Characterization of E. coli Isolates Producing Extended Spectrum Beta-Lactamase SHV-Variants from the Food Chain in Germany
Abstract
1. Introduction
2. Materials and Methods
3. Results
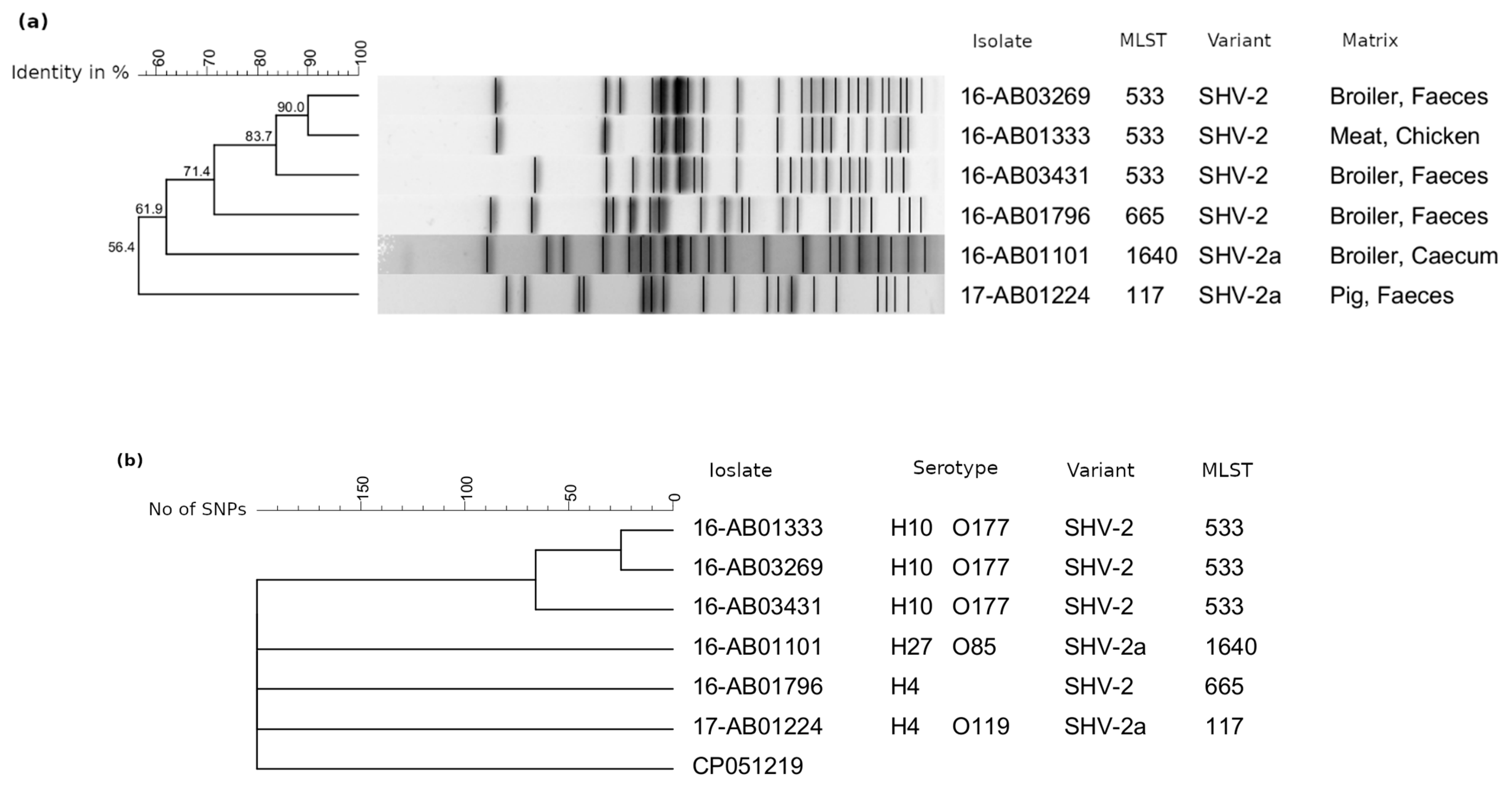
3.1. SVH-2-/SHV-2a-Producing E. coli
3.2. SHV-12-Producing E. coli
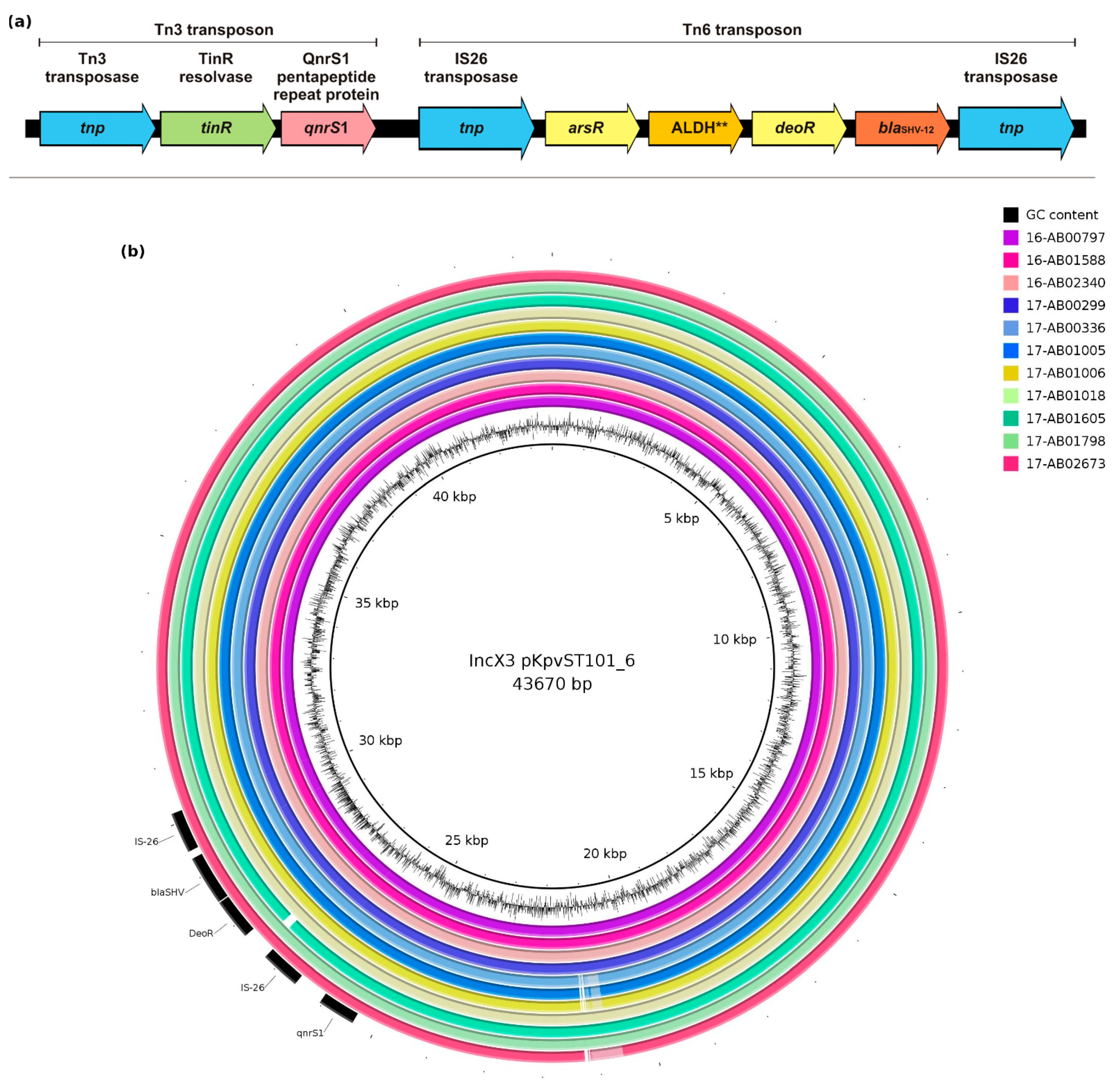
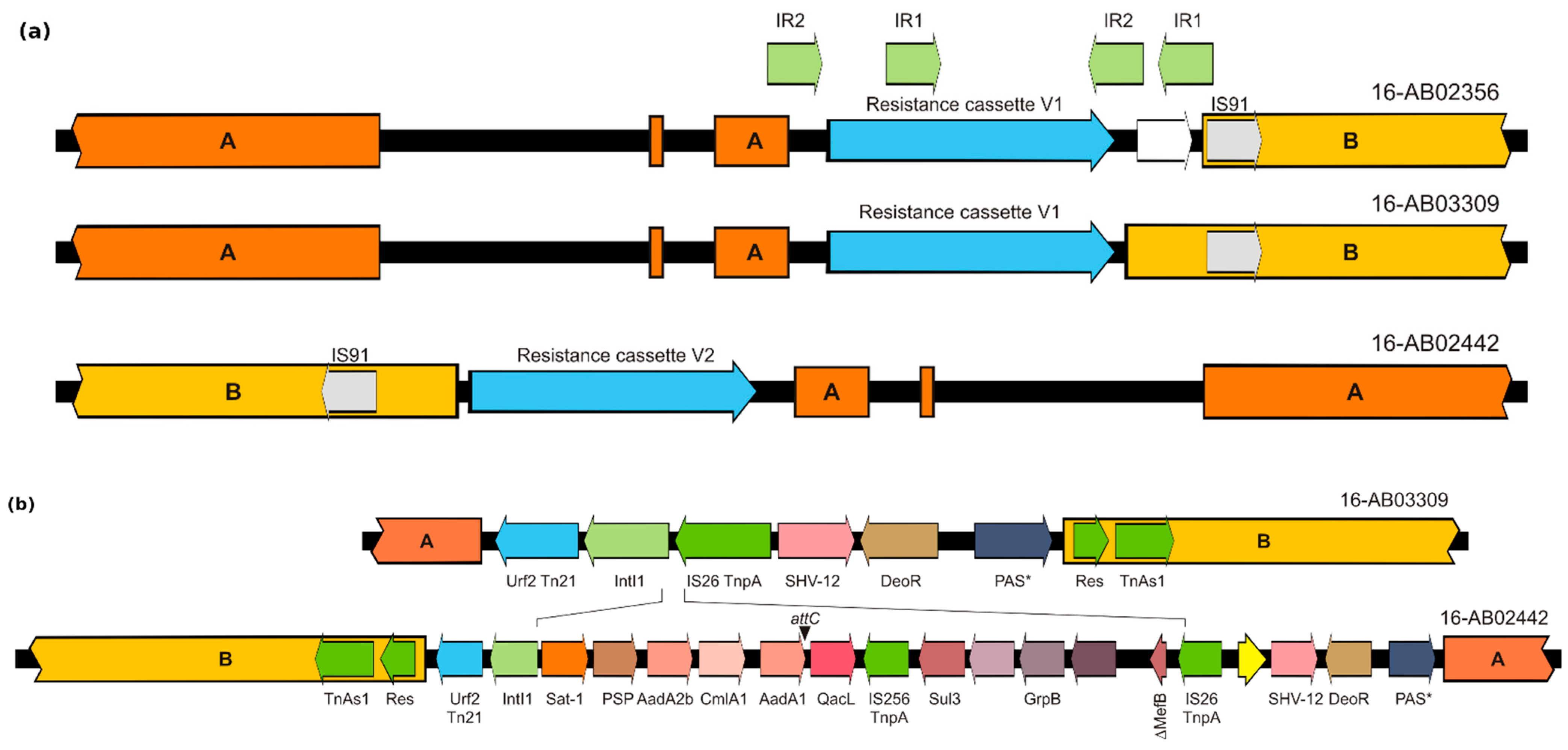
3.3. Genetic Environment of blaSHV-12
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ECDC; EFSA; EMA. ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food-producing animals: Joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA J. 2017, 15, e04872. [Google Scholar] [CrossRef]
- European Medicines Agency; European Surveillance of Veterinary Antimicrobial Consumption. Sales of Veterinary Antimicrobial agents in 31 European Countries in 2018 No. 10. 2020. Available online: https://www.ema.europa.eu/en/documents/report/sales-veterinary-antimicrobial-agents-31-european-countries-2018-trends-2010-2018-tenth-esvac-report_en.pdf (accessed on 27 January 2021).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef]
- Michael, G.B.; Freitag, C.; Wendlandt, S.; Eidam, C.; Feßler, A.T.; Lopes, G.V.; Kadlec, K.; Schwarz, S. Emerging issues in antimicrobial resistance of bacteria from food-producing animals. Future Microbiol. 2015, 10, 427–443. [Google Scholar] [CrossRef]
- Bradford, P.A. Extended-Spectrum -Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef]
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef]
- Rahman, S.U.; Ali, T.; Ali, I.; Khan, N.A.; Han, B.; Gao, J. The Growing Genetic and Functional Diversity of Extended Spectrum Beta-Lactamases. BioMed Res. Int. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017, 15, e04694. [Google Scholar] [CrossRef]
- Liebana, E.; Carattoli, A.; Coque, T.M.; Hasman, H.; Magiorakos, A.-P.; Mevius, D.; Peixe, L.; Poirel, L.; Schuepbach-Regula, G.; Torneke, K.; et al. Public Health Risks of Enterobacterial Isolates Producing Extended-Spectrum -Lactamases or AmpC -Lactamases in Food and Food-Producing Animals: An EU Perspective of Epidemiology, Analytical Methods, Risk Factors, and Control Options. Clin. Infect. Dis. 2013, 56, 1030–1037. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Podglajen, I.; Sougakoff, W.; Gutmann, L.; Nordmann, P. Emergence in Klebsiella pneumoniae of a Chromosome-Encoded SHV β-Lactamase That Compromises the Efficacy of Imipenem. Antimicrob. Agents Chemother. 2003, 47, 755–758. [Google Scholar] [CrossRef]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.; Kirkwood, R.N.; Laird, T.; Saputra, S.; Mitchell, T.; Singh, M.; Linn, B.; Abraham, R.J.; Pang, S.; Gordon, D.M.; et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 2018, 12, 2352–2362. [Google Scholar] [CrossRef]
- Touzain, F.; Le Devendec, L.; De Boisséson, C.; Baron, S.; Jouy, E.; Perrin-Guyomard, A.; Blanchard, Y.; Kempf, I. Characterization of plasmids harboring blaCTX-M and blaCMY genes in E. coli from French broilers. PLoS ONE 2018, 13, e0188768. [Google Scholar] [CrossRef]
- Irrgang, A.; Falgenhauer, L.; Fischer, J.; Ghosh, H.; Guiral, E.; Guerra, B.; Schmoger, S.; Imirzalioglu, C.; Chakraborty, T.; Hammerl, J.A.; et al. CTX-M-15-Producing E. coli Isolates from Food Products in Germany Are Mainly Associated with an IncF-Type Plasmid and Belong to Two Predominant Clonal E. coli Lineages. Front. Microbiol. 2017, 8, 2318. [Google Scholar] [CrossRef]
- Irrgang, A.; Hammerl, J.A.; Falgenhauer, L.; Guiral, E.; Schmoger, S.; Imirzalioglu, C.; Fischer, J.; Guerra, B.; Chakraborty, T.; Käsbohrer, A. Diversity of CTX-M-1-producing E. coli from German food samples and genetic diversity of the bla CTX-M-1 region on IncI1 ST3 plasmids. Vet. Microbiol. 2018, 221, 98–104. [Google Scholar] [CrossRef]
- Roschanski, N.; Fischer, J.; Guerra, B.; Roesler, U. Development of a Multiplex Real-Time PCR for the Rapid Detection of the Predominant Beta-Lactamase Genes CTX-M, SHV, TEM and CIT-Type AmpCs in Enterobacteriaceae. PLoS ONE 2014, 9, e100956. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Irrgang, A.; Tenhagen, B.-A.; Pauly, N.; Schmoger, S.; Kaesbohrer, A.; Hammerl, J.A. Characterization of VIM-1-Producing E. coli Isolated from a German Fattening Pig Farm by an Improved Isolation Procedure. Front. Microbiol. 2019, 10, 2256. [Google Scholar] [CrossRef]
- Irrgang, A.; Pauly, N.; Tenhagen, B.-A.; Grobbel, M.; Kaesbohrer, A.; Hammerl, A.J.A. Spill-Over from Public Health? First Detection of an OXA-48-Producing Escherichia coli in a German Pig Farm. Microorganisms 2020, 8, 855. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Klein, I.; Lanka, E.; Appel, B.; Hertwig, S. Genetic and Functional Properties of the Self-Transmissible Yersinia enterocolitica Plasmid pYE854, Which Mobilizes the Virulence Plasmid pYV. J. Bacteriol. 2008, 190, 991–1010. [Google Scholar] [CrossRef]
- Borowiak, M.; Szabo, I.; Baumann, B.; Junker, E.; Hammerl, J.A.; Kaesbohrer, A.; Malorny, B.; Fischer, J. VIM-1-producing Salmonella Infantis isolated from swine and minced pork meat in Germany. J. Antimicrob. Chemother. 2017, 72, 2131–2133. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.P.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E.; et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Pauly, N.; Hammerl, J.A.; Schwarz, S.; Grobbel, M.; Meemken, D.; Malorny, B.; Tenhagen, B.A.; Käsbohrer, A.; Irrgang, A. Co-occurrence of the blaVIM-1 and blaSHV-12 genes on an IncHI2 plasmid of an Escherichia coli isolate recovered from German livestock. J. Antimicrob. Chemother. 2021, 76, 531–533. [Google Scholar] [CrossRef]
- Ceccarelli, D.; Kant, A.; Van Essen-Zandbergen, A.; Dierikx, C.; Hordijk, J.; Wit, B.; Mevius, D.J.; Veldman, K.T. Diversity of Plasmids and Genes Encoding Resistance to Extended Spectrum Cephalosporins in Commensal Escherichia coli From Dutch Livestock in 2007–2017. Front. Microbiol. 2019, 10, 76. [Google Scholar] [CrossRef]
- Alt, K.; Lorenz, K.; Pfefferkorn, B.; Tenhagen, B.-A.; Wiehle, L. Berichte zur Lebensmittelsicherheit: Zoon-Ose-Monitoring 2017. 2018. Available online: https://www.bvl.bund.de/EN/Home/home_node.html;jsessionid=4CF15F08B391AA08E8296019E365ED8B.2 _cid369 (accessed on 9 July 2021).
- Federal Ministry of Food and Agriculture. Report of the Federal Ministry of Food and Agriculture on the Evaluation of the Antibiotics Minimisation Concept Introduced with the 16th Act to Amend the Medicinal Products Act (16th AMG Amendment): 2. Annex. Available online: https://www.bmel.de/EN/topics/animals/animal-health/Report-16thAMGAmendment.html (accessed on 13 August 2021).
- Konno, T.; Yatsuyanagi, J.; Saito, S. Virulence gene profiling of enteroaggregative Escherichia coli heat-stable enterotoxin 1-harboringE.coli(EAST1EC) derived from sporadic diarrheal patients. FEMS Immunol. Med. Microbiol. 2012, 64, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Veilleux, S.; Dubreuil, J.D. Presence of Escherichia coli carrying the EAST1 toxin gene in farm animals. Vet. Res. 2006, 37, 3–13. [Google Scholar] [CrossRef][Green Version]
- Zajacova, Z.S.; Konstantinova, L.; Alexa, P. Detection of virulence factors of Escherichia coli focused on prevalence of EAST1 toxin in stool of diarrheic and non-diarrheic piglets and presence of adhesion involving virulence factors in astA positive strains. Vet. Microbiol. 2012, 154, 369–375. [Google Scholar] [CrossRef]
- Dubreuil, J.D. EAST1 toxin: An enigmatic molecule associated with sporadic episodes of diarrhea in humans and animals. J. Microbiol. 2019, 57, 541–549. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Pouget, J.G.; Coutinho, F.J.; Reid-Smith, R.J.; Boerlin, P. Characterization ofblaSHVGenes on Plasmids from Escherichia coli and Salmonella enterica Isolates from Canadian Food Animals (2006–2007). Appl. Environ. Microbiol. 2013, 79, 3864–3866. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Nüesch-Inderbinen, M.; Morach, M.; Berner, A.Z.; Hächler, H.; Stephan, R. Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae Isolated from Vegetables Imported from the Dominican Republic, India, Thailand, and Vietnam. Appl. Environ. Microbiol. 2015, 81, 3115–3120. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, X.; An, S.; Zhang, X.; Chen, L.; Li, Y.; Xu, L.; Zhang, Y.; Gao, Z. Genetic environment of β-lactamase genes of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates from patients with lower respiratory tract infection in China. Chin. Med. J. 2014, 127, 2445–2450. [Google Scholar]
- Pons, M.J.; Vubil, D.; Guiral, E.; Jaintilal, D.; Fraile, O.; Soto, S.M.; Sigaúque, B.; Nhampossa, T.; Aide, P.; Alonso, P.L.; et al. Characterisation of extended-spectrum β-lactamases among Klebsiella pneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. J. Glob. Antimicrob. Resist. 2015, 3, 19–25. [Google Scholar] [CrossRef]
- Tóth, A.; Gacs, M.; Márialigeti, K.; Cech, G.; Füzi, M. Occurrence and regional distribution of SHV-type extended-spectrum β-lactamases in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 284–287. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Van Der Goot, J.; Bossers, A.; Betts, J.; Brouwer, M.S.M.; Kant, A.; Smith, H.; Ceccarelli, D.; Mevius, D. Genomic and functional characterisation of IncX3 plasmids encoding blaSHV-12 in Escherichia coli from human and animal origin. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.A.; Michael, G.B.; Li, J.; Somalo, S.; Simón, C.; Wang, Y.; Kaspar, H.; Kadlec, K.; Torres, C.; Schwarz, S. Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 2017, 72, 1589–1596. [Google Scholar] [CrossRef]
- Smith, H.E.; Bossers, A.; Harders, F.; Wu, G.; Woodford, N.; Schwarz, S.; Guerra, B.; Rodríguez, I.; Van Essen-Zandbergen, A.; Brouwer, M.S.M.; et al. Characterization of Epidemic IncI1-Iγ Plasmids Harboring Ambler Class A and C Genes in Escherichia coli and Salmonella enterica from Animals and Humans. Antimicrob. Agents Chemother. 2015, 59, 5357–5365. [Google Scholar] [CrossRef]


| Year | Matrix | # Isolates Investigated | # blaSHV Positive | Ratio in % | ||||
|---|---|---|---|---|---|---|---|---|
| 2016 | Broiler production total | 567 | 126 | 22.2 | ||||
| Broiler, feces | 166 | 33 | 19.9 | |||||
| Broiler, cecum | 184 | 42 | 22.8 | |||||
| Broiler, skin | 5 | 2 | 40.0 | |||||
| Chicken, meat | 212 | 49 | 23.1 | |||||
| 2016 | Turkey production chain total | 296 | 22 | 7.4 | ||||
| Turkey, cecum | 119 | 9 | 7.6 | |||||
| Turkey, meat | 177 | 13 | 7.3 | |||||
| 2017 | Pork production chain total | 344 | 11 | 3.2 | ||||
| Fattening pigs, feces | 325 | 11 | 3.4 | |||||
| Pork | 19 | 0 | 0.0 | |||||
| 2017 | Beef production chain total | 250 | 2 | 0.8 | ||||
| Veal calves, feces | 236 | 2 | 0.8 | |||||
| Beef | 14 | 0 | 0.0 | |||||
| 2016/ 2017 | other samples | Game, meat and feces (wild boar, deer, roe deer); vegetables, sprouts | 60 | 0 | 0.0 | |||
| Total | 1517 | 161 | 10.6 | |||||
| Isolate | Origin | SHV Variant and Localization (Size) | Inc. Group | Phylogenetic Group | MLST | |
|---|---|---|---|---|---|---|
| 16-AB01333 | Meat, chicken | SHV-2 | n.d. | n.d. | B1 | 533 |
| 16-AB01796 | Broiler, feces | SHV-2 | Chromosome | A | 665 | |
| 16-AB03269 | Broiler, feces | SHV-2 | n.d. | n.d. | B1 | 533 |
| 16-AB03431 | Broiler, feces | SHV-2 | n.d. | n.d. | B1 | 533 |
| 16-AB01101 | Broiler, cecum | SHV-2a | Plasmid (87 kb) | B/O | E | 1640 |
| 17-AB01224 | Pig, feces | SHV-2a | Plasmid (91 kb) | B/O | F | 117 |
| Isolate | Origin | SHV Variant | SHV Plasmid Size and Inc Group | Phylogenetic Group | MLST | |
|---|---|---|---|---|---|---|
| 17-AB02384 * | Pig, feces | SHV-12 | 298 kb | HI2 | B1 | 7593 |
| 17-AB01032 | Pig, feces | SHV-12 | 308 kb | HI2 | B1 | n.a. |
| 17-AB01030 | Pig, feces | SHV-12 | 295 kb | HI2 | C | 410 |
| 16-AB00888 | Turkey, meat | SHV-12 | 93 kb | IncI1 | A | n.a. |
| 16-AB00970 | Turkey, cecum | SHV-12 | 100 kb | IncI1 | F | n.a. |
| 16-AB01461 | Turkey, cecum | SHV-12 | 104 kb | IncI1 | D | n.a. |
| 16-AB01700 | Turkey, cecum | SHV-12 | 97 kb | IncI1, ST26 | B2 | 428 |
| 16-AB02356 | Turkey, cecum | SHV-12 | 84 kb | IncI1, ST3 | B1 | 162 |
| 16-AB03339 | Broiler, cecum | SHV-12 | 100 kb * | IncI1 | B1 | n.a. |
| 16-AB02442 * | Turkey, meat | SHV-12 | 110 kb * | IncI1, ST3 | D | 38 |
| 16-AB03438 * | Turkey, meat | SHV-12 | 105 kb 218 Kb | IncI1, ST26 IncFIB/FIC | B2 | 428 |
| 16-AB03529 | Turkey, meat | SHV-12 | 107 kb | IncI1, ST26 | E | 57 |
| 16-AB03530 | Turkey, meat | SHV-12 | 104 kb | IncI1 | F | n.a. |
| 16-AB03534 | Turkey, meat | SHV-12 | 28 kb 100 kb | n.a. IncI1 | A | n.a. |
| 16-AB03309 * | Broiler, cecum | SHV-12 | 91 kb | IncI1, ST3 | B1 | 1196 |
| 17-AB01138 | Calves, feces | SHV-12 | 90 Kb | IncI1 | B1 | n.a. |
| 17-AB01735 | Pig, feces | SHV-12 | 106 kb | n.t. | A1 | 1060 |
| 16-AB00677 | Turkey, cecum | SHV-12 | 39 kb | IncX3 | F | n.a. |
| 16-AB00797 | Broiler, cecum | SHV-12 | 39 kb | IncX3 | A | 10 |
| 16-AB01024 | Turkey, meat | SHV-12 | 41 kb | IncX3 | F | n.a. |
| 16-AB01389 | Broiler, cecum | SHV-12 | 41 kb | IncX3 | A | n.a. |
| 16-AB01588 | Broiler, cecum | SHV-12 | 43 kb | IncX3 | F | 117 |
| 16-AB02021 | Turkey, meat | SHV-12 | 40 kb | IncX3 | C | n.a. |
| 16-AB02026 | Turkey, cecum | SHV-12 | 42 kb | IncX3 | B1 | n.a. |
| 16-AB02340 | Turkey, cecum | SHV-12 | 42 kb | IncX3 | B1 | 9046 |
| 16-AB02352 | Broiler, cecum | SHV-12 | 44 kb | IncX3 | E | n.a. |
| 16-AB02541 | Broiler, cecum | SHV-12 | 43 kb | IncX3 | A | n.a. |
| 16-AB02638 | Turkey, cecum | SHV-12 | 38 kb | IncX3 | B2 | n.a. |
| 17-AB02673 * | Pig, feces | SHV-12 | 37 kb 43 kb | IncN IncX3 | C | 2230 |
| 16-AB02778 | Broiler, cecum | SHV-12 | 43 kb | IncX3 | F | n.a. |
| 16-AB03037 | Broiler, cecum | SHV-12 | 44 kb | IncX3 | F | n.a. |
| 16-AB03425 | Turkey, meat | SHV-12 | 46 kb | IncX3 | F | n.a. |
| 16-AB03444 | Turkey, meat | SHV-12 | 47 kb | IncX3 | D | n.a. |
| 16-AB03515 | Turkey, meat | SHV-12 | 61 kb | IncX3 | A | n.a. |
| 17-AB00299 | Broiler, cecum | SHV-12 | 41 kb | IncX3 | F | 117 |
| 17-AB00308 | Broiler, cecum | SHV-12 | 45 kb | IncX3 | A | n.a. |
| 17-AB00336 | Turkey, cecum | SHV-12 | 220 kb | IncX3 | F | 117 |
| 17-AB01005 | Pig, feces | SHV-12 | 39 kb | IncX3 | A | 1244 |
| 17-AB01006 | Pig, feces | SHV-12 | 40 kb | IncX3 | A | 10 |
| 17-AB01018 | Pig, feces | SHV-12 | 40 kb | IncX3 | C | 88 |
| 17-AB01605 | Pig, feces | SHV-12 | 42 kb | IncX3 | B1 | n.a. |
| 17-AB01798 | Pig, feces | SHV-12 | 42 kb | IncX3 | B1 | 641 |
| 17-AB02071 | Calves, feces | SHV-12 | 41 kb | IncX3 | B1 | 58 |
| 16-AB02401 * | Turkey, meat | SHV-12 | 37 kb | X1 | E | n.t. |
| 16-AB03659 | Turkey, meat | SHV-12 | 30 kb | X1 | F | n.a. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irrgang, A.; Zhao, G.; Juraschek, K.; Kaesbohrer, A.; Hammerl, J.A. Characterization of E. coli Isolates Producing Extended Spectrum Beta-Lactamase SHV-Variants from the Food Chain in Germany. Microorganisms 2021, 9, 1926. https://doi.org/10.3390/microorganisms9091926
Irrgang A, Zhao G, Juraschek K, Kaesbohrer A, Hammerl JA. Characterization of E. coli Isolates Producing Extended Spectrum Beta-Lactamase SHV-Variants from the Food Chain in Germany. Microorganisms. 2021; 9(9):1926. https://doi.org/10.3390/microorganisms9091926
Chicago/Turabian StyleIrrgang, Alexandra, Ge Zhao, Katharina Juraschek, Annemarie Kaesbohrer, and Jens A. Hammerl. 2021. "Characterization of E. coli Isolates Producing Extended Spectrum Beta-Lactamase SHV-Variants from the Food Chain in Germany" Microorganisms 9, no. 9: 1926. https://doi.org/10.3390/microorganisms9091926
APA StyleIrrgang, A., Zhao, G., Juraschek, K., Kaesbohrer, A., & Hammerl, J. A. (2021). Characterization of E. coli Isolates Producing Extended Spectrum Beta-Lactamase SHV-Variants from the Food Chain in Germany. Microorganisms, 9(9), 1926. https://doi.org/10.3390/microorganisms9091926






