Gut Microbiota-Modulated Metabolomic Profiling Shapes the Etiology and Pathogenesis of Autoimmune Diseases
Abstract
:1. Introduction
1.1. The Pathophysiology of T Cell-Modulated Autoimmune Disease
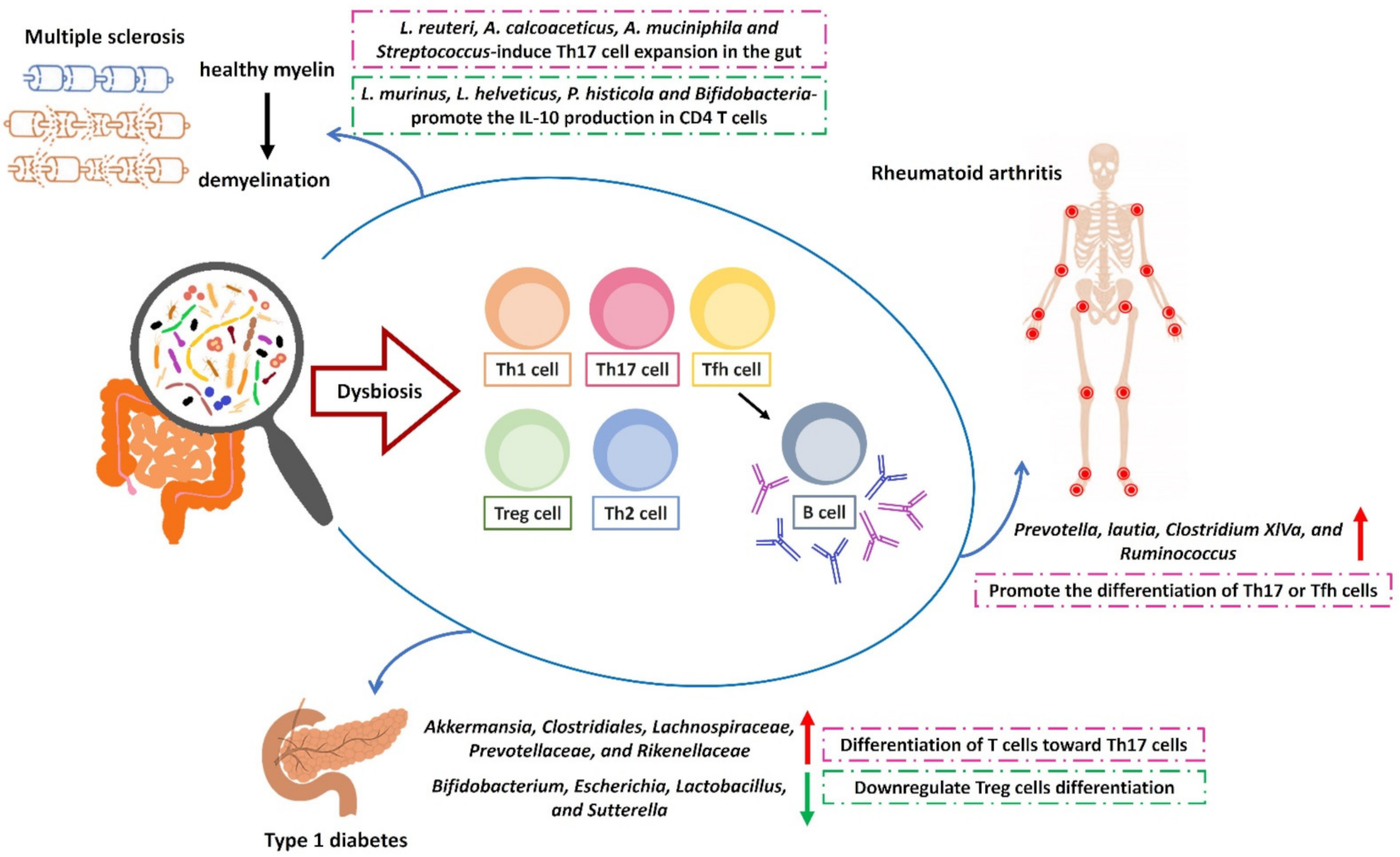
1.2. The Effect of Microbiota on the Development of Autoimmune Diseases
1.3. The Interaction of Microbiota and the Immune System
2. Overview of the Modulatory Effects of Host Microbiota on Autoimmune Diseases
2.1. Type 1 Diabetes
2.1.1. The Introduction of T1D and Gut Microbiota
2.1.2. The Modulatory Effect of Gut Microbiota in T1D
2.1.3. Targeted Therapies for Gut Microbiota in Autoimmune Diabetes
2.2. Multiple Sclerosis
2.2.1. The Role of Gut Microbiota in Central Nervous System-Based Autoimmune Diseases
2.2.2. The Modulatory Effect of Gut Microbiota on the Diet–Microbiota Axis in MS and EAE
2.2.3. Targeted Therapies for Gut Microbiota in MS and EAE
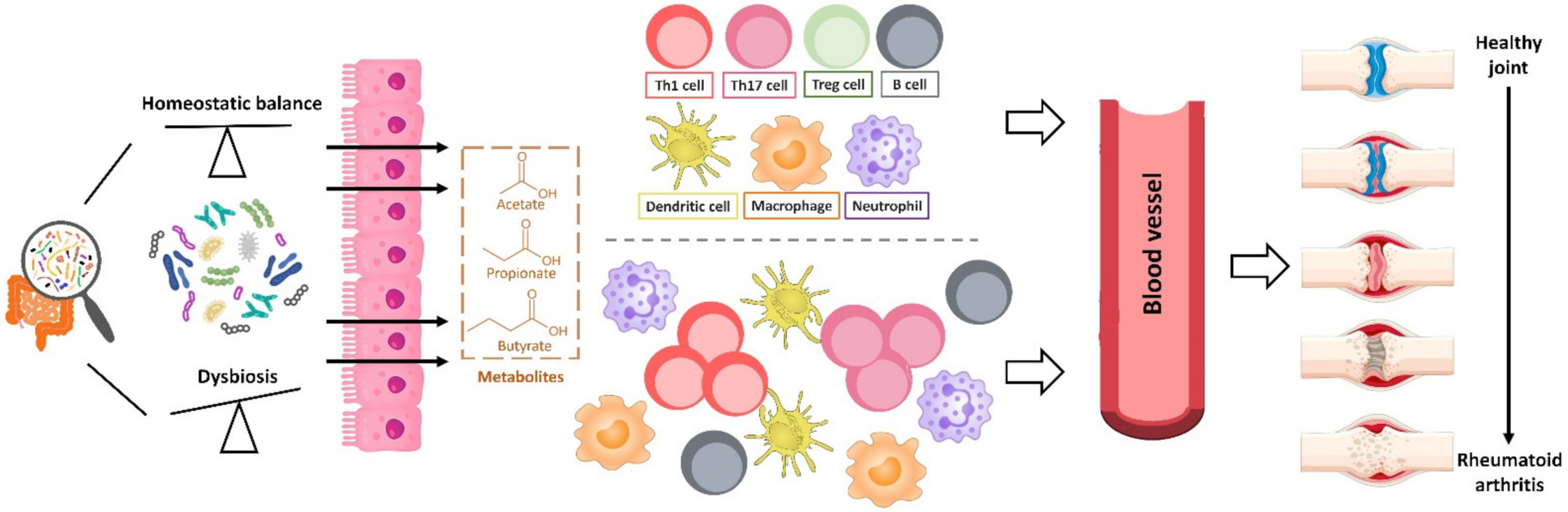
2.3. Rheumatoid Arthritis
2.3.1. The Introduction of Microbiota and RA
2.3.2. The Role of Intestinal Microbiota in Murine Models of Arthritis
2.3.3. The Role of Intestinal Microbiota in Human RA
2.3.4. Dietary Modification and Gut Microbiome as a Potential Therapeutic Target for RA
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Knip, M. Pathogenesis of Type 1 Diabetes: Implications for Incidence Trends. Horm. Res. Paediatr. 2011, 76 (Suppl. 1), 57–64. [Google Scholar] [CrossRef]
- Harjutsalo, V.; Sund, R.; Knip, M.; Groop, P.-H. Incidence of Type 1 Diabetes in Finland. JAMA 2013, 310, 427–428. [Google Scholar] [CrossRef] [Green Version]
- Boyko, A.; Melnikov, M. Prevalence and Incidence of Multiple Sclerosis in Russian Federation: 30 Years of Studies. Brain Sci. 2020, 10, 305. [Google Scholar] [CrossRef]
- Davidson, A.; Diamond, B. Autoimmune Diseases. N. Engl. J. Med. 2001, 345, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.-F. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: An update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef]
- Salliot, C.; Nguyen, Y.; Boutron-Ruault, M.-C.; Seror, R. Environment and Lifestyle: Their Influence on the Risk of RA. J. Clin. Med. 2020, 9, 3109. [Google Scholar] [CrossRef]
- Tierney, B.; Yang, Z.; Luber, J.M.; Beaudin, M.; Wibowo, M.C.; Baek, C.; Mehlenbacher, E.; Patel, C.J.; Kostic, A.D. The Landscape of Genetic Content in the Gut and Oral Human Microbiome. Cell Host Microbe 2019, 26, 283–295.e8. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Hübschmann, T.; Schattenberg, F.; Schumann, J.; Durek, P.; Riedel, R.; Friedrich, M.; Glauben, R.; Siegmund, B.; Radbruch, A.; et al. High-resolution microbiota flow cytometry reveals dynamic colitis-associated changes in fecal bacterial composition. Eur. J. Immunol. 2016, 46, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Kathagen, G.; D’Hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nat. Cell Biol. 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Rubbens, P.; Props, R.; Kerckhof, F.-M.; Boon, N.; Waegeman, W. Cytometric fingerprints of gut microbiota predict Crohn’s disease state. ISME J. 2021, 15, 354–358. [Google Scholar] [CrossRef]
- Koppel, N.; Rekdal, V.M.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Blaser, M. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 2017, 17, 461–463. [Google Scholar] [CrossRef]
- Makki, K.; Deehan, E.C.; Walter, J.; Bäckhed, F. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 2018, 23, 705–715. [Google Scholar] [CrossRef] [Green Version]
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. mBio 2019, 10, e02566-18. [Google Scholar] [CrossRef] [Green Version]
- Wastyk, H.C.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Yu, F.B.; Topf, M.; Gonzalez, C.G.; Van Treuren, W.; Han, S.; et al. Gut-microbiota-targeted diets modulate human immune status. Cell 2021, 184, 4137–4153.e14. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [Green Version]
- Belkaid, Y.; Hand, T.W. Role of the Microbiota in Immunity and Inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.F.; Wang, H. Environmental Exposures and Autoimmune Diseases: Contribution of Gut Microbiome. Front. Immunol. 2020, 10, 3094. [Google Scholar] [CrossRef]
- De Luca, F.; Shoenfeld, Y. The microbiome in autoimmune diseases. Clin. Exp. Immunol. 2019, 195, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, W.; Allsopp, P.J.; Watson, G.E.; Magee, P.J.; Strain, J.; Armstrong, D.J.; Ball, E.; McSorley, E.M. Mercury as an environmental stimulus in the development of autoimmunity—A systematic review. Autoimmun. Rev. 2017, 16, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Imam, T.; Park, S.; Kaplan, M.H.; Olson, M.R. Effector T Helper Cell Subsets in Inflammatory Bowel Diseases. Front. Immunol. 2018, 9, 1212. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of Effector CD4 T Cell Populations. Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Round, J.L.; Lee, S.M.; Li, J.; Tran, G.; Jabri, B.; Chatila, T.; Mazmanian, S.K. The Toll-Like Receptor 2 Pathway Establishes Colonization by a Commensal of the Human Microbiota. Science 2011, 332, 974–977. [Google Scholar] [CrossRef] [Green Version]
- Telesford, K.M.; Yan, W.; Ochoa-Re, J.; Pant, A.; Kircher, C.; Christy, M.A.; Begum-Haque, S.; Kasper, D.L.; Kasper, L.H. A commensal symbiotic factor derived fromBacteroides fragilispromotes human CD39+Foxp3+T cells and Tregfunction. Gut Microbes 2015, 6, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abt, M.C.; Artis, D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr. Opin. Microbiol. 2013, 16, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef]
- Holers, V.M.; Demoruelle, M.K.; Kuhn, K.A.; Buckner, J.H.; Robinson, W.H.; Okamoto, Y.; Norris, J.M.; Deane, K.D. Rheumatoid arthritis and the mucosal origins hypothesis: Protection turns to destruction. Nat. Rev. Rheumatol. 2018, 14, 542–557. [Google Scholar] [CrossRef]
- Alpizar-Rodriguez, D.; Lesker, T.R.; Gronow, A.; Gilbert, B.; Raemy, E.; Lamacchia, C.; Gabay, C.; Finckh, A.; Strowig, T. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2019, 78, 590–593. [Google Scholar] [CrossRef]
- Atkinson, M.A.; Eisenbarth, G.S.; Michels, A.W. Type 1 diabetes. Lancet 2014, 383, 69–82. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.S.; Bluestone, J.A. THE NOD MOUSE: A Model of Immune Dysregulation. Annu. Rev. Immunol. 2005, 23, 447–485. [Google Scholar] [CrossRef]
- Turley, S.; Poirot, L.; Hattori, M.; Benoist, C.; Mathis, D. Physiological β Cell Death Triggers Priming of Self-reactive T Cells by Dendritic Cells in a Type-1 Diabetes Model. J. Exp. Med. 2003, 198, 1527–1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehuen, A.; Diana, J.; Zaccone, P.; Cooke, A. Immune cell crosstalk in type 1 diabetes. Nat. Rev. Immunol. 2010, 10, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Knip, M.; Siljander, H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2016, 12, 154–167. [Google Scholar] [CrossRef]
- Hooper, L.V.; Littman, D.R.; MacPherson, A.J. Interactions Between the Microbiota and the Immune System. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- Kemppainen, K.M.; Ardissone, A.N.; Davis-Richardson, A.G.; Fagen, J.R.; Gano, K.A.; Leon-Novelo, L.G.; Vehik, K.; Casella, G.; Simell, O.; Ziegler, A.G.; et al. Early Childhood Gut Microbiomes Show Strong Geographic Differences Among Subjects at High Risk for Type 1 Diabetes. Diabetes Care 2014, 38, 329–332. [Google Scholar] [CrossRef] [Green Version]
- Patterson, C.C.; Dahlquist, G.G.; Gyürüs, E.; Green, A.; Soltész, G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–2020: A multicentre prospective registration study. Lancet 2009, 373, 2027–2033. [Google Scholar] [CrossRef]
- The TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study: Study design. Pediatr. Diabetes 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Vatanen, T.; Franzosa, E.A.; Schwager, R.; Tripathi, S.; Arthur, T.D.; Vehik, K.; Lernmark, A.; Hagopian, W.A.; Rewers, M.J.; She, J.-X.; et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 2018, 562, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Land, B.V.; Engen, P.A.; Naqib, A.; Green, S.J.; Nato, A.; Leusink-Muis, T.; Garssen, J.; Keshavarzian, A.; Stahl, B.; et al. Human milk oligosaccharides protect against the development of autoimmune diabetes in NOD-mice. Sci. Rep. 2018, 8, 3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Hertzen, L.; Beutler, B.; Bienenstock, J.; Blaser, M.; Cani, P.D.; Eriksson, J.; Färkkilä, M.; Haahtela, T.; Hanski, I.; Jenmalm, M.C.; et al. Helsinki alert of biodiversity and health. Ann. Med. 2015, 47, 218–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quercia, S.; Candela, M.; Giuliani, C.; Turroni, S.; Luiselli, D.; Rampelli, S.; Brigidi, P.; Franceschi, C.; Bacalini, M.G.; Garagnani, P.; et al. From lifetime to evolution: Timescales of human gut microbiota adaptation. Front. Microbiol. 2014, 5, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renz, H.; Brandtzaeg, P.; Hornef, M.W. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 2011, 12, 9–23. [Google Scholar] [CrossRef]
- Sommer, F.; Bäckhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Dunne, J.L.; Triplett, E.W.; Gevers, D.; Xavier, R.; Insel, R.; Danska, J.; Atkinson, M.A. The intestinal microbiome in type 1 diabetes. Clin. Exp. Immunol. 2014, 177, 30–37. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.-U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Janeway, C. Approaching the Asymptote? Evolution and Revolution in Immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54 Pt 1, 1–13. [Google Scholar] [CrossRef]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [Green Version]
- Wen, L.; Ley, R.; Volchkov, P.Y.; Stranges, P.; Avanesyan, L.; Stonebraker, A.C.; Hu, C.; Wong, F.S.; Szot, G.L.; Bluestone, J.A.; et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nat. Cell Biol. 2008, 455, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chien, C.-C.; Paster, B.J.; Ericson, R.L.; Orcutt, R.P.; Schauer, D.B.; Fox, J.G. Phylogeny of the Defined Murine Microbiota: Altered Schaedler Flora. Appl. Environ. Microbiol. 1999, 65, 3287–3292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paun, A.; Yau, C.; Danska, J.S. The Influence of the Microbiome on Type 1 Diabetes. J. Immunol. 2017, 198, 590–595. [Google Scholar] [CrossRef]
- Tai, N.; Peng, J.; Liu, F.; Gulden, E.; Hu, Y.; Zhang, X.; Chen, L.; Wong, F.S.; Ningwen, T. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J. Exp. Med. 2016, 213, 2129–2146. [Google Scholar] [CrossRef] [PubMed]
- Nanjundappa, R.H.; Ronchi, F.; Wang, J.; Clemente-Casares, X.; Yamanouchi, J.; Umeshappa, C.S.; Yang, Y.; Blanco, J.; Bassolas-Molina, H.; Salas, A.; et al. A Gut Microbial Mimic that Hijacks Diabetogenic Autoreactivity to Suppress Colitis. Cell 2017, 171, 655–667.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gülden, E.; Chao, C.; Tai, N.; Pearson, J.; Peng, J.; Majewska-Szczepanik, M.; Zhou, Z.; Wong, F.S.; Wen, L. TRIF deficiency protects non-obese diabetic mice from type 1 diabetes by modulating the gut microbiota and dendritic cells. J. Autoimmun. 2018, 93, 57–65. [Google Scholar] [CrossRef]
- King, C.; Sarvetnick, N. The Incidence of Type-1 Diabetes in NOD Mice Is Modulated by Restricted Flora Not Germ-Free Conditions. PLoS ONE 2011, 6, e17049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegel, M.; Sefik, E.; Hill, J.A.; Wu, H.-J.; Benoist, C.; Mathis, D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc. Natl. Acad. Sci. USA 2011, 108, 11548–11553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hensgens, M.; Keessen, E.; Squire, M.; Riley, T.; Koene, M.; de Boer, E.; Lipman, L.; Kuijper, E. Clostridium difficile infection in the community: A zoonotic disease? Clin. Microbiol. Infect. 2012, 18, 635–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, M.C.G.; Oliveira, R.P.; Torres, L.; Aguiar, S.L.F.; Pinheiro-Rosa, N.; Lemos, L.; Guimarães, M.A.; Reis, D.; Silveira, T.; Ferreira, E.; et al. Frontline Science: Abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J. Leukoc. Biol. 2019, 106, 513–529. [Google Scholar] [CrossRef]
- Simon, M.-C.; Reinbeck, A.L.; Wessel, C.; Heindirk, J.; Jelenik, T.; Kaul, K.; Arreguin-Cano, J.; Strom, A.; Blaut, M.; Bäckhed, F.; et al. Distinct alterations of gut morphology and microbiota characterize accelerated diabetes onset in nonobese diabetic mice. J. Biol. Chem. 2020, 295, 969–980. [Google Scholar] [CrossRef]
- Mullaney, J.; Stephens, J.E.; Geeling, B.E.; Hamilton-Williams, E.E. Early-life exposure to gut microbiota from disease-protected mice does not impact disease outcome in type 1 diabetes susceptible NOD mice. Immunol. Cell Biol. 2019, 97, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Hansen, C.H.F.; Krych, L.; Nielsen, D.S.; Vogensen, F.; Hansen, L.H.; Sørensen, S.; Buschard, K.; Hansen, A.K. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012, 55, 2285–2294. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Jin, P.; Peng, J.; Zhang, X.; Wong, F.S.; Wen, L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J. Autoimmun. 2016, 72, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Livanos, A.; Greiner, T.U.; Vangay, P.; Pathmasiri, W.; Stewart, D.; McRitchie, S.; Li, H.; Chung, J.; Sohn, J.; Kim, S.; et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 2016, 1, 1–13. [Google Scholar] [CrossRef]
- Sofi, M.H.; Johnson, B.M.; Gudi, R.R.; Jolly, A.; Gaudreau, M.-C.; Vasu, C. Polysaccharide A–Dependent Opposing Effects of Mucosal and Systemic Exposures to Human Gut CommensalBacteroides fragilisin Type 1 Diabetes. Diabetes 2019, 68, 1975–1989. [Google Scholar] [CrossRef] [PubMed]
- Joller, N.; Lozano, E.; Burkett, P.R.; Patel, B.; Xiao, S.; Zhu, C.; Xia, J.; Tan, T.G.; Sefik, E.; Yajnik, V.; et al. Treg Cells Expressing the Coinhibitory Molecule TIGIT Selectively Inhibit Proinflammatory Th1 and Th17 Cell Responses. Immunity 2014, 40, 569–581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhag, D.A.; Kumar, M.; Al Khodor, S. Exploring the Triple Interaction between the Host Genome, the Epigenome, and the Gut Microbiome in Type 1 Diabetes. Int. J. Mol. Sci. 2020, 22, 125. [Google Scholar] [CrossRef] [PubMed]
- Craig, M.E.; Kim, K.W.; Isaacs, S.; Penno, M.; Hamilton-Williams, E.; Couper, J.J.; Rawlinson, W.D. Early-life factors contributing to type 1 diabetes. Diabetologia 2019, 62, 1823–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamanouchi, J.; Rainbow, D.; Serra, P.; Howlett, S.; Hunter, K.; Garner, V.E.; Gonzalez-Munoz, A.; Clark, J.; Veijola, R.; Cubbon, R.; et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat. Genet. 2007, 39, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Hunter, K.; Rainbow, D.; Plagnol, V.; Todd, J.A.; Peterson, L.B.; Wicker, L.S. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. J. Immunol. 2007, 179, 8341–8349. [Google Scholar] [CrossRef] [Green Version]
- Mullaney, J.A.; Stephens, J.E.; Costello, M.-E.; Fong, C.; Geeling, B.E.; Gavin, P.; Wright, C.M.; Spector, T.D.; Brown, M.A.; Hamilton-Williams, E.E. Type 1 diabetes susceptibility alleles are associated with distinct alterations in the gut microbiota. Microbiome 2018, 6, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorburn, A.N.; Macia, L.; Mackay, C.R. Diet, Metabolites, and “Western-Lifestyle” Inflammatory Diseases. Immunity 2014, 40, 833–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Pearson, J.A.; Peng, J.; Hu, Y.; Sha, S.; Xing, Y.; Huang, G.; Li, X.; Hu, F.; Xie, Z.; et al. Gut microbial metabolites alter IgA immunity in type 1 diabetes. JCI Insight 2020, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Yap, Y.A.; McLeod, K.H.; McKenzie, C.I.; Gavin, P.G.; Davalos-Salas, M.; Richards, J.L.; Moore, R.J.; Lockett, T.J.; Clarke, J.M.; Eng, V.V.; et al. An acetate-yielding diet imprints an immune and anti-microbial programme against enteric infection. Clin. Transl. Immunol. 2021, 10, e1233. [Google Scholar] [CrossRef] [PubMed]
- Mariño, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.-A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562. [Google Scholar] [CrossRef]
- Brauner, H.; Lüthje, P.; Grünler, J.; Ekberg, N.R.; Dallner, G.; Brismar, K. Markers of innate immune activity in patients with type 1 and type 2 diabetes mellitus and the effect of the anti-oxidant coenzyme Q10 on inflammatory activity. Clin. Exp. Immunol. 2014, 177, 478–482. [Google Scholar] [CrossRef]
- Sun, J.; Furio, L.; Mecheri, R.; van der Does, A.; Lundeberg, E.; Saveanu, L.; Chen, Y.; van Endert, P.; Agerberth, B.; Diana, J. Pancreatic β-Cells Limit Autoimmune Diabetes via an Immunoregulatory Antimicrobial Peptide Expressed under the Influence of the Gut Microbiota. Immunity 2015, 43, 304–317. [Google Scholar] [CrossRef] [Green Version]
- Miani, M.; Le Naour, J.; Waeckel-Enée, E.; Verma, S.C.; Straube, M.; Emond, P.; Ryffel, B.; van Endert, P.; Sokol, H.; Diana, J. Gut Microbiota-Stimulated Innate Lymphoid Cells Support β-Defensin 14 Expression in Pancreatic Endocrine Cells, Preventing Autoimmune Diabetes. Cell Metab. 2018, 28, 557–572.e6. [Google Scholar] [CrossRef] [Green Version]
- Warshauer, J.T.; Bluestone, J.A.; Anderson, M.S. New Frontiers in the Treatment of Type 1 Diabetes. Cell Metab. 2020, 31, 46–61. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, L.; Zhang, S.; Zhao, X.; Gang, X.; Wang, G. Evaluating the Causal Role of Gut Microbiota in Type 1 Diabetes and Its Possible Pathogenic Mechanisms. Front. Endocrinol. 2020, 11, 125. [Google Scholar] [CrossRef]
- Al Theyab, A.; Almutairi, T.; Al-Suwaidi, A.M.; Bendriss, G.; McVeigh, C.; Chaari, A. Epigenetic Effects of Gut Metabolites: Exploring the Path of Dietary Prevention of Type 1 Diabetes. Front. Nutr. 2020, 7, 563605. [Google Scholar] [CrossRef] [PubMed]
- de Goffau, M.; Luopajärvi, K.; Knip, M.; Ilonen, J.; Ruohtula, T.; Härkönen, T.; Orivuori, L.; Hakala, S.; Welling, G.W.; Harmsen, H.J.; et al. Fecal Microbiota Composition Differs Between Children With -Cell Autoimmunity and Those Without. Diabetes 2013, 62, 1238–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, K.; Benitez, P.; Ardissone, A.; Wilson, T.D.; Collins, E.L.; Lorca, G.; Li, N.; Sankar, D.; Wasserfall, C.; Neu, J.; et al. Inhibition of Type 1 Diabetes Correlated to a Lactobacillus johnsonii N6.2-Mediated Th17 Bias. J. Immunol. 2011, 186, 3538–3546. [Google Scholar] [CrossRef] [Green Version]
- Calabrese, M.; Magliozzi, R.; Ciccarelli, O.; Geurts, J.J.G.; Reynolds, R.; Martin, R. Exploring the origins of grey matter damage in multiple sclerosis. Nat. Rev. Neurosci. 2015, 16, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Glatigny, S.; Bettelli, E. Experimental Autoimmune Encephalomyelitis (EAE) as Animal Models of Multiple Sclerosis (MS). Cold Spring Harb. Perspect. Med. 2018, 8, a028977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ransohoff, R.M. Animal models of multiple sclerosis: The good, the bad and the bottom line. Nat. Neurosci. 2012, 15, 1074–1077. [Google Scholar] [CrossRef]
- Tsunoda, I.; Fujinami, R.S. Neuropathogenesis of Theiler’s Murine Encephalomyelitis Virus Infection, An Animal Model for Multiple Sclerosis. J. Neuroimmune Pharmacol. 2009, 5, 355–369. [Google Scholar] [CrossRef] [Green Version]
- Pachner, A.R. Experimental models of multiple sclerosis. Curr. Opin. Neurol. 2011, 24, 291–299. [Google Scholar] [CrossRef] [Green Version]
- Dendrou, C.A.; Fugger, L.; Friese, M.A. Immunopathology of multiple sclerosis. Nat. Rev. Immunol. 2015, 15, 545–558. [Google Scholar] [CrossRef]
- Wlodarczyk, A.; Løbner, M.; Cédile, O.; Owens, T. Comparison of microglia and infiltrating CD11c+ cells as antigen presenting cells for T cell proliferation and cytokine response. J. Neuroinflam. 2014, 11, 57. [Google Scholar] [CrossRef] [Green Version]
- Codarri, L.; Gyülvészi, G.; Tosevski, V.; Hesske, L.; Fontana, A.; Magnenat, L.; Suter, T.; Becher, B. RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat. Immunol. 2011, 12, 560–567. [Google Scholar] [CrossRef]
- Grifka-Walk, H.M.; Giles, D.A.; Segal, B.M. IL-12-polarized Th1 cells produce GM-CSF and induce EAE independent of IL-23. Eur. J. Immunol. 2015, 45, 2780–2786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baecher-Allan, C.; Kaskow, B.; Weiner, H.L. Multiple Sclerosis: Mechanisms and Immunotherapy. Neuron 2018, 97, 742–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filippi, M.; Bar-Or, A.; Piehl, F.; Preziosa, P.; Solari, A.; Vukusic, S.; Rocca, M.A. Multiple sclerosis. Nat. Rev. Dis. Prim. 2018, 4, 43. [Google Scholar] [CrossRef]
- Ochoa-Repáraz, J.; Mielcarz, D.W.; Ditrio, L.E.; Burroughs, A.R.; Foureau, D.M.; Haque-Begum, S.; Kasper, L.H. Role of Gut Commensal Microflora in the Development of Experimental Autoimmune Encephalomyelitis. J. Immunol. 2009, 183, 6041–6050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.K.; Menezes, J.S.; Umesaki, Y.; Mazmanian, S.K. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2011, 108, 4615–4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz Heijtz, R.; Wang, S.; Anuar, F.; Qian, Y.; Björkholm, B.; Samuelsson, A.; Hibberd, M.L.; Forssberg, H.; Pettersson, S. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. USA 2011, 108, 3047–3052. [Google Scholar] [CrossRef] [Green Version]
- Ang, Q.Y.; Alexander, M.; Newman, J.C.; Tian, Y.; Cai, J.; Upadhyay, V.; Turnbaugh, J.A.; Verdin, E.; Hall, K.D.; Leibel, R.L.; et al. Ketogenic Diets Alter the Gut Microbiome Resulting in Decreased Intestinal Th17 Cells. Cell 2020, 181, 1263–1275.e16. [Google Scholar] [CrossRef] [PubMed]
- Duc, D.; Vigne, S.; Bernier-Latmani, J.; Yersin, Y.; Ruiz, F.; Gaïa, N.; Leo, S.; Lazarevic, V.; Schrenzel, J.; Petrova, T.V.; et al. Disrupting Myelin-Specific Th17 Cell Gut Homing Confers Protection in an Adoptive Transfer Experimental Autoimmune Encephalomyelitis. Cell Rep. 2019, 29, 378–390.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosorich, I.; Dalla-Costa, G.; Sorini, C.; Ferrarese, R.; Messina, M.J.; Dolpady, J.; Radice, E.; Mariani, A.; Testoni, P.A.; Canducci, F.; et al. High frequency of intestinal T H 17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci. Adv. 2017, 3, e1700492. [Google Scholar] [CrossRef] [Green Version]
- Erny, D.; De Angelis, A.L.; Jaitin, D.; Wieghofer, P.; Staszewski, O.; David, E.; Keren-Shaul, H.; Mahlakoiv, T.; Jakobshagen, K.; Buch, T.; et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015, 18, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.; Mayo, L.; Chao, C.-C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhang, J. Dietary Modulation of Intestinal Microbiota: Future Opportunities in Experimental Autoimmune Encephalomyelitis and Multiple Sclerosis. Front. Microbiol. 2019, 10, 740. [Google Scholar] [CrossRef]
- Ii, D.M.J.; Goertz, J.E.; Marin, I.A.; Costello, J.; Overall, C.C.; Gaultier, A. Experimental autoimmune encephalomyelitis is associated with changes of the microbiota composition in the gastrointestinal tract. Sci. Rep. 2020, 10, 15183. [Google Scholar] [CrossRef]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mähler, A.; Balogh, A.; Markó, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Farez, M.F.; Fiol, M.P.; Gaitán, M.I.; Quintana, F.J.; Correale, J. Sodium intake is associated with increased disease activity in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 2015, 86, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, M.; Ukibe, K.; Matsubara, Y.; Hosoya, T.; Sakai, F.; Kon, S.; Arima, Y.; Murakami, M.; Nakagawa, H.; Miyazaki, T. Lactobacillus helveticus SBT2171 Attenuates Experimental Autoimmune Encephalomyelitis in Mice. Front. Microbiol. 2018, 8, 2596. [Google Scholar] [CrossRef]
- Montgomery, T.L.; Künstner, A.; Kennedy, J.J.; Fang, Q.; Asarian, L.; Culp-Hill, R.; D’Alessandro, A.; Teuscher, C.; Busch, H.; Krementsov, D.N. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc. Natl. Acad. Sci. USA 2020, 117, 27516–27527. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, E.; Kim, S.-W.; Suda, W.; Kawasumi, M.; Onawa, S.; Taguchi-Atarashi, N.; Morita, H.; Taylor, T.D.; Hattori, M.; Ohno, H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nat. Cell Biol. 2020, 585, 102–106. [Google Scholar] [CrossRef]
- Vamanu, E.; Rai, S. The Link between Obesity, Microbiota Dysbiosis, and Neurodegenerative Pathogenesis. Diseases 2021, 9, 45. [Google Scholar] [CrossRef]
- Al-Assal, K.; Martinez, A.C.; Torrinhas, R.S.; Cardinelli, C.; Waitzberg, D. Gut microbiota and obesity. Clin. Nutr. Exp. 2018, 20, 60–64. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Muscogiuri, G.; Cantone, E.; Cassarano, S.; Tuccinardi, D.; Barrea, L.; Savastano, S.; Colao, A. Gut microbiota: A new path to treat obesity. Int. J. Obes. Suppl. 2019, 9, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [Green Version]
- Ji, Z.; Wu, S.; Xu, Y.; Qi, J.; Su, X.; Shen, L. Obesity Promotes EAE Through IL-6 and CCL-2-Mediated T Cells Infiltration. Front. Immunol. 2019, 10, 1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munger, K.L.; Chitnis, T.; Ascherio, A. Body size and risk of MS in two cohorts of US women. Neurology 2009, 73, 1543–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedström, A.K.; Olsson, T.; Alfredsson, L. High body mass index before age 20 is associated with increased risk for multiple sclerosis in both men and women. Mult. Scler. J. 2012, 18, 1334–1336. [Google Scholar] [CrossRef]
- Wesnes, K.; Riise, T.; Casetta, I.; Drulovic, J.; Granieri, E.; Holmøy, T.; Kampman, M.T.; Landtblom, A.-M.; Lauer, K.; Lossius, A.; et al. Body size and the risk of multiple sclerosis in Norway and Italy: The EnvIMS study. Mult. Scler. J. 2014, 21, 388–395. [Google Scholar] [CrossRef]
- Haghikia, A.; Jörg, S.; Duscha, A.; Berg, J.; Manzel, A.; Waschbisch, A.; Hammer, A.; Lee, D.-H.; May, C.; Wilck, N.; et al. Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 2015, 43, 817–829. [Google Scholar] [CrossRef] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Mizuno, M.; Noto, D.; Kaga, N.; Chiba, A.; Miyake, S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PLoS ONE 2017, 12, e0173032. [Google Scholar] [CrossRef] [Green Version]
- Aharoni, R.; Schottlender, N.; Bar-Lev, D.D.; Eilam, R.; Sela, M.; Tsoory, M.; Arnon, R. Cognitive impairment in an animal model of multiple sclerosis and its amelioration by glatiramer acetate. Sci. Rep. 2019, 9, 4140. [Google Scholar] [CrossRef]
- Kim, M.; Lee, S.; Lim, H.; Lee, J.; Park, J.-Y.; Kwon, H.-J.; Lee, I.-C.; Ryu, Y.-B.; Kim, J.; Shin, T.; et al. Oleanolic Acid Acetate Alleviates Symptoms of Experimental Autoimmune Encephalomyelitis in Mice by Regulating Toll-Like Receptor 2 Signaling. Front. Pharmacol. 2020, 11, 556391. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Noto, D.; Hoshino, Y.; Mizuno, M.; Miyake, S. Butyrate suppresses demyelination and enhances remyelination. J. Neuroinflam. 2019, 16, 65. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Wang, Q.; Wu, Q.; Mao-Draayer, Y.; Kim, C.H. Bidirectional regulatory potentials of short-chain fatty acids and their G-protein-coupled receptors in autoimmune neuroinflammation. Sci. Rep. 2019, 9, 8837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sonner, J.K.; Keil, M.; Falk-Paulsen, M.; Mishra, N.; Rehman, A.; Kramer, M.; Deumelandt, K.; Röwe, J.; Sanghvi, K.; Wolf, L.; et al. Dietary tryptophan links encephalogenicity of autoreactive T cells with gut microbial ecology. Nat. Commun. 2019, 10, 4877. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Raes, J.; Bork, P. The Human Gut Microbiome: From Association to Modulation. Cell 2018, 172, 1198–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berer, K.; Gerdes, L.A.; Cekanaviciute, E.; Jia, X.; Xiao, L.; Xia, Z.; Liu, C.; Klotz, L.; Stauffer, U.; Baranzini, S.; et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 10719–10724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangalam, A.; Shahi, S.K.; Luckey, D.; Karau, M.; Marietta, E.; Luo, N.; Choung, R.S.; Ju, J.; Sompallae, R.; Gibson-Corley, K.; et al. Human Gut-Derived Commensal Bacteria Suppress CNS Inflammatory and Demyelinating Disease. Cell Rep. 2017, 20, 1269–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borody, T.; Leis, S.; Campbell, J.; Torres, M.; Nowak, A. Fecal Microbiota Transplantation (FMT) in Multiple Sclerosis (MS). Am. J. Gastroenterol. 2011, 106, S352. [Google Scholar] [CrossRef]
- Makkawi, S.; Camara-Lemarroy, C.; Metz, L. Fecal microbiota transplantation associated with 10 years of stability in a patient with SPMS. Neurol.-Neuroimmunol. Neuroinflamm. 2018, 5, e459. [Google Scholar] [CrossRef] [Green Version]
- Junker, A.; Hohlfeld, R.; Meinl, E. The emerging role of microRNAs in multiple sclerosis. Nat. Rev. Neurol. 2010, 7, 56–59. [Google Scholar] [CrossRef]
- Juźwik, C.A.; Drake, S.; Lécuyer, M.-A.; Johnson, R.M.; Morquette, B.; Zhang, Y.; Charabati, M.; Sagan, S.M.; Bar-Or, A.; Prat, A.; et al. Neuronal microRNA regulation in Experimental Autoimmune Encephalomyelitis. Sci. Rep. 2018, 8, 13437. [Google Scholar] [CrossRef]
- Liu, S.; Rezende, R.M.; Moreira, T.G.; Tankou, S.K.; Cox, L.M.; Wu, M.; Song, A.; Dhang, F.H.; Wei, Z.; Costamagna, G.; et al. Oral Administration of miR-30d from Feces of MS Patients Suppresses MS-like Symptoms in Mice by Expanding Akkermansia muciniphila. Cell Host Microbe 2019, 26, 779–794.e8. [Google Scholar] [CrossRef] [PubMed]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- Karpouzas, G.A.; Ormseth, S.R.; Hernandez, E.; Budoff, M.J. Impact of Cumulative Inflammation, Cardiac Risk Factors, and Medication Exposure on Coronary Atherosclerosis Progression in Rheumatoid Arthritis. Arthritis Rheumatol. 2019, 72, 400–408. [Google Scholar] [CrossRef]
- Taneja, V. Cytokines pre-determined by genetic factors are involved in pathogenesis of Rheumatoid arthritis. Cytokine 2015, 75, 216–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, D.; Nishimura, K.; Tamaki, K.; Tsuji, G.; Nakazawa, T.; Morinobu, A.; Kumagai, S. Impact of smoking as a risk factor for developing rheumatoid arthritis: A meta-analysis of observational studies. Ann. Rheum. Dis. 2009, 69, 70–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, P.M.; Williams, F.M.; Matey-Hernandez, M.; Menni, C.; Steves, C. ‘RA and the microbiome: Do host genetic factors provide the link? J. Autoimmun. 2019, 99, 104–115. [Google Scholar] [CrossRef]
- MacGregor, A.J.; Snieder, H.; Rigby, A.S.; Koskenvuo, M.; Kaprio, J.; Aho, K.; Silman, A.J. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheumatol. 2000, 43, 30–37. [Google Scholar] [CrossRef]
- Maeda, Y.; Kurakawa, T.; Umemoto, E.; Motooka, D.; Ito, Y.; Gotoh, K.; Hirota, K.; Matsushita, M.; Furuta, Y.; Narazaki, M.; et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016, 68, 2646–2661. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, N.; Takahashi, T.; Hata, H.; Nomura, T.; Tagami, T.; Yamazaki, S.; Sakihama, T.; Matsutani, T.; Negishi, I.; Nakatsuru, S.; et al. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nat. Cell Biol. 2003, 426, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Rogier, R.; Ederveen, T.H.A.; Boekhorst, J.; Wopereis, H.; Scher, J.U.; Manasson, J.; Frambach, S.J.C.M.; Knol, J.; Garssen, J.; Van Der Kraan, P.M.; et al. Aberrant intestinal microbiota due to IL-1 receptor antagonist deficiency promotes IL-17- and TLR4-dependent arthritis. Microbiome 2017, 5, 1–15. [Google Scholar] [CrossRef]
- Abdollahi-Roodsaz, S.; Joosten, L.A.; Koenders, M.; Devesa, I.; Roelofs, M.F.; Radstake, T.R.; Heuvelmans-Jacobs, M.; Akira, S.; Nicklin, M.; Ribeiro-Dias, F.; et al. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Investig. 2008, 118, 205–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehaume, L.M.; Mondot, S.; de Cárcer, D.A.; Velasco, J.; Benham, H.; Hasnain, S.; Bowman, J.; Ruutu, M.; Hansbro, P.; McGuckin, M.; et al. ZAP-70 Genotype Disrupts the Relationship Between Microbiota and Host, Leading to Spondyloarthritis and Ileitis in SKG Mice. Arthritis Rheumatol. 2014, 66, 2780–2792. [Google Scholar] [CrossRef] [Green Version]
- Jubair, W.K.; Hendrickson, J.D.; Severs, E.L.; Schulz, H.M.; Adhikari, S.; Ir, D.; Pagan, J.D.; Anthony, R.M.; Robertson, C.E.; Frank, D.N.; et al. Modulation of Inflammatory Arthritis in Mice by Gut Microbiota Through Mucosal Inflammation and Autoantibody Generation. Arthritis Rheumatol. 2018, 70, 1220–1233. [Google Scholar] [CrossRef]
- Espina, M.D.T.; Gabarrini, G.; Harmsen, H.J.M.; Westra, J.; Van Winkelhoff, A.J.; Van Dijl, J.M. Talk to your gut: The oral-gut microbiome axis and its immunomodulatory role in the etiology of rheumatoid arthritis. FEMS Microbiol. Rev. 2019, 43, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, X.-J.; Cao, N.-W.; Zhou, H.-Y.; Meng, X.; Guo, B.; Zhang, H.-Y.; Li, B.-Z. The oral and gut microbiome in rheumatoid arthritis patients: A systematic review. Rheumatology 2021, 60, 1054–1066. [Google Scholar] [CrossRef]
- Vaahtovuo, J.; Munukka, E.; Korkeamäki, M.; Luukkainen, R.; Toivanen, P. Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 2008, 35, 1500–1505. [Google Scholar]
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2013, 2, e01202. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, S.-X.; Yin, X.-F.; Zhang, M.-X.; Qiao, J.; Xin, X.-H.; Chang, M.-J.; Gao, C.; Li, Y.-F.; Li, X.-F. The Gut Microbiota and Its Relevance to Peripheral Lymphocyte Subpopulations and Cytokines in Patients with Rheumatoid Arthritis. J. Immunol. Res. 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Wells, P.M.; Adebayo, A.S.; Bowyer, R.C.E.; Freidin, M.B.; Finckh, A.; Strowig, T.; Lesker, T.R.; Alpizar-Rodriguez, D.; Gilbert, B.; Kirkham, B.; et al. Associations between gut microbiota and genetic risk for rheumatoid arthritis in the absence of disease: A cross-sectional study. Lancet Rheumatol. 2020, 2, e418–e427. [Google Scholar] [CrossRef]
- Alpízar-Rodríguez, D.; Finckh, A.; Gilbert, B. The Role of Nutritional Factors and Intestinal Microbiota in Rheumatoid Arthritis Development. Nutrients 2020, 13, 96. [Google Scholar] [CrossRef]
- Gan, R.W.; Demoruelle, M.K.; Deane, K.D.; Weisman, M.H.; Buckner, J.H.; Gregersen, P.K.; Mikuls, T.R.; O’Dell, J.R.; Keating, R.M.; Fingerlin, T.E.; et al. Omega-3 fatty acids are associated with a lower prevalence of autoantibodies in shared epitope-positive subjects at risk for rheumatoid arthritis. Ann. Rheum. Dis. 2017, 76, 147–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Giuseppe, D.; Crippa, A.; Orsini, N.; Wolk, A. Fish consumption and risk of rheumatoid arthritis: A dose-response meta-analysis. Arthritis Res. Ther. 2014, 16, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatraman, B.S.R.A. Increased Permeability in Dextran Sulphate Colitis in Rats: Time Course of Development and Effect of Butyrate. Scand. J. Gastroenterol. 2000, 35, 1053–1059. [Google Scholar] [CrossRef] [PubMed]
- Tajik, N.; Frech, M.; Schulz, O.; Schälter, F.; Lucas, S.; Azizov, V.; Dürholz, K.; Steffen, F.; Omata, Y.; Rings, A.; et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat. Commun. 2020, 11, 1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef] [Green Version]
- Fasano, A.; Not, T.; Wang, W.; Uzzau, S.; Berti, I.; Tommasini, A.; Goldblum, S.E. Zonulin, a newly discovered modulator of intestinal permeability, and its expression in coeliac disease. Lancet 2000, 355, 1518–1519. [Google Scholar] [CrossRef]
- Häger, J.; Bang, H.; Hagen, M.; Frech, M.; Träger, P.; Sokolova, M.; Steffen, U.; Tascilar, K.; Sarter, K.; Schett, G.; et al. The Role of Dietary Fiber in Rheumatoid Arthritis Patients: A Feasibility Study. Nutrients 2019, 11, 2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, Y.-W.; Dong, J.-L.; Jian, Y.-J.; Fu, S.-H.; Chien, M.-W.; Liu, Y.-W.; Hsu, C.-Y.; Sytwu, H.-K. Gut Microbiota-Modulated Metabolomic Profiling Shapes the Etiology and Pathogenesis of Autoimmune Diseases. Microorganisms 2021, 9, 1930. https://doi.org/10.3390/microorganisms9091930
Tsai Y-W, Dong J-L, Jian Y-J, Fu S-H, Chien M-W, Liu Y-W, Hsu C-Y, Sytwu H-K. Gut Microbiota-Modulated Metabolomic Profiling Shapes the Etiology and Pathogenesis of Autoimmune Diseases. Microorganisms. 2021; 9(9):1930. https://doi.org/10.3390/microorganisms9091930
Chicago/Turabian StyleTsai, Yi-Wen, Jia-Ling Dong, Yun-Jie Jian, Shin-Huei Fu, Ming-Wei Chien, Yu-Wen Liu, Chao-Yuan Hsu, and Huey-Kang Sytwu. 2021. "Gut Microbiota-Modulated Metabolomic Profiling Shapes the Etiology and Pathogenesis of Autoimmune Diseases" Microorganisms 9, no. 9: 1930. https://doi.org/10.3390/microorganisms9091930
APA StyleTsai, Y.-W., Dong, J.-L., Jian, Y.-J., Fu, S.-H., Chien, M.-W., Liu, Y.-W., Hsu, C.-Y., & Sytwu, H.-K. (2021). Gut Microbiota-Modulated Metabolomic Profiling Shapes the Etiology and Pathogenesis of Autoimmune Diseases. Microorganisms, 9(9), 1930. https://doi.org/10.3390/microorganisms9091930





