [17,18] |
| Pediatric Asthma | Corynebacterium [19] | Puerto-Ricans (PRs), African Americans (AAs) [20] | parental asthma, prenatal environmental tobacco smoke, having cats and prematurity [21,22] |
| Chronic Rhinosinusitis (CRS) | Corynebacterium, Burkholderia [23,24] | Latino Americans (LAs) [24] | Asthma, genetics, GERD, rheumatoid arthritis, migraine, cigarette smoking [25] |
| COVID-19 | Salmonella, Scardovia, Serratia and Pectobacteriaceae [26,27] | AAs and Asian Americans (AS) [28] | Hypertension, coronary artery disease, history of stroke, diabetes, obesity, severe obesity, chronic kidney disease, asthma, and chronic obstructive pulmonary disease [29] |
| Granulomatosis with Polyangiitis (GPA) | Staphylococcus aureus [30] | Not Distinguishable [31] | Animal (horses) exposure, history of bronchiectasis, autoimmune disease, chronic renal impairment, Pulmonary fibrosis [32,33] |
| Acute Otitis Media (AOM) | Haemophilus, Moraxella, and Neisseria [16] | CAs [34] | Out-of-home daycare, multiple children living in the home, and mother’s multiparity [34] |
| Influenza B virus (IBV) | Streptococcus species and Prevotella salivae [35] | Diabetes, chronic respiratory disease [36] | |
| Influenza A virus (IAV) | Staphylococcus aureus, Staphylococcu pneumoniae, Klebsiella and Aerococcus [37] | ||
| Acute Respiratory Distress Syndrome (ARDS) | CAs [38] | abuse of alcohol and tobacco, malnutrition and obesity [39] | |
| Obstructive Sleep Apnea (OSA) | Streptococcus, Prevotella and Veillonella [40] | Obesity, rhinitis, adenoid hypertrophy [41] | |
| Parkinson’s Disease (PD) | Actinobacter [42,43] | OSA, Head injury, family history of trauma and depression, family history of PD [41,44] | |
| Alzheimer’s Disease (AZ) | OSA, diet, the immune system, mitochondrial function, metal exposure, and infection [41,45] | ||
| Essential Tremors (ET) | AAs, LAs [46] | Exposure to neurotoxic compounds such as β-carboline alkaloids and ethanol. Exposure to pesticide and lead. Tobacco exposure [47,48] | |
| Atopic Dermatitis (AD) | AAs [49] | Viral and bacterial skin infections, neuropsychiatric diseases, family history, smoking, allergy, maternal asthma, and dogs [50] | |
| Psoriasis | CAs, Eastern African [51,52] | Stress, diabetes mellitus, obesity, smoking, air pollution arthritis, inflammatory bowel disease, alcohol, drugs, cardiovascular disease, infection, sun exposure, hypertension [53,54] |
| Disease | Varying Factor | Higher Prevalence | Lower Prevalence |
|---|---|---|---|
| Adult- Onset Asthma | Citizenship | Turkish, Moroccan and South-Asian Surinamese [99] | Dutch, Ghanaian and African Surinamese origin groups [99] |
| Allergies (asthma, hay fever, atopic eczema, self-reported rhinitis and atopic sensitization,) | Citizenship | Finland [100] | Russia (High Actinobacter) [100] |
| sinonasal carcinomas & paranasal sinus squamous cell carcinomas | Ethnicity | AAs [101,102] | CAs [101,102] |
| Endoscopic Sinus Surgery | Ethnicity, Stage at presentation | AAs, LAs [104] | Cas [104] |
| Allergies | Amount of Exposure to Environment | Long Term Western Migrants (S. aureus and S. pneumoniae) [105] | Recent Western Migrants [105] |
| Asthma | Ethnicity, Genetics | PR LAs [106] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peace, O.; Rachakonda, K.; Kress, M.; Villalta, F.; Rachakonda, G. Respiratory and Neurological Disease across Different Ethnic Groups Is Influenced by the Microbiome. Microorganisms 2021, 9, 1965. https://doi.org/10.3390/microorganisms9091965
Peace O, Rachakonda K, Kress M, Villalta F, Rachakonda G. Respiratory and Neurological Disease across Different Ethnic Groups Is Influenced by the Microbiome. Microorganisms. 2021; 9(9):1965. https://doi.org/10.3390/microorganisms9091965
Chicago/Turabian StylePeace, Odiase, Kartik Rachakonda, Miller Kress, Fernando Villalta, and Girish Rachakonda. 2021. "Respiratory and Neurological Disease across Different Ethnic Groups Is Influenced by the Microbiome" Microorganisms 9, no. 9: 1965. https://doi.org/10.3390/microorganisms9091965
APA StylePeace, O., Rachakonda, K., Kress, M., Villalta, F., & Rachakonda, G. (2021). Respiratory and Neurological Disease across Different Ethnic Groups Is Influenced by the Microbiome. Microorganisms, 9(9), 1965. https://doi.org/10.3390/microorganisms9091965








