Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments
Abstract
1. Introduction
2. Persistent Strains of L. monocytogenes
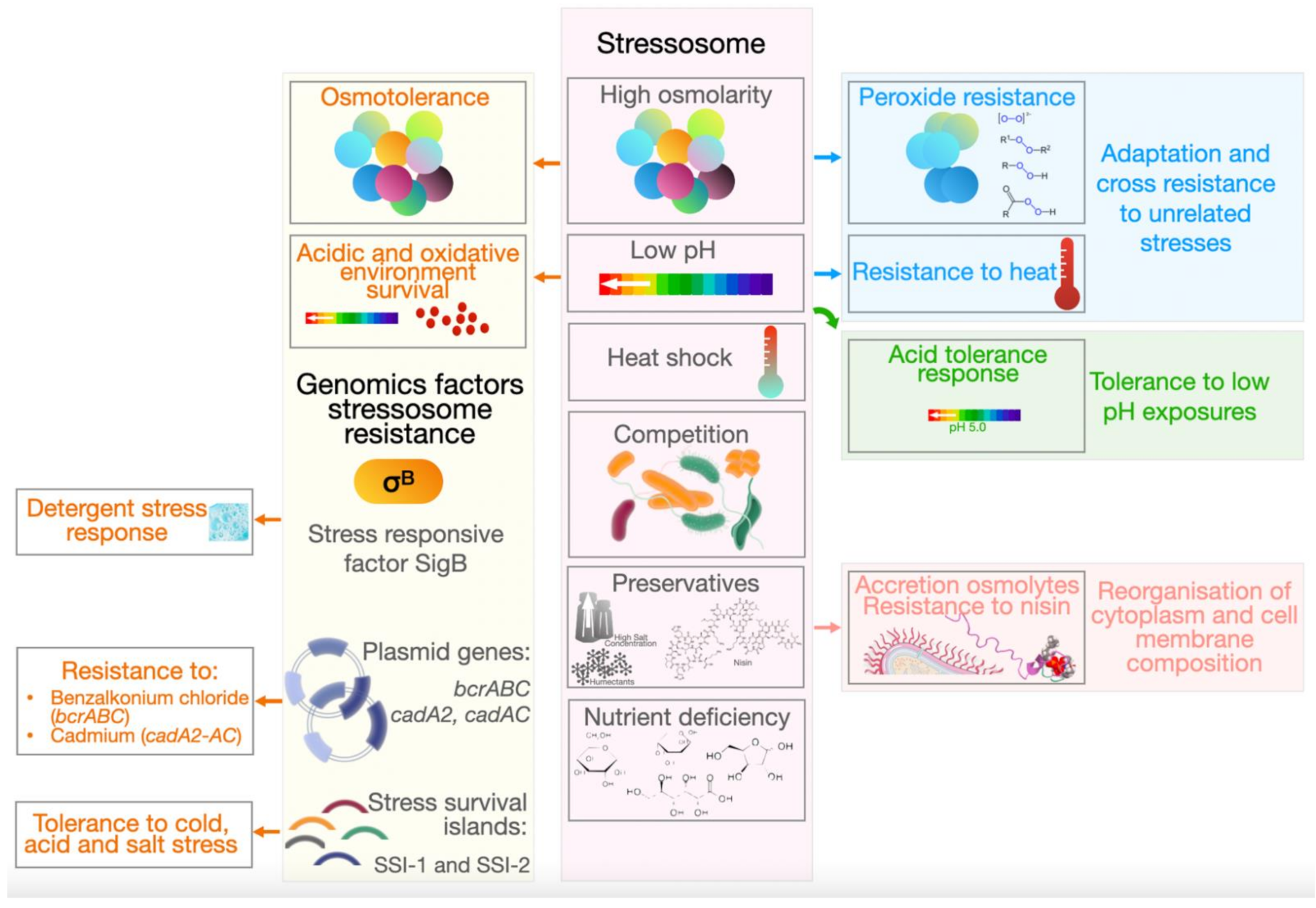
3. Stress Resistance
4. Biofilm Formation
5. Biocide Susceptibility
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Buchrieser, C.; Rusniok, C.; Kunst, F.; Cossart, P.; Glaser, P.; Listeria, C. Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: Clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 2003, 35, 207–213. [Google Scholar] [CrossRef]
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991, 55, 476–511. [Google Scholar] [CrossRef]
- Quereda, J.J.; Leclercq, A.; Moura, A.; Vales, G.; Gomez-Martin, A.; Garcia-Munoz, A.; Thouvenot, P.; Tessaud-Rita, N.; Bracq-Dieye, H.; Lecuit, M. Listeria valentina sp. nov., isolated from a water trough and the faeces of healthy sheep. Int. J. Syst. Evol. Microbiol. 2020, 70, 5868–5879. [Google Scholar] [CrossRef]
- Seeliger, H.P. Modern taxonomy of the Listeria group relationship to its pathogenicity. Clin. Investig. Med. 1984, 7, 217–221. [Google Scholar]
- Guillet, C.; Join-Lambert, O.; Le Monnier, A.; Leclercq, A.; Mechai, F.; Mamzer-Bruneel, M.F.; Bielecka, M.K.; Scortti, M.; Disson, O.; Berche, P.; et al. Human listeriosis caused by Listeria ivanovii. Emerg. Infect. Dis. 2010, 16, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Cummins, J.; Casey, P.G.; Joyce, S.A.; Gahan, C.G. A mariner transposon-based signature-tagged mutagenesis system for the analysis of oral infection by Listeria monocytogenes. PLoS ONE 2013, 8, e75437. [Google Scholar] [CrossRef]
- Alexander, A.V.; Walker, R.L.; Johnson, B.J.; Charlton, B.R.; Woods, L.W. Bovine abortions attributable to Listeria ivanovii: Four cases (1988–1990). J. Am. Vet. Med. Assoc. 1992, 200, 711–714. [Google Scholar]
- Chand, P.; Sadana, J.R. Outbreak of Listeria ivanovii abortion in sheep in India. Vet. Rec. 1999, 145, 83–84. [Google Scholar] [CrossRef]
- Graves, L.M.; Helsel, L.O.; Steigerwalt, A.G.; Morey, R.E.; Daneshvar, M.I.; Roof, S.E.; Orsi, R.H.; Fortes, E.D.; Milillo, S.R.; den Bakker, H.C.; et al. Listeria marthii sp. nov., isolated from the natural environment, Finger Lakes National Forest. Int. J. Syst. Evol. Microbiol. 2010, 60, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Chiara, M.; Caruso, M.; D’Erchia, A.M.; Manzari, C.; Fraccalvieri, R.; Goffredo, E.; Latorre, L.; Miccolupo, A.; Padalino, I.; Santagada, G.; et al. Comparative Genomics of Listeria sensu lato: Genus-wide differences in evolutionary dynamics and the progressive gain of complex, potentially pathogenicity-related traits through lateral gene transfer. Genome Biol. Evol. 2015, 7, 2154–2172. [Google Scholar] [CrossRef]
- Rocourt, J.; Seeliger, H.P. Distribution of species of the genus Listeria. Zentralbl. Bakteriol. Mikrobiol. Hyg. A 1985, 259, 317–330. [Google Scholar]
- Huang, B.; Eglezos, S.; Heron, B.A.; Smith, H.; Graham, T.; Bates, J.; Savill, J. Comparison of multiplex PCR with conventional biochemical methods for the identification of Listeria spp. isolates from food and clinical samples in Queensland, Australia. J. Food Prot. 2007, 70, 1874–1880. [Google Scholar] [CrossRef]
- Nayak, D.N.; Savalia, C.V.; Kalyani, I.H.; Kumar, R.; Kshirsagar, D.P. Isolation, identification, and characterization of Listeria spp. from various animal origin foods. Vet. World 2015, 8, 695–701. [Google Scholar] [CrossRef]
- Dahshan, H.; Merwad, A.M.; Mohamed, T.S. Listeria species in broiler poultry farms: Potential public health hazards. J. Microbiol. Biotechnol. 2016, 26, 1551–1556. [Google Scholar] [CrossRef]
- Husu, J.R. Epidemiological studies on the occurrence of Listeria monocytogenes in the feces of dairy cattle. J. Vet. Med. B 1990, 37, 276–282. [Google Scholar] [CrossRef]
- Yin, Y.; Yao, H.; Doijad, S.; Kong, S.; Shen, Y.; Cai, X.; Tan, W.; Wang, Y.; Feng, Y.; Ling, Z.; et al. A hybrid sub-lineage of Listeria monocytogenes comprising hypervirulent isolates. Nat. Commun. 2019, 10, 4283. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yao, H.; Chen, S.; Sun, X.; Yin, Y.; Jiao, X. Rapid detection of hypervirulent serovar 4h Listeria monocytogenes by multiplex PCR. Front. Microbiol. 2020, 11, 1309. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Piffaretti, J.C.; Kressebuch, H.; Aeschbacher, M.; Bille, J.; Bannerman, E.; Musser, J.M.; Selander, R.K.; Rocourt, J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc. Natl. Acad. Sci. USA 1989, 86, 3818–3822. [Google Scholar] [CrossRef]
- Vines, A.; Swaminathan, B. Identification and characterization of nucleotide sequence differences in three virulence-associated genes of Listeria monocytogenes strains representing clinically important serotypes. Curr. Microbiol. 1998, 36, 309–318. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef]
- Vazquez-Boland, J.A.; Dominguez-Bernal, G.; Gonzalez-Zorn, B.; Kreft, J.; Goebel, W. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 2001, 3, 571–584. [Google Scholar] [CrossRef]
- Gudmundsdottir, S.; Gudbjornsdottir, B.; Lauzon, H.L.; Einarsson, H.; Kristinsson, K.G.; Kristjansson, M. Tracing Listeria monocytogenes isolates from cold-smoked salmon and its processing environment in Iceland using pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2005, 101, 41–51. [Google Scholar] [CrossRef]
- Guerini, M.N.; Brichta-Harhay, D.M.; Shackelford, T.S.; Arthur, T.M.; Bosilevac, J.M.; Kalchayanand, N.; Wheeler, T.L.; Koohmaraie, M. Listeria prevalence and Listeria monocytogenes serovar diversity at cull cow and bull processing plants in the United States. J. Food Prot. 2007, 70, 2578–2582. [Google Scholar] [CrossRef][Green Version]
- Zhu, L.; Feng, X.; Zhang, L.; Zhu, R.; Luo, X. Prevalence and serotypes of Listeria monocytogenes contamination in Chinese beef processing plants. Foodborne Pathog. Dis. 2012, 9, 556–560. [Google Scholar] [CrossRef]
- Almeida, G.; Magalhaes, R.; Carneiro, L.; Santos, I.; Silva, J.; Ferreira, V.; Hogg, T.; Teixeira, P. Foci of contamination of Listeria monocytogenes in different cheese processing plants. Int. J. Food Microbiol. 2013, 167, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.C.; Dias, M.R.; Cossi, M.V.; Lanna, F.G.; Cavicchioli, V.Q.; Vallim, D.C.; Pinto, P.S.; Hofer, E.; Nero, L.A. Serotypes and pulsotypes diversity of Listeria monocytogenes in a beef-processing environment. Foodborne Pathog. Dis. 2015, 12, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Tau, N.P.; Smouse, S.L.; Allam, M.; Ismail, A.; Ramalwa, N.R.; Disenyeng, B.; Ngomane, M.; Thomas, J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: Laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodborne Pathog. Dis. 2019, 16, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Painset, A.; Bjorkman, J.T.; Kiil, K.; Guillier, L.; Mariet, J.F.; Felix, B.; Amar, C.; Rotariu, O.; Roussel, S.; Perez-Reche, F.; et al. LiSEQ—Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 2019, 5. [Google Scholar] [CrossRef]
- Rietberg, K.; Lloyd, J.; Melius, B.; Wyman, P.; Treadwell, R.; Olson, G.; Kang, M.G.; Duchin, J.S. Outbreak of Listeria monocytogenes infections linked to a pasteurized ice cream product served to hospitalized patients. Epidemiol. Infect. 2016, 144, 2728–2731. [Google Scholar] [CrossRef][Green Version]
- EFSA; ECDC. The European Union One Health 2019 Zoonosesn Report. EFSA J. 2021, 19, 286. [Google Scholar]
- Deurenberg, R.H.; Bathoorn, E.; Chlebowicz, M.A.; Couto, N.; Ferdous, M.; Garcia-Cobos, S.; Kooistra-Smid, A.M.D.; Raangs, E.C.; Rosema, S.; Veloo, A.C.M.; et al. Reprint of “Application of next generation sequencing in clinical microbiology and infection prevention”. J. Biotechnol. 2017, 250, 2–10. [Google Scholar] [CrossRef]
- Moran-Gilad, J. Whole genome sequencing (WGS) for food-borne pathogen surveillance and control—Taking the pulse. Eurosurveillance 2017, 22, 30547. [Google Scholar] [CrossRef] [PubMed]
- Das, A. Listeriosis in Australia—January to july 2018. Global Biosecurity 2019, 1, 1. [Google Scholar] [CrossRef]
- Kwong, J.C.; Mercoulia, K.; Tomita, T.; Easton, M.; Li, H.Y.; Bulach, D.M.; Stinear, T.P.; Seemann, T.; Howden, B.P. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 2016, 54, 333–342. [Google Scholar] [CrossRef]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef]
- Reddy, S.; Lawrence, M.L. Virulence characterization of Listeria monocytogenes. Methods Mol. Biol. 2014, 1157, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Rolhion, N.; Cossart, P. How the study of Listeria monocytogenes has led to new concepts in biology. Future Microbiol. 2017, 12, 621–638. [Google Scholar] [CrossRef]
- Pizarro-Cerda, J.; Cossart, P. Listeria monocytogenes: Cell biology of invasion and intracellular growth. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes virulence, antimicrobial resistance and environmental persistence: A review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, M. Listeria monocytogenes, a model in infection biology. Cell Microbiol. 2020, 22, e13186. [Google Scholar] [CrossRef] [PubMed]
- Disson, O.; Moura, A.; Lecuit, M. Making sense of the biodiversity and virulence of Listeria monocytogenes. Trends Microbiol. 2021, 29, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Nightingale, K.K.; Windham, K.; Martin, K.E.; Yeung, M.; Wiedmann, M. Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 2005, 71, 8764–8772. [Google Scholar] [CrossRef]
- Hingston, P.; Chen, J.; Dhillon, B.K.; Laing, C.; Bertelli, C.; Gannon, V.; Tasara, T.; Allen, K.; Brinkman, F.S.; Truelstrup Hansen, L.; et al. Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 2017, 8, 369. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Author correction: Hypervirulent Listeria monocytogenes clones’ adaptation to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 3619. [Google Scholar] [CrossRef] [PubMed]
- Pistor, S.; Chakraborty, T.; Niebuhr, K.; Domann, E.; Wehland, J. The ActA protein of Listeria monocytogenes acts as a nucleator inducing reorganization of the actin cytoskeleton. EMBO J. 1994, 13, 758–763. [Google Scholar] [CrossRef]
- Cheng, M.I.; Chen, C.; Engstrom, P.; Portnoy, D.A.; Mitchell, G. Actin-based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell Microbiol. 2018, 20, e12854. [Google Scholar] [CrossRef]
- Travier, L.; Lecuit, M. Listeria monocytogenes ActA: A new function for a ‘classic’ virulence factor. Curr. Opin. Microbiol. 2014, 17, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Dominguez, C.; Vazquez-Boland, J.A.; Carrasco-Marin, E.; Lopez-Mato, P.; Leyva-Cobian, F. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 1997, 65, 78–88. [Google Scholar] [CrossRef]
- Marquis, H.; Goldfine, H.; Portnoy, D.A. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 1997, 137, 1381–1392. [Google Scholar] [CrossRef]
- Gandhi, A.J.; Perussia, B.; Goldfine, H. Listeria monocytogenes phosphatidylinositol (PI)-specific phospholipase C has low activity on glycosyl-PI-anchored proteins. J. Bacteriol. 1993, 175, 8014–8017. [Google Scholar] [CrossRef][Green Version]
- Camilli, A.; Tilney, L.G.; Portnoy, D.A. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 1993, 8, 143–157. [Google Scholar] [CrossRef]
- Marquis, H.; Doshi, V.; Portnoy, D.A. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 1995, 63, 4531–4534. [Google Scholar] [CrossRef]
- Scortti, M.; Monzo, H.J.; Lacharme-Lora, L.; Lewis, D.A.; Vazquez-Boland, J.A. The PrfA virulence regulon. Microbes Infect. 2007, 9, 1196–1207. [Google Scholar] [CrossRef] [PubMed]
- Gedde, M.M.; Higgins, D.E.; Tilney, L.G.; Portnoy, D.A. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 2000, 68, 999–1003. [Google Scholar] [CrossRef]
- Dramsi, S.; Cossart, P. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 2003, 71, 3614–3618. [Google Scholar] [CrossRef]
- Hamon, M.A.; Cossart, P. K+ efflux is required for histone H3 dephosphorylation by Listeria monocytogenes listeriolysin O and other pore-forming toxins. Infect. Immun. 2011, 79, 2839–2846. [Google Scholar] [CrossRef]
- Stavru, F.; Cossart, P. Listeria infection modulates mitochondrial dynamics. Commun. Integr. Biol. 2011, 4, 364–366. [Google Scholar] [CrossRef]
- Dominguez-Bernal, G.; Muller-Altrock, S.; Gonzalez-Zorn, B.; Scortti, M.; Herrmann, P.; Monzo, H.J.; Lacharme, L.; Kreft, J.; Vazquez-Boland, J.A. A spontaneous genomic deletion in Listeria ivanovii identifies LIPI-2, a species-specific pathogenicity island encoding sphingomyelinase and numerous internalins. Mol. Microbiol. 2006, 59, 415–432. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016, 48, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Leong, D.; Alvarez-Ordonez, A.; Jordan, K. Monitoring occurrence and persistence of Listeria monocytogenes in foods and food processing environments in the Republic of Ireland. Front. Microbiol. 2014, 5, 436. [Google Scholar] [CrossRef]
- Harvey, J.; Gilmour, A. Application of multilocus enzyme electrophoresis and restriction fragment length polymorphism analysis to the typing of Listeria monocytogenes strains isolated from raw milk, nondairy foods, and clinical and veterinary sources. Appl. Environ. Microbiol. 1994, 60, 1547–1553. [Google Scholar] [CrossRef] [PubMed]
- Melero, B.; Manso, B.; Stessl, B.; Hernandez, M.; Wagner, M.; Rovira, J.; Rodriguez-Lazaro, D. Distribution and persistence of Listeria monocytogenes in a heavily contaminated poultry processing facility. J. Food Prot. 2019, 82, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Cherifi, T.; Arsenault, J.; Pagotto, F.; Quessy, S.; Cote, J.C.; Neira, K.; Fournaise, S.; Bekal, S.; Fravalo, P. Distribution, diversity and persistence of Listeria monocytogenes in swine slaughterhouses and their association with food and human listeriosis strains. PLoS ONE 2020, 15, e0236807. [Google Scholar] [CrossRef]
- Castro, H.; Jaakkonen, A.; Hakkinen, M.; Korkeala, H.; Lindstrom, M. Occurrence, persistence, and contamination routes of Listeria monocytogenes genotypes on three finnish dairy cattle farms: A longitudinal study. Appl. Environ. Microbiol. 2018, 84, e02000-17. [Google Scholar] [CrossRef]
- Leong, D.; NicAogain, K.; Luque-Sastre, L.; McManamon, O.; Hunt, K.; Alvarez-Ordonez, A.; Scollard, J.; Schmalenberger, A.; Fanning, S.; O’Byrne, C.; et al. A 3-year multi-food study of the presence and persistence of Listeria monocytogenes in 54 small food businesses in Ireland. Int. J. Food Microbiol. 2017, 249, 18–26. [Google Scholar] [CrossRef]
- D’Arrigo, M.; Mateo-Vivaracho, L.; Guillamon, E.; Fernandez-Leon, M.F.; Bravo, D.; Peiroten, A.; Medina, M.; Garcia-Lafuente, A. Characterization of persistent Listeria monocytogenes strains from ten dry-cured ham processing facilities. Food Microbiol. 2020, 92, 103581. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Hill, C.; Gahan, C.G. Acid stress responses in Listeria monocytogenes. Adv. Appl. Microbiol. 2008, 65, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Ricci, A.; Allende, A.; Bolton, D.; Chemaly, M.; Davies, R.; Fernandez Escamez, P.S.; Girones, R.; Herman, L.; Koutsoumanis, K.; Norrung, B.; et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018, 16, e05134. [Google Scholar] [CrossRef] [PubMed]
- EFSA; ECDC. Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections probably linked to frozen corn. EFSA Support. Publ. 2018, 15. [Google Scholar] [CrossRef]
- Pouillot, R.; Klontz, K.C.; Chen, Y.; Burall, L.S.; Macarisin, D.; Doyle, M.; Bally, K.M.; Strain, E.; Datta, A.R.; Hammack, T.S.; et al. Infectious dose of Listeria monocytogenes in outbreak linked to ice cream, United States, 2015. Emerg. Infect. Dis. 2016, 22, 2113–2119. [Google Scholar] [CrossRef]
- Angelo, K.M.; Conrad, A.R.; Saupe, A.; Dragoo, H.; West, N.; Sorenson, A.; Barnes, A.; Doyle, M.; Beal, J.; Jackson, K.A.; et al. Multistate outbreak of Listeria monocytogenes infections linked to whole apples used in commercially produced, prepackaged caramel apples: United States, 2014–2015. Epidemiol. Infect. 2017, 145, 848–856. [Google Scholar] [CrossRef]
- CDC. Sprouts and Investigation of Human Listeriosis Cases (Final Update). Available online: https://www.cdc.gov/listeria/outbreaks/bean-sprouts-11-14/key-resources.html (accessed on 15 April 2021).
- U.S. Department of Agriculture. FSIS Comparative Risk Assessment for Listeria monocytogenes in Ready-to-Eat Meat and Poultry Deli meats; U.S. Department of Agriculture: Washington, DC, USA, 2010.
- Stoller, A.; Stevens, M.J.A.; Stephan, R.; Guldimann, C. Characteristics of Listeria monocytogenes strains persisting in a meat processing facility over a 4-year period. Pathogens 2019, 8, 32. [Google Scholar] [CrossRef]
- Guidi, F.; Orsini, M.; Chiaverini, A.; Torresi, M.; Centorame, P.; Acciari, V.A.; Salini, R.; Palombo, B.; Brandi, G.; Amagliani, G.; et al. Hypo- and hyper-virulent Listeria monocytogenes clones persisting in two different food processing plants of central Italy. Microorganisms 2021, 9, 376. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Int. J. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Cherifi, T.; Carrillo, C.; Lambert, D.; Miniai, I.; Quessy, S.; Lariviere-Gauthier, G.; Blais, B.; Fravalo, P. Genomic characterization of Listeria monocytogenes isolates reveals that their persistence in a pig slaughterhouse is linked to the presence of benzalkonium chloride resistance genes. BMC Microbiol. 2018, 18, 220. [Google Scholar] [CrossRef]
- Hill, C.; Cotter, P.D.; Sleator, R.D.; Gahan, C.G. Bacterial stress response in Listeria monocytogenes: Jumping the hurdles imposed by minimal processing. Int. Dairy J. 2002, 12, 273–283. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Bowen, B.; Wiedmann, M.; Boor, K.J. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 2012, 78, 2602–2612. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y.; Yousef, A.E. Resistance of Listeria monocytogenes to heat after adaptation to environmental stresses. J. Food Prot. 1996, 59, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Bucur, F.I.; Grigore-Gurgu, L.; Crauwels, P.; Riedel, C.U.; Nicolau, A.I. Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 2018, 9, 2700. [Google Scholar] [CrossRef] [PubMed]
- Chaturongakul, S.; Raengpradub, S.; Wiedmann, M.; Boor, K.J. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol. 2008, 16, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Wiedmann, M. Physiology and genetics of Listeria monocytogenes survival and growth at cold temperatures. Crit. Rev. Food Sci. Nutr. 2009, 49, 237–253. [Google Scholar] [CrossRef]
- Abdel Karem, H.; Mattar, Z. Heat resistance and growth of Salmonella enteritidis, Listeria monocytogenes and Aeromonas hydrophila in whole liquid egg. Acta Microbiol. Pol. 2001, 50, 27–35. [Google Scholar]
- Huang, L. Thermal resistance of Listeria monocytogenes, Salmonella heidelberg, and Escherichia coli O157:H7 at elevated temperatures. J. Food Prot. 2004, 67, 1666–1670. [Google Scholar] [CrossRef]
- Sallami, L.; Marcotte, M.; Naim, F.; Ouattara, B.; Leblanc, C.; Saucier, L. Heat inactivation of Listeria monocytogenes and Salmonella enterica serovar Typhi in a typical bologna matrix during an industrial cooking-cooling cycle. J. Food Prot. 2006, 69, 3025–3030. [Google Scholar] [CrossRef]
- Hill, D.; Sugrue, I.; Arendt, E.; Hill, C.; Stanton, C.; Ross, R.P. Recent advances in microbial fermentation for dairy and health. F1000Research 2017, 6, 751. [Google Scholar] [CrossRef]
- Caplice, E.; Fitzgerald, G.F. Food fermentations: Role of microorganisms in food production and preservation. Int. J. Food Microbiol. 1999, 50, 131–149. [Google Scholar] [CrossRef]
- Davis, M.J.; Coote, P.J.; O’Byrne, C.P. Acid tolerance in Listeria monocytogenes: The adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 1996, 142, 2975–2982. [Google Scholar] [CrossRef]
- Chorianopoulos, N.; Giaouris, E.; Grigoraki, I.; Skandamis, P.; Nychas, G.J. Effect of acid tolerance response (ATR) on attachment of Listeria monocytogenes Scott A to stainless steel under extended exposure to acid or/and salt stress and resistance of sessile cells to subsequent strong acid challenge. Int. J. Food Microbiol. 2011, 145, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knochel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela Saldinger, S.; Tresse, O. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.; Liu, C.; Zhang, T.; Jones, M.; Peterson, S.N.; Wang, C. Global gene expression of Listeria monocytogenes to salt stress. J. Food Prot. 2012, 75, 906–912. [Google Scholar] [CrossRef]
- Duche, O.; Tremoulet, F.; Glaser, P.; Labadie, J. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 2002, 68, 1491–1498. [Google Scholar] [CrossRef]
- Cleveland, J.; Montville, T.J.; Nes, I.F.; Chikindas, M.L. Bacteriocins: Safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 2001, 71, 1–20. [Google Scholar] [CrossRef]
- Wiedemann, I.; Breukink, E.; van Kraaij, C.; Kuipers, O.P.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Daeschel, M.A. Correlation of cellular phospholipid content with nisin resistance of Listeria monocytogenes Scott A. J. Food Prot. 1995, 58, 416–420. [Google Scholar] [CrossRef]
- Verheul, A.; Russell, N.J.; Van, T.H.R.; Rombouts, F.M.; Abee, T. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 1997, 63, 3451–3457. [Google Scholar] [CrossRef]
- Kuenne, C.; Voget, S.; Pischimarov, J.; Oehm, S.; Goesmann, A.; Daniel, R.; Hain, T.; Chakraborty, T. Comparative analysis of plasmids in the genus Listeria. PLoS ONE 2010, 5, e0012511. [Google Scholar] [CrossRef]
- Diaz Ricci, J.C.; Hernandez, M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000, 20, 79–108. [Google Scholar] [CrossRef]
- Elhanafi, D.; Dutta, V.; Kathariou, S. Genetic characterization of plasmid-associated benzalkonium chloride resistance determinants in a Listeria monocytogenes strain from the 1998–1999 outbreak. Appl. Environ. Microbiol. 2010, 76, 8231–8238. [Google Scholar] [CrossRef]
- Rakic-Martinez, M.; Drevets, D.A.; Dutta, V.; Katic, V.; Kathariou, S. Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 2011, 77, 8714–8721. [Google Scholar] [CrossRef] [PubMed]
- Katharios-Lanwermeyer, S.; Rakic-Martinez, M.; Elhanafi, D.; Ratani, S.; Tiedje, J.M.; Kathariou, S. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 2012, 78, 7549–7556. [Google Scholar] [CrossRef]
- Lebrun, M.; Loulergue, J.; Chaslus-Dancla, E.; Audurier, A. Plasmids in Listeria monocytogenes in relation to cadmium resistance. Appl. Environ. Microbiol. 1992, 58, 3183–3186. [Google Scholar] [CrossRef] [PubMed]
- Franciosa, G.; Maugliani, A.; Scalfaro, C.; Floridi, F.; Aureli, P. Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int. J. Immunopathol. Pharmacol. 2009, 22, 183–193. [Google Scholar] [CrossRef]
- Piercey, M.J.; Hingston, P.A.; Truelstrup Hansen, L. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 degrees C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef] [PubMed]
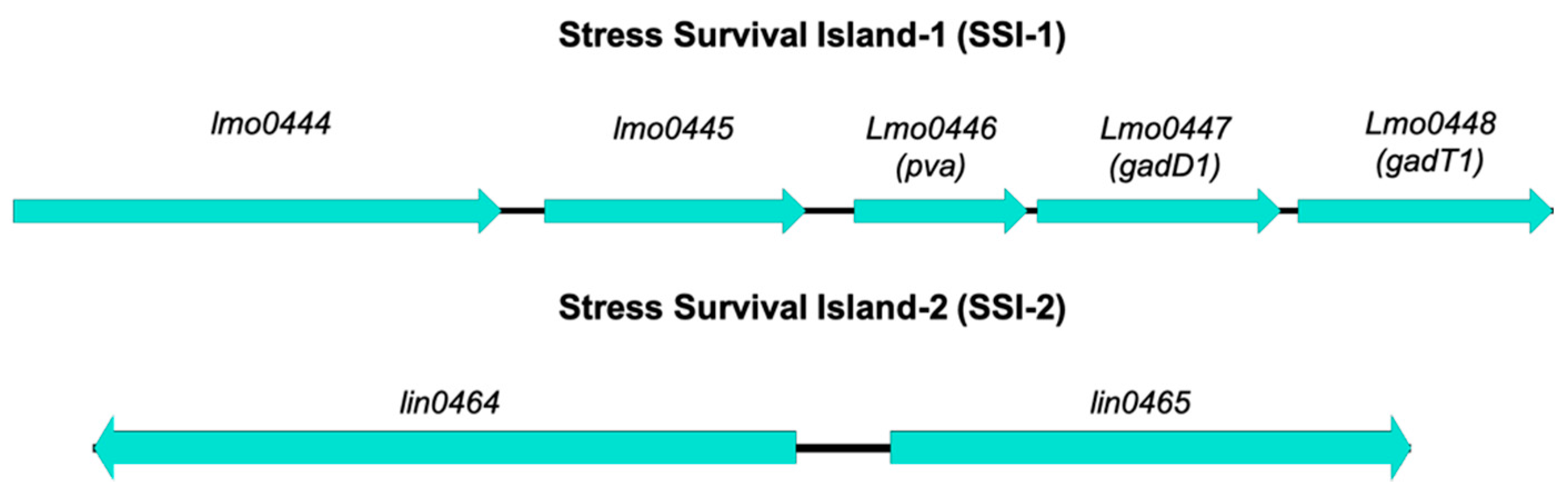
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef]
- Arguedas-Villa, C.; Kovacevic, J.; Allen, K.J.; Stephan, R.; Tasara, T. Cold growth behaviour and genetic comparison of Canadian and Swiss Listeria monocytogenes strains associated with the food supply chain and human listeriosis cases. Food Microbiol. 2014, 40, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Karatzas, K.A.; Wouters, J.A.; Gahan, C.G.; Hill, C.; Abee, T.; Bennik, M.H. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 2003, 49, 1227–1238. [Google Scholar] [CrossRef]
- Schwartz, B.; Ciesielski, C.A.; Broome, C.V.; Gaventa, S.; Brown, G.R.; Gellin, B.G.; Hightower, A.W.; Mascola, L. Association of sporadic listeriosis with consumption of uncooked hot dogs and undercooked chicken. Lancet 1998, 2, 779–782. [Google Scholar]
- Graves, L.M.; Hunter, S.B.; Ong, A.R.; Schoonmaker-Bopp, D.; Hise, K.; Kornstein, L.; DeWitt, W.E.; Hayes, P.S.; Dunne, E.; Mead, P.; et al. Microbiological aspects of the investigation that traced the 1998 outbreak of listeriosis in the United States to contaminated hot dogs and establishment of molecular subtyping-based surveillance for Listeria monocytogenes in the PulseNet network. J. Clin. Microbiol. 2005, 43, 2350–2355. [Google Scholar] [CrossRef][Green Version]
- Mead, P.S.; Dunne, E.F.; Graves, L.; Wiedmann, M.; Patrick, M.; Hunter, S.; Salehi, E.; Mostashari, F.; Craig, A.; Mshar, P.; et al. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiol. Infect. 2006, 134, 744–751. [Google Scholar] [CrossRef]
- Gilot, P.; Genicot, A.; Andre, P. Serotyping and esterase typing for analysis of Listeria monocytogenes populations recovered from foodstuffs and from human patients with listeriosis in Belgium. J. Clin. Microbiol. 1996, 34, 1007–1010. [Google Scholar] [CrossRef]
- Baek, S.Y.; Lim, S.Y.; Lee, D.H.; Min, K.H.; Kim, C.M. Incidence and characterization of Listeria monocytogenes from domestic and imported foods in Korea. J. Food Prot. 2000, 63, 186–189. [Google Scholar] [CrossRef]
- Choi, Y.C.; Cho, S.Y.; Park, B.K.; Chung, D.H.; Oh, D.H. Incidence and characterization of Listeria spp. from foods available in Korea. J. Food Prot. 2001, 64, 554–558. [Google Scholar] [CrossRef] [PubMed]
- Gilbreth, S.E.; Call, J.E.; Wallace, F.M.; Scott, V.N.; Chen, Y.; Luchansky, J.B. Relatedness of Listeria monocytogenes Isolates recovered from selected ready-to-eat foods and listeriosis patients in the United States. Appl. Environ. Microbiol. 2005, 71, 8115–8122. [Google Scholar] [CrossRef][Green Version]
- Thevenot, D.; Delignette-Muller, M.L.; Christieans, S.; Vernozy-Rozand, C. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. Int. J. Food Microbiol. 2005, 102, 85–94. [Google Scholar] [CrossRef]
- Hein, I.; Klinger, S.; Dooms, M.; Flekna, G.; Stessl, B.; Leclercq, A.; Hill, C.; Allerberger, F.; Wagner, M. Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 2011, 77, 2169–2173. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Muller, A.; Stessl, B.; Wagner, M. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 2015, 6, 380. [Google Scholar] [CrossRef]
- Holch, A.; Webb, K.; Lukjancenko, O.; Ussery, D.; Rosenthal, B.M.; Gram, L. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl. Environ. Microbiol. 2013, 79, 2944–2951. [Google Scholar] [CrossRef]
- Fox, E.M.; Leonard, N.; Jordan, K. Physiological and transcriptional characterization of persistent and nonpersistent Listeria monocytogenes isolates. Appl. Environ. Microbiol. 2011, 77, 6559–6569. [Google Scholar] [CrossRef]
- Begley, M.; Sleator, R.D.; Gahan, C.G.; Hill, C. Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 2005, 73, 894–904. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ryan, S.; Gahan, C.G.; Hill, C. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 2005, 71, 2832–2839. [Google Scholar] [CrossRef]
- Harter, E.; Wagner, E.M.; Zaiser, A.; Halecker, S.; Wagner, M.; Rychli, K. Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 2017, 83, e00827-17. [Google Scholar] [CrossRef]
- Casey, A.; Fox, E.M.; Schmitz-Esser, S.; Coffey, A.; McAuliffe, O.; Jordan, K. Transcriptome analysis of Listeria monocytogenes exposed to biocide stress reveals a multi-system response involving cell wall synthesis, sugar uptake, and motility. Front. Microbiol. 2014, 5, 68. [Google Scholar] [CrossRef] [PubMed]
- Suo, Y.; Liu, Y.; Zhou, X.; Huang, Y.; Shi, C.; Matthews, K.; Shi, X. Impact of Sod on the expression of stress-related genes in Listeria monocytogenes 4b G with/without paraquat treatment. J. Food Sci. 2014, 79, M1745–M1749. [Google Scholar] [CrossRef]
- Muller, A.; Rychli, K.; Zaiser, A.; Wieser, C.; Wagner, M.; Schmitz-Esser, S. The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 2014, 361, 166–173. [Google Scholar] [CrossRef]
- Muller, A.; Rychli, K.; Muhterem-Uyar, M.; Zaiser, A.; Stessl, B.; Guinane, C.M.; Cotter, P.D.; Wagner, M.; Schmitz-Esser, S. Tn6188—A novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS ONE 2013, 8, e76835. [Google Scholar] [CrossRef]
- Hoffmann, R.F.; McLernon, S.; Feeney, A.; Hill, C.; Sleator, R.D. A single point mutation in the listerial betL sigma(A)-dependent promoter leads to improved osmo- and chill-tolerance and a morphological shift at elevated osmolarity. Bioengineered 2013, 4, 401–407. [Google Scholar] [CrossRef][Green Version]
- Read, T.D.; Massey, R.C. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: A new direction for bacteriology. Genome Med. 2014, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Falush, D. Bacterial genomics: Microbial GWAS coming of age. Nat. Microbiol. 2016, 1, 16059. [Google Scholar] [CrossRef]
- Kazmierczak, M.J.; Mithoe, S.C.; Boor, K.J.; Wiedmann, M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 2003, 185, 5722–5734. [Google Scholar] [CrossRef]
- Voelker, U.; Dufour, A.; Haldenwang, W.G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of sigma B. J. Bacteriol. 1995, 177, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, L.; Felten, A.; Palma, F.; Mariet, J.F.; Radomski, N.; Mistou, M.Y.; Augustin, J.C.; Guillier, L. Insights from genome-wide approaches to identify variants associated to phenotypes at pan-genome scale: Application to L. monocytogenes’ ability to grow in cold conditions. Int. J. Food Microbiol. 2019, 291, 181–188. [Google Scholar] [CrossRef]
- Huang, H.W.; Hsu, C.P.; Yang, B.B.; Wang, C.Y. Potential utility of high-pressure processing to address the risk of food allergen concerns. Compr. Rev. Food Sci. Food Saf. 2014, 13, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Van Boeijen, I.K.; Moezelaar, R.; Abee, T.; Zwietering, M.H. Inactivation kinetics of three Listeria monocytogenes strains under high hydrostatic pressure. J. Food Prot. 2008, 71, 2007–2013. [Google Scholar] [CrossRef]
- Góngora-Nieto, M.M.; Sepúlveda, D.R.; Pedrow, P.; Barbosa-Cánovas, G.V.; Swanson, B.G. Food processing by pulsed electric fields: Treatment delivery, inactivation level, and regulatory aspects. LWT Food Sci. Technol. 2002, 35, 375–388. [Google Scholar] [CrossRef]
- Lado, B.H.; Yousef, A.E. Alternative food-preservation technologies: Efficacy and mechanisms. Microbes Infect. 2002, 4, 433–440. [Google Scholar] [CrossRef]
- Gómez-López, V.M.; Ragaert, P.; Debevere, J.; Devlieghere, F. Pulsed light for food decontamination: A review. Trends Food Sci. Technol. 2007, 18, 464–473. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Richa; Kumar, A.; Tyagi, M.B.; Sinha, R.P. Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef]
- Beauchamp, S.; Lacroix, M. Resistance of the genome of Escherichia coli and Listeria monocytogenes to irradiation evaluated by the induction of cyclobutane pyrimidine dimers and 6-4 photoproducts using gamma and UV-C radiations. Radiat. Phys. Chem. 2012, 81, 1193–1197. [Google Scholar] [CrossRef]
- Chmielewski, R.; Frank, J. Biofilm formation and control in food processing facilities. Compr. Rev. Food Sci. Food Saf. 2003, 2, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Heir, E.; Hebraud, M.; Chorianopoulos, N.; Langsrud, S.; Moretro, T.; Habimana, O.; Desvaux, M.; Renier, S.; Nychas, G.J. Attachment and biofilm formation by foodborne bacteria in meat processing environments: Causes, implications, role of bacterial interactions and control by alternative novel methods. Meat Sci. 2014, 97, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef]
- Esbelin, J.; Santos, T.; Hebraud, M. Desiccation: An environmental and food industry stress that bacteria commonly face. Food Microbiol. 2018, 69, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Price, R.; Jayeola, V.; Niedermeyer, J.; Parsons, C.; Kathariou, S. The Listeria monocytogenes key virulence determinants hly and prfA are involved in biofilm formation and aggregation but not colonization of fresh produce. Pathogens 2018, 7, 18. [Google Scholar] [CrossRef]
- Rodriguez-Lopez, P.; Rodriguez-Herrera, J.J.; Vazquez-Sanchez, D.; Lopez Cabo, M. Current knowledge on Listeria monocytogenes biofilms in food-related environments: Incidence, resistance to biocides, ecology and biocontrol. Foods 2018, 7, 85. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rammer, N.; Pospisilova, L.; Alispahic, M.; Wagner, M.; Rychli, K. Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef] [PubMed]
- Blackman, I.C.; Frank, J.F. Growth of Listeria monocytogenes as a biofilm on various food-processing surfaces. J. Food Prot. 1996, 59, 827–831. [Google Scholar] [CrossRef]
- Chae, M.S.; Schraft, H.; Truelstrup Hansen, L.; Mackereth, R. Effects of physicochemical surface characteristics of Listeria monocytogenes strains on attachment to glass. Food Microbiol. 2006, 23, 250–259. [Google Scholar] [CrossRef]
- Chavant, P.; Martinie, B.; Meylheuc, T.; Bellon-Fontaine, M.N.; Hebraud, M. Listeria monocytogenes LO28: Surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 2002, 68, 728–737. [Google Scholar] [CrossRef]
- Spurlock, A.T.; Zottola, E.A. Growth and attachment of Listeria monocytogenes to cast iron. J. Food Prot. 1991, 54, 925–929. [Google Scholar] [CrossRef]
- Manso, B.; Melero, B.; Stessl, B.; Fernandez-Natal, I.; Jaime, I.; Hernandez, M.; Wagner, M.; Rovira, J.; Rodriguez-Lazaro, D. Characterization of virulence and persistence abilities of Listeria monocytogenes strains isolated from food processing premises. J. Food Prot. 2019, 82, 1922–1930. [Google Scholar] [CrossRef]
- Lee, B.H.; Cole, S.; Badel-Berchoux, S.; Guillier, L.; Felix, B.; Krezdorn, N.; Hebraud, M.; Bernardi, T.; Sultan, I.; Piveteau, P. Biofilm formation of Listeria monocytogenes strains under food processing environments and pan-genome-wide association study. Front. Microbiol 2019, 10, 2698. [Google Scholar] [CrossRef]
- Stepanovic, S.; Cirkovic, I.; Ranin, L.; Svabic-Vlahovic, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Mureddu, A.; Mazza, R.; Fois, F.; Meloni, D.; Bacciu, R.; Piras, F.; Mazzette, R. Listeria monocytogenes persistence in ready-to-eat sausages and in processing plants. Ital. J. Food Saf. 2014, 3, 1697. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, F.; Palma, F.; Guillier, L.; Lucchi, A.; De Cesare, A.; Manfreda, G. Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: Persistence and ecophysiology. Front. Microbiol. 2018, 9, 596. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm formation ability of Listeria monocytogenes isolates from raw ready-to-eat seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef] [PubMed]
- Borucki, M.K.; Peppin, J.D.; White, D.; Loge, F.; Call, D.R. Variation in biofilm formation among strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2003, 69, 7336–7342. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, J.; Borges, S.; Camilo, R.; Magalhaes, R.; Ferreira, V.; Santos, I.; Silva, J.; Almeida, G.; Teixeira, P. Biofilm formation among clinical and food isolates of Listeria monocytogenes. Int. J. Microbiol. 2013, 2013, 524975. [Google Scholar] [CrossRef] [PubMed]
- Djordjevic, D.; Wiedmann, M.; McLandsborough, L.A. Microtiter plate assay for assessment of Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2002, 68, 2950–2958. [Google Scholar] [CrossRef]
- Keeney, K.; Trmcic, A.; Zhu, Z.; Delaquis, P.; Wang, S. Stress survival islet 1 contributes to serotype-specific differences in biofilm formation in Listeria monocytogenes. Lett. Appl. Microbiol. 2018, 67, 530–536. [Google Scholar] [CrossRef]
- Gueriri, I.; Cyncynatus, C.; Dubrac, S.; Arana, A.T.; Dussurget, O.; Msadek, T. The DegU orphan response regulator of Listeria monocytogenes autorepresses its own synthesis and is required for bacterial motility, virulence and biofilm formation. Microbiology 2008, 154, 2251–2264. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Parvathi, A.; George, J.; Krohne, G.; Karunasagar, I.; Karunasagar, I. A study on the effects of some laboratory derived genetic mutations on biofilm formation by Listeria monocytogenes. World J. Microbiol. Biotechnol. 2009, 25, 527–531. [Google Scholar] [CrossRef]
- Knudsen, G.M.; Olsen, J.E.; Dons, L. characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 2006, 240, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.; Bauer, S.; Beier, D.; Kuhn, M. Construction and characterisation of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 2005, 73, 3152–3159. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.U.; Monk, I.R.; Casey, P.G.; Waidmann, M.S.; Gahan, C.G.; Hill, C. AgrD-dependent quorum sensing affects biofilm formation, invasion, virulence and global gene expression profiles in Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1177–1189. [Google Scholar] [CrossRef]
- Rieu, A.; Weidmann, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. Agr system of Listeria monocytogenes EGD-e: Role in adherence and differential expression pattern. Appl. Environ. Microbiol. 2007, 73, 6125–6133. [Google Scholar] [CrossRef]
- Jordan, S.J.; Perni, S.; Glenn, S.; Fernandes, I.; Barbosa, M.; Sol, M.; Tenreiro, R.P.; Chambel, L.; Barata, B.; Zilhao, I.; et al. Listeria monocytogenes biofilm-associated protein (BapL) may contribute to surface attachment of L. monocytogenes but is absent from many field isolates. Appl. Environ. Microbiol. 2008, 74, 5451–5456. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, S.; Abee, T. Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 2010, 76, 7854–7860. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Gu, W.; Fischer, N.; McLandsborough, L. Identification of genes involved in Listeria monocytogenes biofilm formation by mariner-based transposon mutagenesis. Appl. Microbiol. Biotechnol. 2012, 93, 2051–2062. [Google Scholar] [CrossRef]
- Alonso, A.N.; Perry, K.J.; Regeimbal, J.M.; Regan, P.M.; Higgins, D.E. Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS ONE 2014, 9, e113696. [Google Scholar] [CrossRef]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef]
- Mauder, N.; Williams, T.; Fritsch, F.; Kuhn, M.; Beier, D. Response regulator DegU of Listeria monocytogenes controls temperature-responsive flagellar gene expression in its unphosphorylated state. J. Bacteriol 2008, 190, 4777–4781. [Google Scholar] [CrossRef][Green Version]
- McGann, P.; Wiedmann, M.; Boor, K.J. The alternative sigma factor sigma B and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl. Environ. Microbiol. 2007, 73, 2919–2930. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Graham, J.E.; Bigelow, L.; Morse, P.D., 2nd; Wilkinson, B.J. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 2002, 68, 1697–1705. [Google Scholar] [CrossRef]
- McDonnell, G.; Russell, A.D. Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Harbarth, S.; Tuan Soh, S.; Horner, C.; Wilcox, M.H. Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J. Hosp. Infect. 2014, 87, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.; Fernández-Fuentes, M.A.; Grande Burgos, M.J.; Abriouel, H.; Pérez Pulido, R.; Gálvez, A. Biocide tolerance in bacteria. Int. J. Food Microbiol. 2013, 162, 13–25. [Google Scholar] [CrossRef]
- Cramer, M.M. Food Plant. Sanitation. Design, Maintenance, and Good Manufacturing Practices; CRC Press: Boca Raton, FL, USA, 2006. [Google Scholar]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- McBain, A.J.; Ledder, R.G.; Moore, L.E.; Catrenich, C.E.; Gilbert, P. Effects of quaternary-ammonium-based formulations on bacterial community dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 2004, 70, 3449–3456. [Google Scholar] [CrossRef]
- Kovacevic, J.; Ziegler, J.; Walecka-Zacharska, E.; Reimer, A.; Kitts, D.D.; Gilmour, M.W. Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 2016, 82, 939–953. [Google Scholar] [CrossRef]
- Dutta, V.; Elhanafi, D.; Kathariou, S. Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 2013, 79, 6067–6074. [Google Scholar] [CrossRef] [PubMed]
- Tamburro, M.; Ripabelli, G.; Vitullo, M.; Dallman, T.J.; Pontello, M.; Amar, C.F.; Sammarco, M.L. Gene expression in Listeria monocytogenes exposed to sublethal concentration of benzalkonium chloride. Comp. Immunol. Microbiol. Infect. Dis. 2015, 40, 31–39. [Google Scholar] [CrossRef]
- Mereghetti, L.; Quentin, R.; Marquet-Van Der Mee, N.; Audurier, A. Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl. Environ. Microbiol. 2000, 66, 5083–5086. [Google Scholar] [CrossRef]
- Romanova, N.A.; Wolffs, P.F.; Brovko, L.Y.; Griffiths, M.W. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl. Environ. Microbiol. 2006, 72, 3498–3503. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, S.; Lopez, V.; Martinez-Suarez, J.V. The influence of subminimal inhibitory concentrations of benzalkonium chloride on biofilm formation by Listeria monocytogenes. Int. J. Food Microbiol. 2014, 189, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Tezel, U.; Pavlostathis, S.G. Quaternary ammonium disinfectants: Microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 2015, 33, 296–304. [Google Scholar] [CrossRef]
- Moretro, T.; Schirmer, B.C.T.; Heir, E.; Fagerlund, A.; Hjemli, P.; Langsrud, S. Tolerance to quaternary ammonium compound disinfectants may enhance growth of Listeria monocytogenes in the food industry. Int. J. Food Microbiol. 2017, 241, 215–224. [Google Scholar] [CrossRef]
- Martinez-Suarez, J.V.; Ortiz, S.; Lopez-Alonso, V. Potential impact of the resistance to quaternary ammonium disinfectants on the persistence of Listeria monocytogenes in food processing environments. Front. Microbiol. 2016, 7, 638. [Google Scholar] [CrossRef]
- Cooper, A.L.; Carrillo, C.D.; DeschEnes, M.; Blais, B.W. Genomic markers for quaternary ammonium compound resistance as a persistence indicator for Listeria monocytogenes contamination in food manufacturing environments. J. Food Prot. 2021, 84, 389–398. [Google Scholar] [CrossRef]
- Piddock, L.J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef]
- Aase, B.; Sundheim, G.; Langsrud, S.; Rorvik, L.M. Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 2000, 62, 57–63. [Google Scholar] [CrossRef]
- Lunden, J.M.; Autio, T.J.; Sjoberg, A.M.; Korkeala, H.J. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 2003, 66, 2062–2069. [Google Scholar] [CrossRef]
- Ortiz, S.; Lopez-Alonso, V.; Rodriguez, P.; Martinez-Suarez, J.V. The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate iberian pork processing plants: Evidence from comparative genome analysis. Appl. Environ. Microbiol. 2016, 82, 308–317. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Assessment of the possible effect of the four antimicrobial treatment substances on the emergence of antimicrobial resistance, Scientific Opinion of the Panel on Biological Hazards. EFSA J. 2008, 6, 659. [Google Scholar] [CrossRef]
- Pan, Y.; Breidt, F., Jr.; Kathariou, S. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 2006, 72, 7711–7717. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Takakura, K.; Sone, Y.; Itano, Y.; Nishikawa, Y. Biofilm formation and resistance to benzalkonium chloride in Listeria monocytogenes isolated from a fish processing plant. J. Food Prot. 2013, 76, 1179–1186. [Google Scholar] [CrossRef]
- Kremer, P.H.; Lees, J.A.; Koopmans, M.M.; Ferwerda, B.; Arends, A.W.; Feller, M.M.; Schipper, K.; Valls Seron, M.; van der Ende, A.; Brouwer, M.C.; et al. Benzalkonium tolerance genes and outcome in Listeria monocytogenes meningitis. Clin. Microbiol. Infect. 2017, 23, 265.e1–265.e7. [Google Scholar] [CrossRef]
- Xu, D.; Nie, Q.; Wang, W.; Shi, L.; Yan, H. Characterization of a transferable bcrABC and cadAC genes-harboring plasmid in Listeria monocytogenes strain isolated from food products of animal origin. Int. J. Food Microbiol. 2016, 217, 117–122. [Google Scholar] [CrossRef]
- Nguyen, U.T.; Burrows, L.L. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 2014, 187, 26–32. [Google Scholar] [CrossRef]
- Gray, J.A.; Chandry, P.S.; Kaur, M.; Kocharunchitt, C.; Bowman, J.P.; Fox, E.M. Novel biocontrol methods for Listeria monocytogenes biofilms in food production facilities. Front. Microbiol. 2018, 9, 605. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Lopez, P.; Carballo-Justo, A.; Draper, L.A.; Cabo, M.L. Removal of Listeria monocytogenes dual-species biofilms using combined enzyme-benzalkonium chloride treatments. Biofouling 2017, 33, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Nuesch-Inderbinen, M.; Bloemberg, G.V.; Muller, A.; Stevens, M.J.A.; Cernela, N.; Kolloffel, B.; Stephan, R. Listeriosis caused by persistence of Listeria monocytogenes serotype 4b sequence type 6 in cheese production environment. Emerg. Infect. Dis. 2021, 27, 284–288. [Google Scholar] [CrossRef]
- Thomas, J.; Govender, N.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; Sekwadi, P.; Ntshoe, G.; et al. Outbreak of Listeriosis in South Africa associated with processed meat. N. Engl. J. Med. 2020, 382, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Halbedel, S.; Wilking, H.; Holzer, A.; Kleta, S.; Fischer, M.A.; Luth, S.; Pietzka, A.; Huhulescu, S.; Lachmann, R.; Krings, A.; et al. Large nationwide outbreak of invasive Listeriosis associated with blood sausage, Germany, 2018–2019. Emerg. Infect. Dis. 2020, 26, 1456–1464. [Google Scholar] [CrossRef]
| Island | Gene | Product | Function |
|---|---|---|---|
| LIPI-1 | actA | Actin assembly-inducing protein |
|
| mpI | Metalloprotease | Process the PC-PLC into its mature and active form [49] | |
| plcA | Phosphatidylinositol-specific Phospholipase C | ||
| plcB | phosphatidylcholine-specific Phospholipase C or lecithinase | ||
| prfA | PrfA (Pleiotropic regulatory factor) | Belongs to CAP (for catabolite gene activator protein)/FNR family, Dimeric protein and bindis binds to its DNA target sequences in dimeric form [54] | |
| hly | Listeriolysin O |
| |
| LIPI-2 | i-inlB2 i-inlL i-inlK i-inlB1 i-inlJ i-inlI i-inlH i-inlG smcL i-inlF i-inlE surF3 | Inernalins and SMase | |
| LIPI-3 | llsAGHXBYDP | Listeriolysin S | Haemolysin that is post-translationally modified and belongs to a family of modified virulence peptides, including streptolysin S and several as-yet uncharacterized members of the same family in other pathogens. LLS demonstrated to play a role in the survival of L. monocytogenes in PMNs and also contributes to its virulence in the mouse model [60] |
| LIPI-4 | lm4b_02324 | Maltose-6′-P-glucosidase | Putative 6-phospho-beta-glucosidase [61] |
| lm4b_02325 | Transcriptional antiterminator | Putative transcription antiterminator BglG family [61] | |
| lm4b_02326 | Uncharacterized protein associated to PTS systems | Unknown [61] | |
| lm4b_02327 | Membrane permease EIIA | Putative PTS system, cellobiose-specific enzyme component [61] | |
| lm4b_02328 | Membrane permease EIIB | Putative PTS system, cellobiose-specific enzyme component [61] | |
| lm4b_02329 | Membrane permease EIIC | Putative PTS system, cellobiose-specific enzyme component [61] |
| Persistence Definition | Time Frame | Sample Number | Persisters Identified | Reference |
|---|---|---|---|---|
| PFGE type detected repeatedly for longer than 6 months | 1 year | |||
| 319 | ST9, ST121 | [64] | ||
| PFGE type isolated at least 3 occasions over the 16-month sampling period | 16 months | 2496 | LS1, LS2, LS4, LS5, LS7, LS25, LS35, LS45 | [65] |
| Genotypes isolated at least on 3 occasions with a minimum interval of 6 months between first and last isolation | 3 years | 1702 | Serogroup IIa | [66] |
| PFGE type isolated at least six months apart | 3 years | 5869 | P59, P6, P10, P32, P44 | [67] |
| PFGE type isolated repeatedly, at least 4 times | 2 years | 1801 | 26 pulsotypes | [68] |
| CTs isolated at least 3 times with a minimum of 1 year between the first and last isolation | 4 years | 100 | CT1526, CT1828, CT1833, CT1834, CT1836, CT1839 | [69] |
| Island | Acronym | Gene | Product | Function | Reference |
|---|---|---|---|---|---|
| Stress Survival Island-1 | SSI-1 | lmo0444 | Hypothetical protein | Unknown | Ryan et al. [110] |
| lmo0445 | Transcriptional regulator | Regulation of transcription | |||
| lmo0446 (pva) | Penicillin acylase | Conversion of penicillin to 6-amino-penicillinate and phenylacetate—reduced susceptibility to penicillin V Survival in bile salts | Begley et al. [125] | ||
| lmo0447 (gadD1) | Glutamate decarboxylase | Growth in mildly acidic pHs | Cotter et al. [126] | ||
| lmo0448 (gadT1) | Amino acid antiporter | ||||
| Stress Survival Island-2 | SSI-2 | lin0464 | Transcriptional factor | Involvement in alkaline and oxidative stress responses | Harter et al. [127] |
| lin0465 | Pfpl protease |
| Method | Duration | Temp. | Substrate | Strains | Summary | Reference |
|---|---|---|---|---|---|---|
| MPA stained with crystal violet | 24 h | 37 °C and 10 °C | Bacterial biomass | 19 persistent, 20 prevalent, 19 rare (27 genotypes) | Persistence/prevalence did not correspond to a higher biofilm formation, supplementation with NaCl in nutrient deprived cells improved biofilms, production ~5 times greater at 37 °C than at 10 °C | [159] |
| MPA/stainless steel sheets stained with crystal violet | 24 h | 37 °C | Bacterial biomass | ST1, ST2, ST5, ST8, ST9, ST87, ST121, ST199, ST321, ST388 | All strains formed moderate biofilms on microtiter plates but not on stainless steel | [158] |
| MPA stained with crystal violet | 16, 24 and 36 h | 30 °C | Bacterial biomass | Range of serotypes from food/clinical samples | Lineage I isolates produced more biofilm, no significant difference in biofilm formation found between food/clinical isolates | [163] |
| MPA stained with crystal violet | 224 h | 37 °C | Bacterial biomass | Range of serotypes | Lineage II (persistent) isolates produced more biofilms, biofilm formation correlated with phylogenetic division but not serotype | [164] |
| MPA stained with crystal violet | 24 h (37C) and 5 days (4C) | 37 °C and 4 °C | Bacterial biomass | Range of serotypes from food/clinical samples | Higher biofilm formation at 37 °C with food isolates producing slightly more biofilm at both temperatures | [165] |
| Gene | Product | Function | Reference |
|---|---|---|---|
| degU | Putative response regulator | degU is essential for flagellar synthesis and motility in L. monocytogenes. It is required for growth at high temperature, adherence to plastic surfaces and formation of efficient biofilms. It also functions in virulence of L. monocytogenes. | [168,170] |
| flaA | Flagellin A | Involved in initial attachment of L. monocytogenes in biofilm formation. | [169,171] |
| agrBDCA | Peptide-sensing system | Peptide-sensing system involved in quorum-sensing. Involved in early stages of biofilm formation. Also involved in virulence. | [172,173] |
| bapL | Putative cell wall-anchored protein | Surface adherence of L. monocytogenes in L. monocytogenes. | [174] |
| sigB | Major transcriptional regulator of stress response genes | Required to obtain wild-type levels of static and continuous-flow biofilms. Also involved in resistance of planktonic/biofilm cells to benzalkonium chloride and peracetic acid. | [175] |
| dltABCD | D-alanylation pathway | Further work needed to find specific function. | [177] |
| phoPR | Phosphate-sensing two component system | Further work needed to find specific function. | [177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Unrath, N.; McCabe, E.; Macori, G.; Fanning, S. Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments. Microorganisms 2021, 9, 1856. https://doi.org/10.3390/microorganisms9091856
Unrath N, McCabe E, Macori G, Fanning S. Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments. Microorganisms. 2021; 9(9):1856. https://doi.org/10.3390/microorganisms9091856
Chicago/Turabian StyleUnrath, Natalia, Evonne McCabe, Guerrino Macori, and Séamus Fanning. 2021. "Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments" Microorganisms 9, no. 9: 1856. https://doi.org/10.3390/microorganisms9091856
APA StyleUnrath, N., McCabe, E., Macori, G., & Fanning, S. (2021). Application of Whole Genome Sequencing to Aid in Deciphering the Persistence Potential of Listeria monocytogenes in Food Production Environments. Microorganisms, 9(9), 1856. https://doi.org/10.3390/microorganisms9091856







