Assessing the Risks of Potential Bacterial Pathogens Attaching to Different Microplastics during the Summer–Autumn Period in a Mariculture Cage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Data Collection
2.2. Construction of Bacterial Pathogens Database
2.3. Taxonomic Assignment of Bacterial Pathogens
2.4. Statistical Analyses
3. Results
3.1. Overview of Potential Bacterial Pathogens
3.2. Distribution of Pathogens across Different Microplastics and Water Fractions
3.3. The Succession of Pathogens on/in Microplastics and Water Fractions
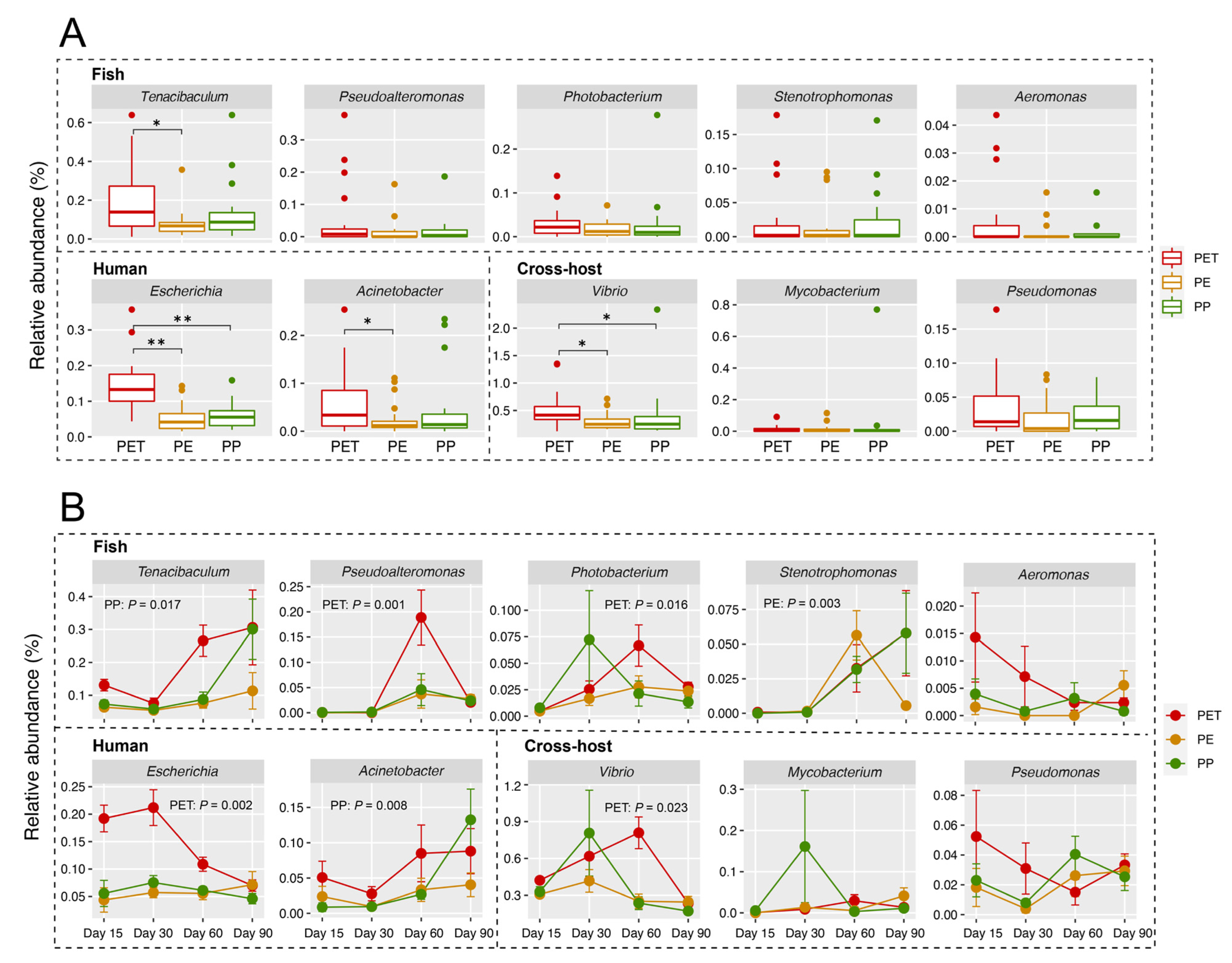
3.4. Representative Pathogens on Three Microplastics
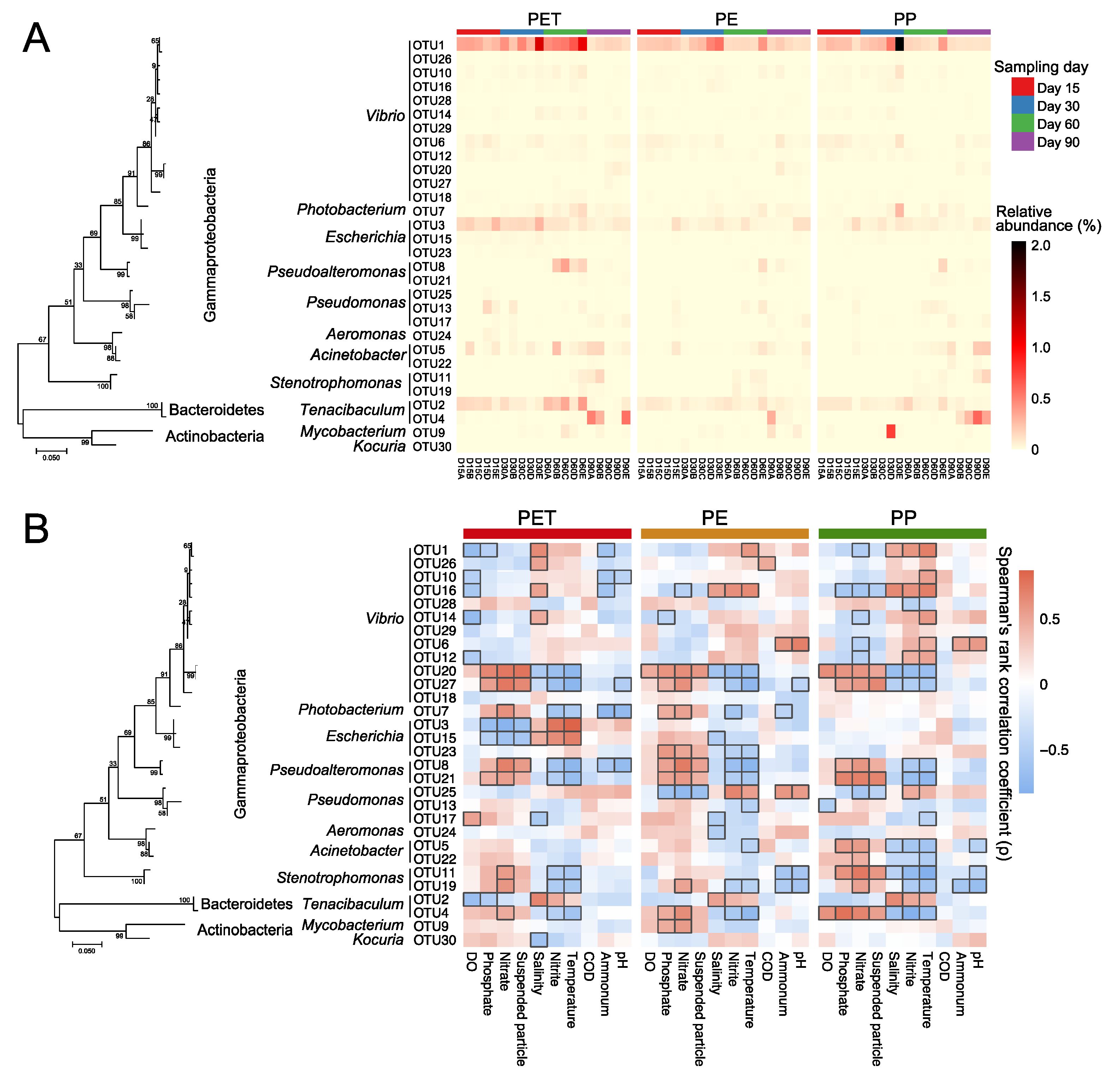
3.5. Correlation between the Microplastic-Attached Pathogens and Environmental Factors
4. Discussion
4.1. Pathogenic Bacteria Were Not Be Enriched on the Microplastics Compared with the Surrounding Environments
4.2. The Colonization and Succession of Pathogens on Microplastics Varied with Different Substrates
4.3. High Temperature and Nitrite in Mariculture May Increase the Risk of Pathogen Attachment on Microplastics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cole, M.; Lindeque, P.; Halsband, C.; Galloway, T.S. Microplastics as contaminants in the marine environment: A review. Mar. Pollut. Bull. 2011, 62, 2588–2597. [Google Scholar] [CrossRef] [PubMed]
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nat. Rev. Microbiol. 2020, 18, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Bowley, J.; Baker-Austin, C.; Porter, A.; Hartnell, R.; Lewis, C. Oceanic hitchhikers—Assessing pathogen risks from marine microplastic. Trends Microbiol. 2021, 29, 107–116. [Google Scholar] [CrossRef]
- Albano, M.; Panarello, G.; Di Paola, D.; Capparucci, F.; Crupi, R.; Gugliandolo, E.; Spanò, N.; Capillo, G.; Savoca, S. The influence of polystyrene microspheres abundance on development and feeding behavior of Artemia salina (Linnaeus, 1758). Appl. Sci. 2021, 11, 3352. [Google Scholar] [CrossRef]
- Albano, M.; Panarello, G.; Di Paola, D.; D’Angelo, G.; Granata, A.; Savoca, S.; Capillo, G. The mauve stinger Pelagia noctiluca (Cnidaria, Scyphozoa) plastics contamination, the Strait of Messina case. Int. J. Environ. Stud. 2021. [Google Scholar] [CrossRef]
- Capillo, G.; Savoca, S.; Panarello, G.; Mancuso, M.; Branca, C.; Romano, V.; D’Angelo, G.; Bottari, T.; Spanò, N. Quali-quantitative analysis of plastics and synthetic microfibers found in demersal species from Southern Tyrrhenian Sea (Central Mediterranean). Mar. Pollut. Bull. 2020, 150, 110596. [Google Scholar] [CrossRef]
- Prata, J.C.; da Costa, J.P.; Lopes, I.; Duarte, A.C.; Rocha-Santos, T. Environmental exposure to microplastics: An overview on possible human health effects. Sci. Total Environ. 2020, 702, 134455. [Google Scholar] [CrossRef]
- Watts, A.J.R.; Urbina, M.A.; Goodhead, R.; Moger, J.; Lewis, C.; Galloway, T.S. Effect of microplastic on the gills of the shore crab Carcinus maenas. Environ. Sci. Technol. 2016, 50, 5364–5369. [Google Scholar] [CrossRef] [Green Version]
- Avio, C.G.; Gorbi, S.; Milan, M.; Benedetti, M.; Fattorini, D.; d’Errico, G.; Pauletto, M.; Bargelloni, L.; Regoli, F. Pollutants bioavailability and toxicological risk from microplastics to marine mussels. Environ. Pollut. 2015, 198, 211–222. [Google Scholar] [CrossRef]
- Hermabessiere, L.; Dehaut, A.; Paul-Pont, I.; Lacroix, C.; Jezequel, R.; Soudant, P.; Duflos, G. Occurrence and effects of plastic additives on marine environments and organisms: A review. Chemosphere 2017, 182, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Mughini-Gras, L.; van der Plaats, R.Q.J.; van der Wielen, P.W.J.J.; Bauerlein, P.S.; de Roda Husman, A.M. Riverine microplastic and microbial community compositions: A field study in the Netherlands. Water Res. 2021, 192, 116852. [Google Scholar] [CrossRef]
- Pinto, M.; Langer, T.M.; Hueffer, T.; Hofmann, T.; Herndl, G.J. The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS ONE 2019, 14, e0217165. [Google Scholar] [CrossRef] [Green Version]
- Zettler, E.R.; Mincer, T.J.; Amaral-Zettler, L.A. Life in the “plastisphere”: Microbial communities on plastic marine debris. Environ. Sci. Technol. 2013, 47, 7137–7146. [Google Scholar] [CrossRef] [PubMed]
- Kirstein, I.V.; Kirmizi, S.; Wichels, A.; Garin-Fernandez, A.; Erler, R.; Loeder, M.; Gerdts, G. Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 2016, 120, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, A.; Oliver, D.M.; McCarron, A.; Quilliam, R.S. Colonisation of plastic pellets (nurdles) by E. coli at public bathing beaches. Mar. Pollut. Bull. 2019, 139, 376–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, J.P.; Schratzberger, M.; Sapp, M.; Osborn, A.M. Rapid bacterial colonization of low-density polyethylene microplastics in coastal sediment microcosms. BMC Microbiol. 2014, 14, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesy, K.; Labrenz, M.; Scales, B.S.; Kreikemeyer, B.; Oberbeckmann, S. Vibrio colonization is highly dynamic in early microplastic-associated biofilms as well as on field-collected microplastics. Microorganisms 2021, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Frere, L.; Maignien, L.; Chalopin, M.; Huvet, A.; Rinnert, E.; Morrison, H.; Kerninon, S.; Cassone, A.-L.; Lambert, C.; Reveillaud, J.; et al. Microplastic bacterial communities in the Bay of Brest: Influence of polymer type and size. Environ. Pollut. 2018, 242, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Jongmans-Hochschulz, E.; Mauder, N.; Imirzalioglu, C.; Wichels, A.; Gerdts, G. The Travelling Particles: Investigating microplastics as possible transport vectors for multidrug resistant E. coli in the Weser estuary (Germany). Sci. Total Environ. 2020, 720, 137603. [Google Scholar] [CrossRef]
- Wang, J.; Qin, X.; Guo, J.; Jia, W.; Wang, Q.; Zhang, M.; Huang, Y. Evidence of selective enrichment of bacterial assemblages and antibiotic resistant genes by microplastics in urban rivers. Water Res. 2020, 183, 1116113. [Google Scholar] [CrossRef]
- Oberbeckmann, S.; Loeder, M.G.J.; Labrenz, M. Marine microplastic-associated biofilms—A review. Environ. Chem. 2015, 12, 551–562. [Google Scholar] [CrossRef]
- Wu, X.; Pan, J.; Li, M.; Li, Y.; Bartlam, M.; Wang, Y. Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 2019, 165, 114979. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, J.; Zhao, Y.; Dai, H.; Jia, J.; Zhang, D. Plastisphere enrich antibiotic resistance genes and potential pathogenic bacteria in sewage with pharmaceuticals. Sci. Total Environ. 2021, 768, 144663. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Zhang, Z.; Zhu, J.; Shi, J.; Wei, H.; Xie, B.; Shi, H. Microplastics act as vectors for antibiotic resistance genes in landfill leachate: The enhanced roles of the long-term aging process. Environ. Pollut. 2021, 270, 116278. [Google Scholar] [CrossRef]
- Bryant, J.A.; Clemente, T.M.; Viviani, D.A.; Fong, A.A.; Thomas, K.A.; Kemp, P.; Karl, D.M.; White, A.E.; DeLong, E.F. Diversity and activity of communities inhabiting plastic debris in the North Pacific Gyre. mSystems 2016, 1, e00024-16. [Google Scholar] [CrossRef] [Green Version]
- Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 2018, 8, 2709. [Google Scholar] [CrossRef] [Green Version]
- Pauly, D.; Zeller, D. Catch reconstructions reveal that global marine fisheries catches are higher than reported and declining. Nat. Commun. 2016, 7, 10244. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, M.; Zhang, L.; Wang, K.; Yu, X.; Zheng, Z.; Zheng, R. Sorption behaviors of phenanthrene on the microplastics identified in a mariculture farm in Xiangshan Bay, southeastern China. Sci. Total Environ. 2018, 628–629, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Andrady, A.L. Microplastics in the marine environment. Mar. Pollut. Bull. 2011, 62, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, I.A.; Thiel, M. Floating marine debris in fjords, gulfs and channels of southern Chile. Mar. Pollut. Bull. 2009, 58, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Love, D.C.; Rochman, C.M.; Neff, R.A. Microplastics in seafood and the implications for human health. Curr. Environ. Health Rep. 2018, 5, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Hou, D.; Hong, M.; Wang, K.; Yan, H.; Wang, Y.; Dong, P.; Li, D.; Liu, K.; Zhou, Z.; Zhang, D. Prokaryotic community succession and assembly on different types of microplastics in a mariculture cage. Environ. Pollut. 2021, 268, 115756. [Google Scholar] [CrossRef]
- AQSIQ. The Specification for Marine Monitoring of China—Part 4: Seawater Analysis (GB 17378.4-2007); General Administration of Quality Supervision, Inspection and Quarantine (AQSIQ) of the People’s Republic of China: Beijing, China, 2007. (In Chinese) [Google Scholar]
- Schweitzer-Natan, O.; Lalzar, M.; Sher, D.; Sukenik, A. Particle-associated microbial community in a subtropical lake during thermal mixing and phytoplankton succession. Front. Microbiol. 2019, 10, 2142. [Google Scholar] [CrossRef]
- Tang, X.; Chao, J.; Gong, Y.; Wang, Y.; Wilhelm, S.W.; Gao, G. Spatiotemporal dynamics of bacterial community composition in large shallow eutrophic Lake Taihu: High overlap between free-living and particle-attached assemblages. Limnol. Oceanogr. 2017, 62, 1366–1382. [Google Scholar] [CrossRef]
- Mestre, M.; Borrull, E.; Sala, M.M.; Gasol, J.M. Patterns of bacterial diversity in the marine planktonic particulate matter continuum. ISME J. 2017, 11, 999–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milici, M.; Tomasch, J.; Wos-Oxley, M.L.; Wang, H.; Jáuregui, R.; Camarinha-Silva, A.; Deng, Z.-L.; Plumeier, I.; Giebel, H.-A.; Wurst, M.; et al. Low diversity of planktonic bacteria in the tropical ocean. Sci. Rep. 2016, 6, 19054. [Google Scholar] [CrossRef] [PubMed]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Lanzen, A.; Davenport, R.J.; Turnbaugh, P.J. Removing noise from pyrosequenced amplicons. BMC Bioinform. 2011, 12, 38. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Dong, P.; Guo, H.; Wang, Y.; Cheng, H.; Wang, K.; Hong, M.; Hou, D.; Wu, Y.; Zhang, D. DPiWE: A curated database for pathogenic bacteria involved in water environment. J. Fish. China 2021. (In Chinese) [CrossRef]
- Austin, B.; Austin, D. Bacterial Fish Pathogens: Diseases of Farmed and Wild Fish, 6th ed.; Springer International Publishing: Dordrecht, The Netherlands, 2016. [Google Scholar]
- Fang, H.; Chen, C.; Zhang, X. Aquacultural Animal Pathogenic Bacteriology; China Agricultural Press: Beijing, China, 2016; pp. 171–693. [Google Scholar]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; An, X.; Li, H.; Su, J.; Ma, Y.; Zhu, Y.-G. Long-term field application of sewage sludge increases the abundance of antibiotic resistance genes in soil. Environ. Int. 2016, 92–93, 1–10. [Google Scholar] [CrossRef]
- Kanehisa, M. Inferring antimicrobial resistance from pathogen genomes in KEGG. Methods Mol. Biol. 2018, 1807, 225–239. [Google Scholar]
- Parte, A.; Carbasse, J.; Meier-Kolthoff, J.; Reimer, L.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Micr. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Parte, A. LPSN—List of Prokaryotic names with Standing in Nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5–7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 5 July 2021).
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 5 July 2021).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, N.; Ferry, M. ggtern: Ternary Diagrams Using ggplot2. J. Stat. Softw. 2018, 87, 1. [Google Scholar] [CrossRef] [Green Version]
- Lyons, M.M.; Ward, J.E.; Gaff, H.; Hicks, R.E.; Drake, J.M.; Dobbs, F.C. Theory of island biogeography on a microscopic scale: Organic aggregates as islands for aquatic pathogens. Aquat. Microb. Ecol. 2010, 60, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Fang, T.; Cui, Q.; Huang, Y.; Dong, P.; Wang, H.; Liu, W.-T.; Ye, Q. Distribution comparison and risk assessment of free-floating and particle-attached bacterial pathogens in urban recreational water: Implications for water quality management. Sci. Total Environ. 2018, 613, 428–438. [Google Scholar] [CrossRef]
- Mammo, F.K.; Amoah, I.D.; Gani, K.M.; Pillay, L.; Ratha, S.K.; Bux, F.; Kumari, S. Microplastics in the environment: Interactions with microbes and chemical contaminants. Sci. Total Environ. 2020, 743, 140518. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Liang, X.; Soupir, M.L.; Jarboe, L.R. Cellular, particle and environmental parameters influencing attachment in surface waters: A review. J. Appl. Microbiol. 2015, 119, 315–330. [Google Scholar] [CrossRef] [Green Version]
- Oberbeckmann, S.; Labrenz, M. Marine microbial assemblages on microplastics: Diversity, adaptation, and role in degradation. Annu. Rev. Mar. Sci. 2020, 12, 209–232. [Google Scholar] [CrossRef] [Green Version]
- Kesy, K.; Oberbeckmann, S.; Kreikemeyer, B.; Labrenz, M. Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in Baltic Sea mesocosms. Front. Microbiol. 2019, 10, 1665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhagwat, G.; Zhu, Q.; O’Connor, W.; Subashchandrabose, S.; Grainge, I.; Knight, R.; Palanisami, T. Exploring the composition and functions of plastic microbiome using whole-genome sequencing. Environ. Sci. Technol. 2021, 55, 4899–4913. [Google Scholar] [CrossRef] [PubMed]
- Amaral-Zettler, L.A.; Zettler, E.R.; Slikas, B.; Boyd, G.D.; Melvin, D.W.; Morrall, C.E.; Proskurowski, G.; Mincer, T.J. The biogeography of the Plastisphere: Implications for policy. Front. Ecol. Environ. 2015, 13, 541–546. [Google Scholar] [CrossRef]
- Laverty, A.L.; Primpke, S.; Lorenz, C.; Gerdts, G.; Dobbs, F.C. Bacterial biofilms colonizing plastics in estuarine waters, with an emphasis on Vibrio spp. and their antibacterial resistance. PLoS ONE 2020, 15, e0237704. [Google Scholar]
- Schmidt, V.T.; Reveillaud, J.; Zettler, E.; Mincer, T.J.; Murphy, L.; Amaral-Zettler, L.A. Oligotyping reveals community level habitat selection within the genus Vibrio. Front. Microbiol. 2014, 5, 563. [Google Scholar] [CrossRef]
- Chatterjee, S. Vibrio related diseases in aquaculture and development of rapid and accurate identification methods. J. Mar. Sci. Res. Dev. 2012, S1, 002. [Google Scholar]
- Fernández-Álvarez, C.; Santos, Y. Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biot. 2018, 102, 9973–9989. [Google Scholar] [CrossRef]
- Piñeiro-Vidal, M.; Riaza, A.; Santos, Y. Tenacibaculum discolor sp. nov. and Tenacibaculum gallaicum sp. nov., isolated from sole (Solea senegalensis) and turbot (Psetta maxima) culture systems. Int. J. Syst. Evol. Microbiol. 2008, 58, 21–25. [Google Scholar] [CrossRef]
- Ashrafudoulla, M.; Mizan, M.F.R.; Park, H.; Byun, K.H.; Lee, N.; Park, S.H.; Ha, S.D. Genetic relationship, virulence factors, drug resistance profile and biofilm formation ability of Vibrio parahaemolyticus isolated from mussel. Front. Microbiol. 2019, 10, 513. [Google Scholar] [CrossRef] [PubMed]
- Froelich, B.A.; Daines, D.A. In hot water: Effects of climate change on Vibrio–human interactions. Environ. Microbiol. 2020, 22, 4101–4111. [Google Scholar] [CrossRef] [Green Version]
- Vezzulli, L.; Previati, M.; Pruzzo, C.; Marchese, A.; Bourne, D.G.; Cerrano, C.; Consortium, t.V. Vibrio infections triggering mass mortality events in a warming Mediterranean Sea. Environ. Microbiol. 2010, 12, 2007–2019. [Google Scholar] [CrossRef] [PubMed]
- Sobrinho, P.d.S.C.; Destro, M.T.; Franco, B.D.G.M.; Landgraf, M. Correlation between environmental factors and prevalence of Vibrio parahaemolyticus in oysters harvested in the southern coastal area of Sao Paulo State, Brazil. Appl. Environ. Microbiol. 2010, 76, 1290–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vezzulli, L.; Brettar, I.; Pezzati, E.; Reid, P.C.; Colwell, R.R.; Höfle, M.G.; Pruzzo, C. Long-term effects of ocean warming on the prokaryotic community: Evidence from the vibrios. ISME J. 2012, 6, 21–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Datta, S. Management of Water Quality in Intensive Aquaculture; Central Institute of Fisheries Education: Mumbai, India, 2012; Volume 5. [Google Scholar]
- Seo, J.S.; Haque, M.N.; Nam, S.-E.; Kim, B.-M.; Rhee, J.-S. Inorganic nitrogen compounds reduce immunity and induce oxidative stress in red seabream. Fish Shellfish Immunol. 2020, 104, 237–244. [Google Scholar] [CrossRef] [PubMed]



| Environmental Factors | PET | PE | PP | LPA | SPA | FL |
|---|---|---|---|---|---|---|
| Temperature | 0.681 | 0.437 | 0.526 | 0.436 | 0.570 | 0.714 |
| Nitrate | 0.647 | 0.443 | 0.489 | 0.491 | 0.619 | 0.715 |
| DO | 0.639 | 0.323 | 0.437 | 0.292 | 0.440 | 0.624 |
| Phosphate | 0.359 | 0.211 | 0.254 | 0.601 | 0.622 | 0.614 |
| Salinity | 0.556 | 0.179 | 0.315 | 0.350 | 0.417 | 0.603 |
| Nitrite | 0.184 | 0.079 | 0.125 | 0.561 | 0.557 | 0.451 |
| pH | 0.026 | 0.098 | −0.110 | 0.203 | 0.184 | 0.101 |
| Ammonium | −0.026 | 0.017 | −0.062 | 0.299 | 0.224 | 0.096 |
| Suspended particle | −0.043 | −0.104 | −0.020 | 0.340 | 0.238 | 0.215 |
| COD | −0.085 | −0.108 | −0.130 | −0.032 | 0.029 | −0.064 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, D.; Hong, M.; Wang, Y.; Dong, P.; Cheng, H.; Yan, H.; Yao, Z.; Li, D.; Wang, K.; Zhang, D. Assessing the Risks of Potential Bacterial Pathogens Attaching to Different Microplastics during the Summer–Autumn Period in a Mariculture Cage. Microorganisms 2021, 9, 1909. https://doi.org/10.3390/microorganisms9091909
Hou D, Hong M, Wang Y, Dong P, Cheng H, Yan H, Yao Z, Li D, Wang K, Zhang D. Assessing the Risks of Potential Bacterial Pathogens Attaching to Different Microplastics during the Summer–Autumn Period in a Mariculture Cage. Microorganisms. 2021; 9(9):1909. https://doi.org/10.3390/microorganisms9091909
Chicago/Turabian StyleHou, Dandi, Man Hong, Yanting Wang, Pengsheng Dong, Huangwei Cheng, Huizhen Yan, Zhiyuan Yao, Daoji Li, Kai Wang, and Demin Zhang. 2021. "Assessing the Risks of Potential Bacterial Pathogens Attaching to Different Microplastics during the Summer–Autumn Period in a Mariculture Cage" Microorganisms 9, no. 9: 1909. https://doi.org/10.3390/microorganisms9091909
APA StyleHou, D., Hong, M., Wang, Y., Dong, P., Cheng, H., Yan, H., Yao, Z., Li, D., Wang, K., & Zhang, D. (2021). Assessing the Risks of Potential Bacterial Pathogens Attaching to Different Microplastics during the Summer–Autumn Period in a Mariculture Cage. Microorganisms, 9(9), 1909. https://doi.org/10.3390/microorganisms9091909







