Characterization of Pseudomonas aeruginosa Quorum Sensing Inhibitors from the Endophyte Lasiodiplodia venezuelensis and Evaluation of Their Antivirulence Effects by Metabolomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Experimental Procedures
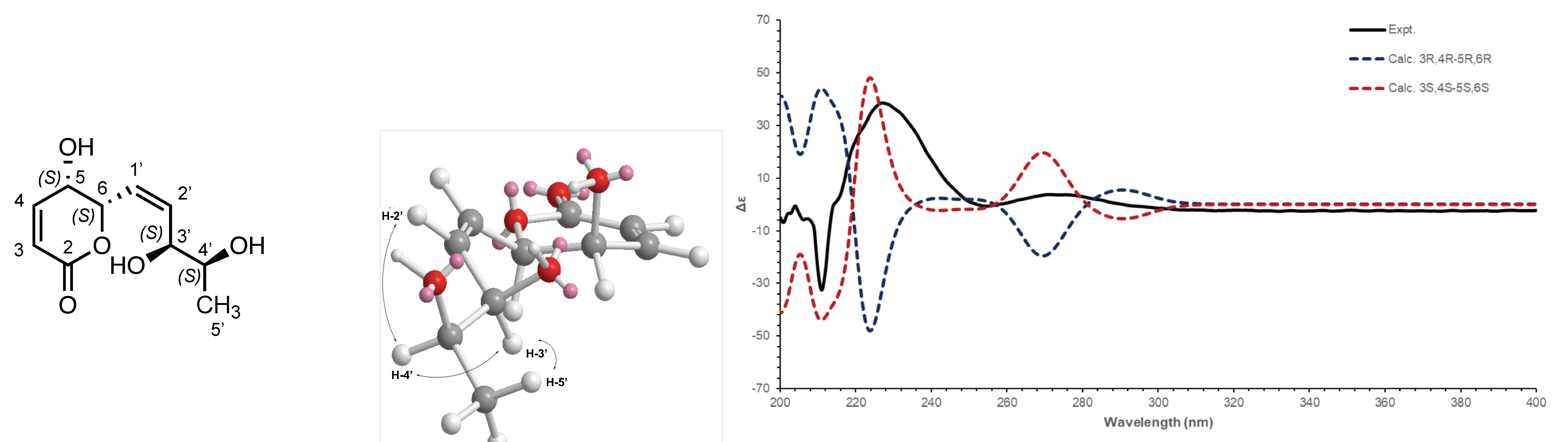
2.2. Electronic Circular Dichroism (ECD) and Optical Rotation (αD)
2.3. Plant Material
2.4. Isolation of Endophytes
2.5. Identification of Fungal Strains
2.6. Culture and Extraction
2.7. Scale Up of the Fungal Culture of Lasiodiplodia Venezuelensis
2.8. HPLC-PDA-ELSD Analyses
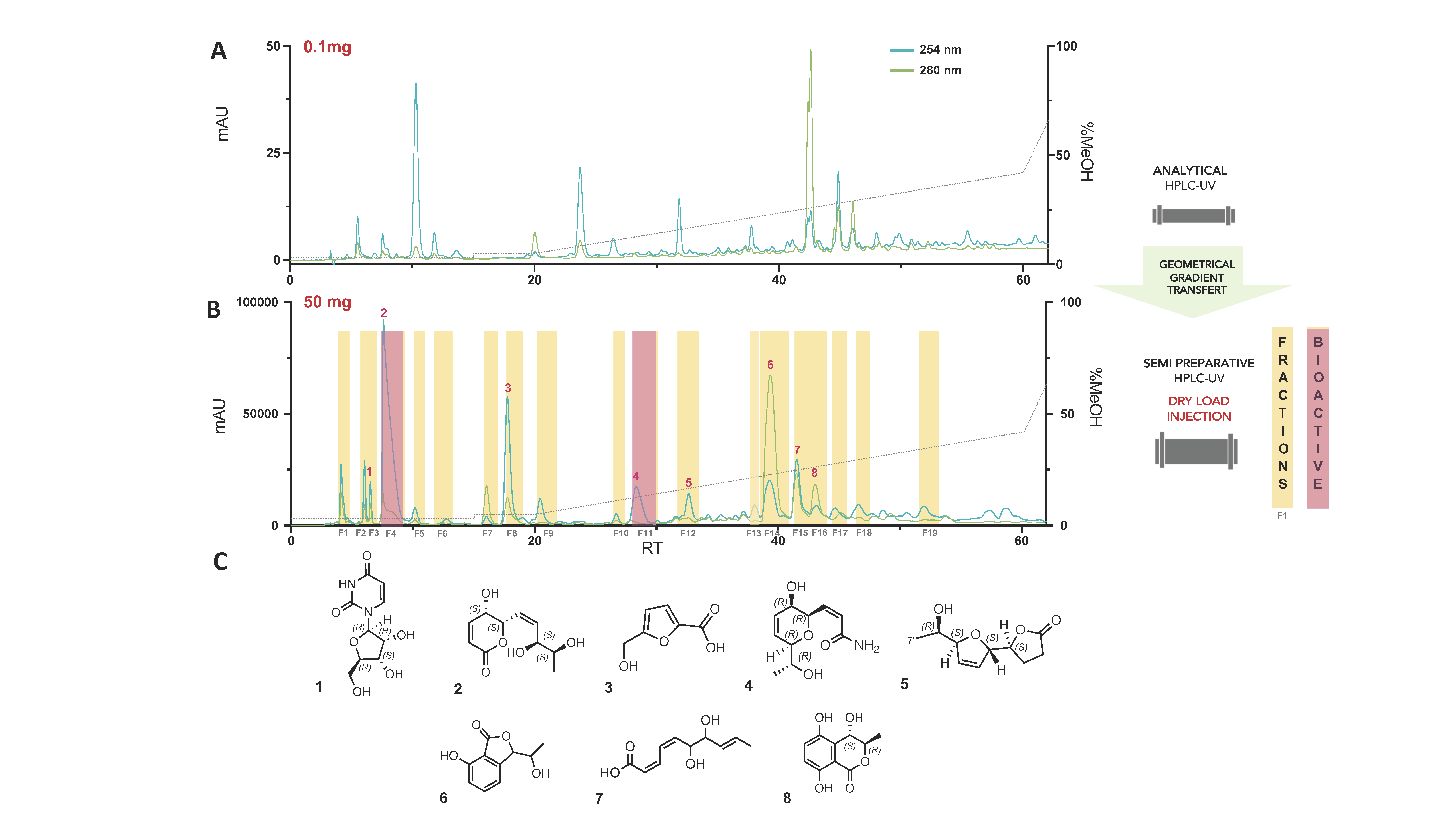
2.9. Semi-Preparative HPLC-UV Purification of the Water Partition of the Strain Lasiodiplodia Venezuelensis (A02W)
2.10. Description of the Isolated Compounds
2.11. ECD Experimental Details
2.12. UHPLC-PDA-CAD-HRMS/MS Analysis
2.13. UHPLC-HRMS/MS Data Processing
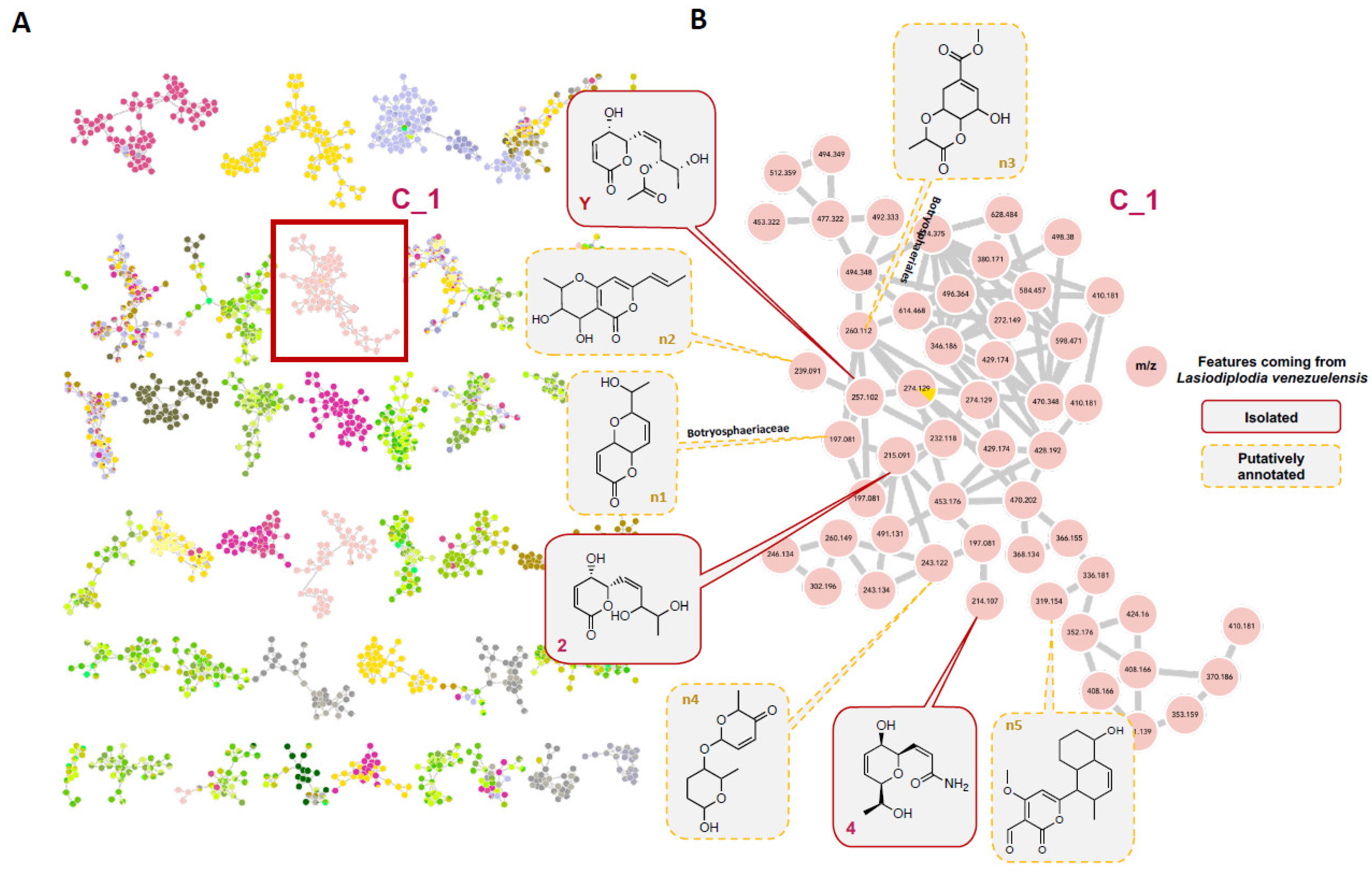
2.14. Molecular Network Analysis
2.15. Antibacterial Assays
2.16. Fluorescence Screening
2.17. qRT-PCR
2.18. Rhamnolipids Assay
2.19. P. aeruginosa Culture for Supernatant Analysis
3. Results
3.1. Biological Activities of Fungal Extracts
3.2. Bioactivity-Guided Fractionation and Isolation of the Bioactive Compounds
3.3. Chemical Characterisation of the Secondary Metabolites Isolated from the Water Extract (A02W)
3.4. Untargeted Metabolomics to Search for Compounds 2 and 4 Analogues
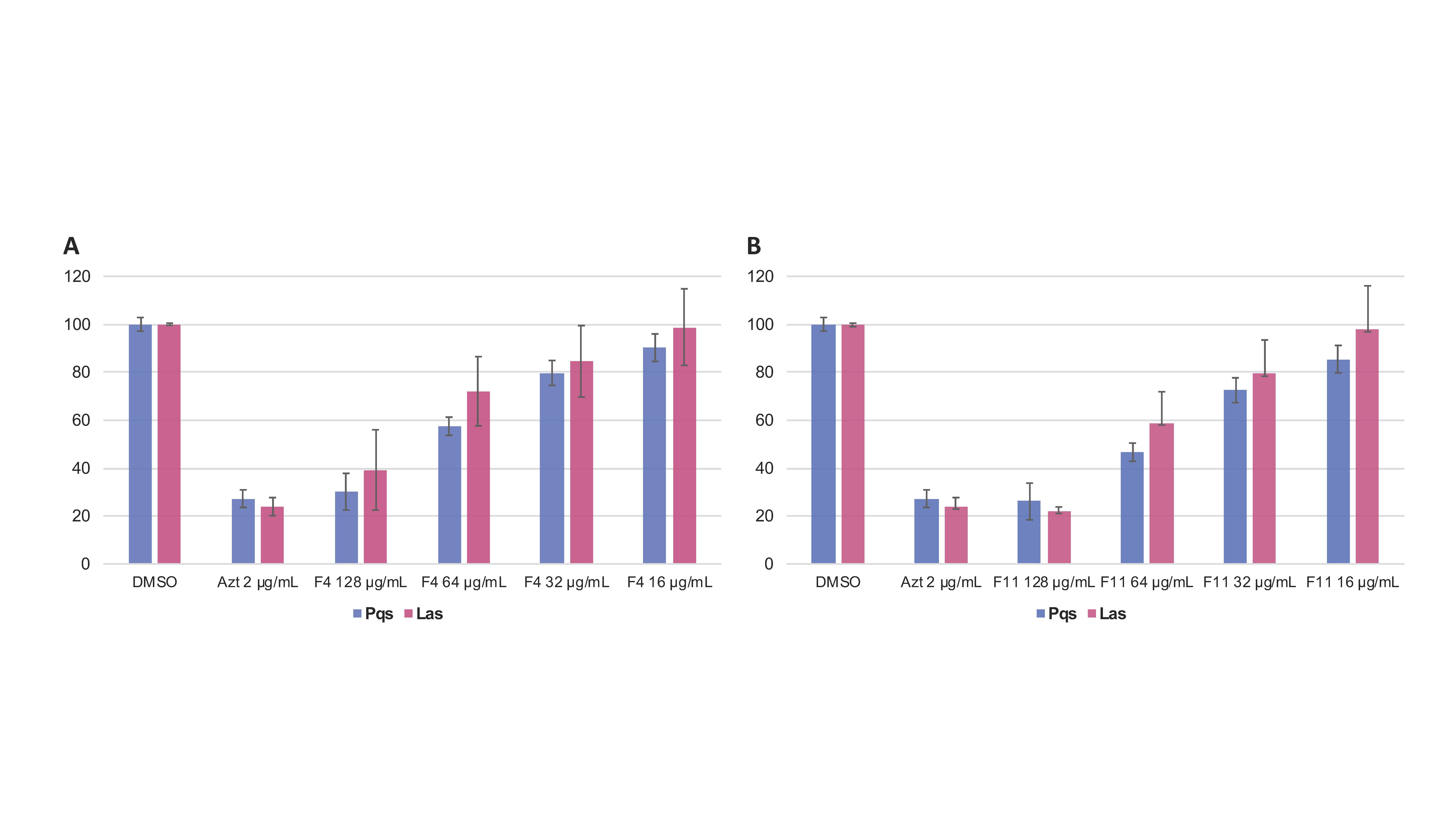
3.5. Effect of Compounds 2 and 4 on the Expression of QS-Regulated Genes
3.6. Effect of the Compounds 2 and 4 on Virulence Factors Production
3.7. Untargeted Metabolomics of P. aeruginosa PAO1 to Assess the Modulation of Virulence-Linked Metabolites When Incubated with 2 or 4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]
- Defoirdt, T. Quorum-Sensing Systems as Targets for Antivirulence Therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net); Annual Epidemiological Report 2019; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2020. [Google Scholar]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical Relevance of the ESKAPE Pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Cegelski, L.; Marshall, G.R.; Eldridge, G.R.; Hultgren, S.J. The Biology and Future Prospects of Antivirulence Therapies. Nat. Rev. Microbiol. 2008, 6, 17–27. [Google Scholar] [CrossRef]
- LaSarre, B.; Federle, M.J. Exploiting Quorum Sensing to Confuse Bacterial Pathogens. Microbiol. Mol. Biol. Rev. 2013, 77, 73–111. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities; WHO Guidelines Approved by the Guidelines Review Committee; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-155017-8. [Google Scholar]
- Obritsch, M.D.; Fish, D.N.; MacLaren, R.; Jung, R. Nosocomial Infections Due to Multidrug-Resistant Pseudomonas Aeruginosa: Epidemiology and Treatment Options. Pharmacotherapy 2005, 25, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Crousilles, A.; Maunders, E.; Bartlett, S.; Fan, C.; Ukor, E.-F.; Abdelhamid, Y.; Baker, Y.; Floto, A.; Spring, D.R.; Welch, M. Which Microbial Factors Really Are Important in Pseudomonas Aeruginosa Infections? Future Microbiol. 2015, 10, 1825–1836. [Google Scholar] [CrossRef] [Green Version]
- Davies, D.G.; Parsek, M.R.; Pearson, J.P.; Iglewski, B.H.; Costerton, J.W.; Greenberg, E.P. The Involvement of Cell-to-Cell Signals in the Development of a Bacterial Biofilm. Science 1998, 280, 295–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearson, J.P.; Passador, L.; Iglewski, B.H.; Greenberg, E.P. A Second N-Acylhomoserine Lactone Signal Produced by Pseudomonas Aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 1490–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef]
- Schuster, M.; Greenberg, E.P. A Network of Networks: Quorum-Sensing Gene Regulation in Pseudomonas Aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. [Google Scholar] [CrossRef]
- Soheili, V.; Tajani, A.S.; Ghodsi, R.; Bazzaz, B.S.F. Anti-PqsR Compounds as next-Generation Antibacterial Agents against Pseudomonas Aeruginosa: A Review. Eur. J. Med. Chem. 2019, 172, 26–35. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The Hierarchy Quorum Sensing Network in Pseudomonas Aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papaioannou, E.; Utari, P.D.; Quax, W.J. Choosing an Appropriate Infection Model to Study Quorum Sensing Inhibition in Pseudomonas Infections. Int. J. Mol. Sci. 2013, 14, 19309–19340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mookherjee, A.; Singh, S.; Maiti, M.K. Quorum Sensing Inhibitors: Can Endophytes Be Prospective Sources? Arch. Microbiol. 2018, 200, 355–369. [Google Scholar] [CrossRef]
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Brock/Springer Series in Contemporary Bioscience; Springer: New York, NY, USA, 1991; pp. 179–197. ISBN 978-1-4612-3168-4. [Google Scholar]
- Arnold, A.E.; Maynard, Z.; Gilbert, G.S.; Coley, P.D.; Kursar, T.A. Are Tropical Fungal Endophytes Hyperdiverse? Ecol. Lett. 2000, 3, 267–274. [Google Scholar] [CrossRef]
- Zhang, H.W.; Song, Y.C.; Tan, R.X. Biology and Chemistry of Endophytes. Nat. Prod. Rep. 2006, 23, 753–771. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Proksch, P. Fungal Endophytes: Unique Plant Inhabitants with Great Promises. Appl. Microbiol. 2011, 90, 1829–1845. [Google Scholar] [CrossRef] [PubMed]
- Krings, M.; Taylor, T.N.; Hass, H.; Kerp, H.; Dotzler, N.; Hermsen, E.J. Fungal Endophytes in a 400-Million-Yr-Old Land Plant: Infection Pathways, Spatial Distribution, and Host Responses. New Phytol. 2007, 174, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Kusari, P.; Kusari, S.; Lamshöft, M.; Sezgin, S.; Spiteller, M.; Kayser, O. Quorum Quenching Is an Antivirulence Strategy Employed by Endophytic Bacteria. Appl. Microbiol. Biotechnol. 2014, 98, 7173–7183. [Google Scholar] [CrossRef] [PubMed]
- Kusari, P.; Kusari, S.; Spiteller, M.; Kayser, O. Implications of Endophyte-Plant Crosstalk in Light of Quorum Responses for Plant Biotechnology. Appl. Microbiol. Biotechnol. 2015, 99, 5383–5390. [Google Scholar] [CrossRef]
- Hong, K.-W.; Koh, C.-L.; Sam, C.-K.; Yin, W.-F.; Chan, K.-G. Quorum Quenching Revisited—From Signal Decays to Signalling Confusion. Sensors 2012, 12, 4661–4696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, A.; Rothballer, M.; Hense, B.A.; Schröder, P. Bacterial Quorum Sensing Compounds Are Important Modulators of Microbe-Plant Interactions. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [Green Version]
- Figueroa, M.; Jarmusch, A.K.; Raja, H.A.; El-Elimat, T.; Kavanaugh, J.S.; Horswill, A.R.; Cooks, R.G.; Cech, N.B.; Oberlies, N.H. Polyhydroxyanthraquinones as Quorum Sensing Inhibitors from the Guttates of Penicillium Restrictum and Their Analysis by Desorption Electrospray Ionization Mass Spectrometry. J. Nat. Prod. 2014, 77, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Bai, Z.-Q.; Lin, X.; Wang, Y.; Wang, J.; Zhou, X.; Yang, B.; Liu, J.; Yang, X.; Wang, Y.; Liu, Y. New Phenyl Derivatives from Endophytic Fungus Aspergillus Flavipes AIL8 Derived of Mangrove Plant Acanthus Ilicifolius. Fitoterapia 2014, 95, 194–202. [Google Scholar] [CrossRef]
- Kahn, F. The Genus Astrocaryum (Arecaceae). Rev. Peru. Biol. 2008, 15, 29–46. [Google Scholar]
- Charles-Dominique, P.; Chave, J.; Dubois, M.-A.; De Granville, J.-J.; Riera, B.; Vezzoli, C. Colonization Front of the Understorey Palm Astrocaryum Sciophilum in a Pristine Rain Forest of French Guiana. Glob. Ecol. Biogeogr. 2003, 12, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Arnold, A.E.; Mejía, L.C.; Kyllo, D.; Rojas, E.I.; Maynard, Z.; Robbins, N.; Herre, E.A. Fungal Endophytes Limit Pathogen Damage in a Tropical Tree. Proc. Natl. Acad. Sci. USA 2003, 100, 15649–15654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthélemy, M.; Elie, N.; Pellissier, L.; Wolfender, J.-L.; Stien, D.; Touboul, D.; Eparvier, V. Structural Identification of Antibacterial Lipids from Amazonian Palm Tree Endophytes through the Molecular Network Approach. Int. J. Mol. Sci. 2019, 20, 2006. [Google Scholar] [CrossRef] [Green Version]
- Donald, J.; Barthélemy, M.; Gazal, N.; Eveno, Y.; Manzi, S.; Eparvier, V.; Stien, D.; Roy, M. Tropical Palm Endophytes Exhibit Low Competitive Structuring When Assessed Using Co-Occurrence and Antipathogen Activity Analysis. Front. For. Glob. Chang. 2019, 2. [Google Scholar] [CrossRef] [Green Version]
- Burgess, T.I.; Barber, P.A.; Mohali, S.; Pegg, G.; de Beer, W.; Wingfield, M.J. Three New Lasiodiplodia Spp. from the Tropics, Recognized Based on DNA Sequence Comparisons and Morphology. Mycologia 2006, 98, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Robert, V.; Stegehuis, G.; Stalpers, J. The MycoBank Engine and Related Databases. Available online: https://www.mycobank.org (accessed on 19 April 2020).
- Correia, K.C.; Silva, M.A.; De Morais, M.A.; Armenglo, J.; Phillips, A.J.L.; Câmara, M.P.S.; Michereff, S.J. Phylogeny, Distribution and Pathogenicity of Lasiodiplodia Species Associated with Dieback of Table Grape in the Main Brazilian Exporting Region. Plant. Pathol. 2016, 65, 92–103. [Google Scholar] [CrossRef]
- Vieira, J.C.B.; Câmara, M.P.S.; Bezerra, J.D.P.; Motta, C.M.S.; Machado, A.R. First Report of Lasiodiplodia Theobromae Causing Rot in Eggplant Fruit in Brazil. Plant Dis. 2018, 102. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Jayawardena, R.S.; Xu, W.; Hu, M.; Li, X.H.; Liu, J.H.; Hyde, K.D.; Yan, J. Lasiodiplodia Theobromae and L. Pseudotheobromae Causing Leaf Necrosis on Camellia Sinensis in Fujian Province, China. Can. J. Plant Pathol. 2019, 41, 277–284. [Google Scholar] [CrossRef]
- De Wet, J.; Slippers, B.; Preisig, O.; Wingfield, B.D.; Wingfield, M.J. Phylogeny of the Botryosphaeriaceae Reveals Patterns of Host Association. Mol. Phylogenet. Evol. 2008, 46, 116–126. [Google Scholar] [CrossRef]
- Smith, H.; Wingfield, M.J.; Coutinho, T.A.; Crous, P.W. Sphaeropsis Sapinea and Botryosphaeria Dothidea Endophytic in Pinus Spp. and Eucalyptus Spp. in South Africa. S Afr J. Bot. 1996, 62, 86–88. [Google Scholar] [CrossRef] [Green Version]
- Guillarme, D.; Nguyen, D.T.T.; Rudaz, S.; Veuthey, J.-L. Method Transfer for Fast Liquid Chromatography in Pharmaceutical Analysis: Application to Short Columns Packed with Small Particle. Part II: Gradient Experiments. Eur. J. Pharm. Biopharm. 2008, 68, 430–440. [Google Scholar] [CrossRef]
- Guoyou, L.; Bogang, L.; Guangye, L.; Guolin, Z. Sterols from Aspergillus Ochraceus 43. Ying Yong Yu Huan Jing Sheng Wu Xue Bao 2005, 11, 67–70. [Google Scholar]
- Klemke, C.; Kehraus, S.; Wright, A.D.; König, G.M. New Secondary Metabolites from the Marine Endophytic Fungus Apiospora Montagnei. J. Nat. Prod. 2004, 67, 1058–1063. [Google Scholar] [CrossRef] [PubMed]
- Pellissier, L.; Koval, A.; Marcourt, L.; Ferreira Queiroz, E.; Lecoultre, N.; Leoni, S.; Quiros-Guerrero, L.-M.; Barthélémy, M.; Duivelshof, B.L.; Guillarme, D.; et al. Isolation and Identification of Isocoumarin Derivatives with Specific Inhibitory Activity against Wnt Pathway and Metabolome Characterization of Lasiodiplodia Venezuelensis. Front. Chem. In press. [CrossRef]
- Cheng, X.; Quintanilla, C.D.; Zhang, L. Total Synthesis and Structure Revision of Diplobifuranylone B. J. Org. Chem. 2019, 84, 11054–11060. [Google Scholar] [CrossRef]
- Xiang Yang, J.; Chen, Y.; Huang, C.; She, Z.; Lin, Y. A New Isochroman Derivative from the Marine Fungus Phomopsis Sp. (No. ZH-111). Chem. Nat. Compd. 2011, 47, 13. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, P.-M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Pham, V.C.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.-L.; Allard, P.-M. Taxonomically Informed Scoring Enhances Confidence in Natural Products Annotation. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Eloff, J.N. A sensitive and quick microplate method to determine the minimal inhibitory concentration of plant extracts for bacteria. Planta Med. 1998, 64, 711–713. [Google Scholar] [CrossRef] [Green Version]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of Quorum Sensing in Pseudomonas Aeruginosa Biofilm Bacteria by a Halogenated Furanone Compound. Microbiology 2002, 148, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Siegmund, I.; Wagner, F. New Method for Detecting Rhamnolipids Excreted ByPseudomonas Species during Growth on Mineral Agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]
- Queiroz, E.F.; Alfattani, A.; Afzan, A.; Marcourt, L.; Guillarme, D.; Wolfender, J.-L. Utility of Dry Load Injection for an Efficient Natural Products Isolation at the Semi-Preparative Chromatographic Scale. J. Chromatogr. A 2019, 1598, 85–91. [Google Scholar] [CrossRef]
- Martínez-Fructuoso, L.; Pereda-Miranda, R.; Rosas-Ramírez, D.; Fragoso-Serrano, M.; Cerda-García-Rojas, C.M.; da Silva, A.S.; Leitão, G.G.; Leitão, S.G. Structure Elucidation, Conformation, and Configuration of Cytotoxic 6-Heptyl-5,6-Dihydro-2H-Pyran-2-Ones from Hyptis Species and Their Molecular Docking to α-Tubulin. J. Nat. Prod. 2019, 82, 520–531. [Google Scholar] [CrossRef]
- Rahman, M.M.; Gray, A.I. A Benzoisofuranone Derivative and Carbazole Alkaloids from Murraya Koenigii and Their Antimicrobial Activity. Phytochemistry 2005, 66, 1601–1606. [Google Scholar] [CrossRef]
- Meepagala, K.M.; Briscoe, W.E.; Techen, N.; Johnson, R.D.; Clausen, B.M.; Duke, S.O. Isolation of a Phytotoxic Isocoumarin from Diaporthe Eres-Infected Hedera Helix (English Ivy) and Synthesis of Its Phytotoxic Analogs. Pest. Manag. Sci. 2018, 74, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Zezzi do Valle Gomes, M.; Nitschke, M. Evaluation of Rhamnolipid and Surfactin to Reduce the Adhesion and Remove Biofilms of Individual and Mixed Cultures of Food Pathogenic Bacteria. Food Control. 2012, 25, 441–447. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Lépine, F.; Déziel, E. Rhamnolipids: Diversity of Structures, Microbial Origins and Roles. Appl. Microbiol. Biotechnol. 2010, 86, 1323–1336. [Google Scholar] [CrossRef] [Green Version]
- Gdaniec, B.G.; Allard, P.-M.; Queiroz, E.F.; Wolfender, J.-L.; van Delden, C.; Köhler, T. Surface Sensing Triggers a Broad-Spectrum Antimicrobial Response in Pseudomonas Aeruginosa. Environ. Microbiol. 2020, 22, 3572–3587. [Google Scholar] [CrossRef] [PubMed]
- Davenport, P.W.; Griffin, J.L.; Welch, M. Quorum Sensing Is Accompanied by Global Metabolic Changes in the Opportunistic Human Pathogen Pseudomonas Aeruginosa. J. Bacteriol. 2015, 197, 2072–2082. [Google Scholar] [CrossRef] [Green Version]
- Depke, T.; Thöming, J.G.; Kordes, A.; Häussler, S.; Brönstrup, M. Untargeted LC-MS Metabolomics Differentiates Between Virulent and Avirulent Clinical Strains of Pseudomonas Aeruginosa. Biomolecules 2020, 10, 1041. [Google Scholar] [CrossRef] [PubMed]
- Lybbert, A.C.; Williams, J.L.; Raghuvanshi, R.; Jones, A.D.; Quinn, R.A. Mining Public Mass Spectrometry Data to Characterize the Diversity and Ubiquity of P. Aeruginosa Specialized Metabolites. Metabolites 2020, 10, 445. [Google Scholar] [CrossRef]
- Szamosvári, D.; Prothiwa, M.; Dieterich, C.L.; Böttcher, T. Profiling Structural Diversity and Activity of 2-Alkyl-4(1H)-Quinolone N-Oxides of Pseudomonas and Burkholderia. Chem. Commun. 2020, 56, 6328–6331. [Google Scholar] [CrossRef]
- Fletcher, M.P.; Diggle, S.P.; Cámara, M.; Williams, P. Biosensor-Based Assays for PQS, HHQ and Related 2-Alkyl-4-Quinolone Quorum Sensing Signal Molecules. Nat. Protoc. 2007, 2, 1254–1262. [Google Scholar] [CrossRef] [Green Version]
- Munir, S.; Shah, A.A.; Shahid, M.; Manzoor, I.; Aslam, B.; Rasool, M.H.; Saeed, M.; Ayaz, S.; Khurshid, M. Quorum Sensing Interfering Strategies and Their Implications in the Management of Biofilm-Associated Bacterial Infections. Braz. Arch. Biol. Technol. 2020, 63. [Google Scholar] [CrossRef]
- Boucard, V.; Broustal, G.; Campagne, J.M. Synthetic Approaches to α,β-Unsaturated δ-Lactones and Lactols. Eur. J. Org. Chem. 2007, 2007, 225–236. [Google Scholar] [CrossRef]
- Stierle, D.B.; Stierle, A.A.; Ganser, B. New Phomopsolides from a Penicillium Sp. J. Nat. Prod. 1997, 60, 1207–1209. [Google Scholar] [CrossRef]
- Grove, J.F. Metabolic Products of Phomopsis Oblonga. Part 2. Phomopsolide A and B, Tiglic Esters of Two 6-Substituted 5,6-Dihydro-5-Hydroxypyran-2-Ones. J. Chem. Soc. Perkin Trans. 1 1985, 865–869. [Google Scholar] [CrossRef]
- Matsumoto, M.; Nago, H. (R)-2-Octeno-δ-Lactone and Other Volatiles Produced by Lasiodiplodia Theobromae. Biosci. Biotechnol. Biochem. 1994, 58, 1262–1266. [Google Scholar] [CrossRef]
- Rajesh, P.S.; Ravishankar Rai, V. Hydrolytic Enzymes and Quorum Sensing Inhibitors from Endophytic Fungi of Ventilago Madraspatana Gaertn. Biocatal. Agric. Biotechnol. 2013, 2, 120–124. [Google Scholar] [CrossRef]
- Taufiq, M.M.J.; Darah, I. Antibacterial and Antibiofilm Activities of Crude Extract of Lasiodiplodia Pseudotheobromae IBRL OS-64 against Foodborne Bacterium, Yersinia Enterocolitica. J. Pharm Res. Int. 2020, 87–102. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Brameyer, S.; Kresovic, D.; Hitkova, I.; Kopp, Y.; Manske, C.; Schubert, K.; Bode, H.B.; Heermann, R. Pyrones as Bacterial Signaling Molecules. Nat. Chem. Biol. 2013, 9, 573–578. [Google Scholar] [CrossRef]
- Kong, F.; Singh, M.P.; Carter, G.T. Pseudopyronines A and B, α-Pyrones Produced by a Marine Pseudomonas Sp. F92S91, and Evidence for the Conversion of 4-Hydroxy-α-Pyrone to 3-Furanone. J. Nat. Prod. 2005, 68, 920–923. [Google Scholar] [CrossRef] [PubMed]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The Role of Pyocyanin in Pseudomonas Aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef]
- Cornelis, P.; Dingemans, J. Pseudomonas Aeruginosa Adapts Its Iron Uptake Strategies in Function of the Type of Infections. Front. Cell. Infect. Microbiol. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Bielecki, P.; Puchałka, J.; Wos-Oxley, M.L.; Loessner, H.; Glik, J.; Kawecki, M.; Nowak, M.; Tümmler, B.; Weiss, S.; Santos, V.A.P.M. dos In-Vivo Expression Profiling of Pseudomonas Aeruginosa Infections Reveals Niche-Specific and Strain-Independent Transcriptional Programs. PLoS ONE 2011, 6, e24235. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, M.R.; Fleuchot, B.; Lauciello, L.; Jafari, P.; Applegate, L.A.; Raffoul, W.; Que, Y.-A.; Perron, K. Effect of Human Burn Wound Exudate on Pseudomonas Aeruginosa Virulence. mSphere 1 2016, 1, e00111-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukarieh, F.; Williams, P.; Stocks, M.J.; Cámara, M. Pseudomonas Aeruginosa Quorum Sensing Systems as Drug Discovery Targets: Current Position and Future Perspectives. J. Med. Chem. 2018, 61, 10385–10402. [Google Scholar] [CrossRef] [Green Version]
- McInnis, C.E.; Blackwell, H.E. Design, Synthesis, and Biological Evaluation of Abiotic, Non-Lactone Modulators of LuxR-Type Quorum Sensing. Bioorg. Med. Chem. 2011, 19, 4812–4819. [Google Scholar] [CrossRef] [Green Version]
- Park, S.; Kim, H.-S.; Ok, K.; Kim, Y.; Park, H.-D.; Byun, Y. Design, Synthesis and Biological Evaluation of 4-(Alkyloxy)-6-Methyl-2H-Pyran-2-One Derivatives as Quorum Sensing Inhibitors. Bioorganic Med. Chem. Lett. 2015, 25, 2913–2917. [Google Scholar] [CrossRef]
- Cathcart, G.R.A.; Quinn, D.; Greer, B.; Harriott, P.; Lynas, J.F.; Gilmore, B.F.; Walker, B. Novel Inhibitors of the Pseudomonas Aeruginosa Virulence Factor LasB: A Potential Therapeutic Approach for the Attenuation of Virulence Mechanisms in Pseudomonal Infection. Antimicrob. Agents Chemother. 2011, 55, 2670–2678. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellissier, L.; Leoni, S.; Marcourt, L.; Ferreira Queiroz, E.; Lecoultre, N.; Quiros-Guerrero, L.-M.; Barthélémy, M.; Eparvier, V.; Chave, J.; Stien, D.; et al. Characterization of Pseudomonas aeruginosa Quorum Sensing Inhibitors from the Endophyte Lasiodiplodia venezuelensis and Evaluation of Their Antivirulence Effects by Metabolomics. Microorganisms 2021, 9, 1807. https://doi.org/10.3390/microorganisms9091807
Pellissier L, Leoni S, Marcourt L, Ferreira Queiroz E, Lecoultre N, Quiros-Guerrero L-M, Barthélémy M, Eparvier V, Chave J, Stien D, et al. Characterization of Pseudomonas aeruginosa Quorum Sensing Inhibitors from the Endophyte Lasiodiplodia venezuelensis and Evaluation of Their Antivirulence Effects by Metabolomics. Microorganisms. 2021; 9(9):1807. https://doi.org/10.3390/microorganisms9091807
Chicago/Turabian StylePellissier, Léonie, Sara Leoni, Laurence Marcourt, Emerson Ferreira Queiroz, Nicole Lecoultre, Luis-Manuel Quiros-Guerrero, Morgane Barthélémy, Véronique Eparvier, Jérôme Chave, Didier Stien, and et al. 2021. "Characterization of Pseudomonas aeruginosa Quorum Sensing Inhibitors from the Endophyte Lasiodiplodia venezuelensis and Evaluation of Their Antivirulence Effects by Metabolomics" Microorganisms 9, no. 9: 1807. https://doi.org/10.3390/microorganisms9091807
APA StylePellissier, L., Leoni, S., Marcourt, L., Ferreira Queiroz, E., Lecoultre, N., Quiros-Guerrero, L.-M., Barthélémy, M., Eparvier, V., Chave, J., Stien, D., Gindro, K., Perron, K., & Wolfender, J.-L. (2021). Characterization of Pseudomonas aeruginosa Quorum Sensing Inhibitors from the Endophyte Lasiodiplodia venezuelensis and Evaluation of Their Antivirulence Effects by Metabolomics. Microorganisms, 9(9), 1807. https://doi.org/10.3390/microorganisms9091807








