The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origin of AP_SAP11-Like_PM19 DNA
2.2. Transient Protein Expression in Planta
2.3. Generation of Transgenic A. thaliana Lines
2.4. RT-qPCR
2.5. Yeast Two-Hybrid Analysis
2.6. TCP Destabilization Assay
3. Results
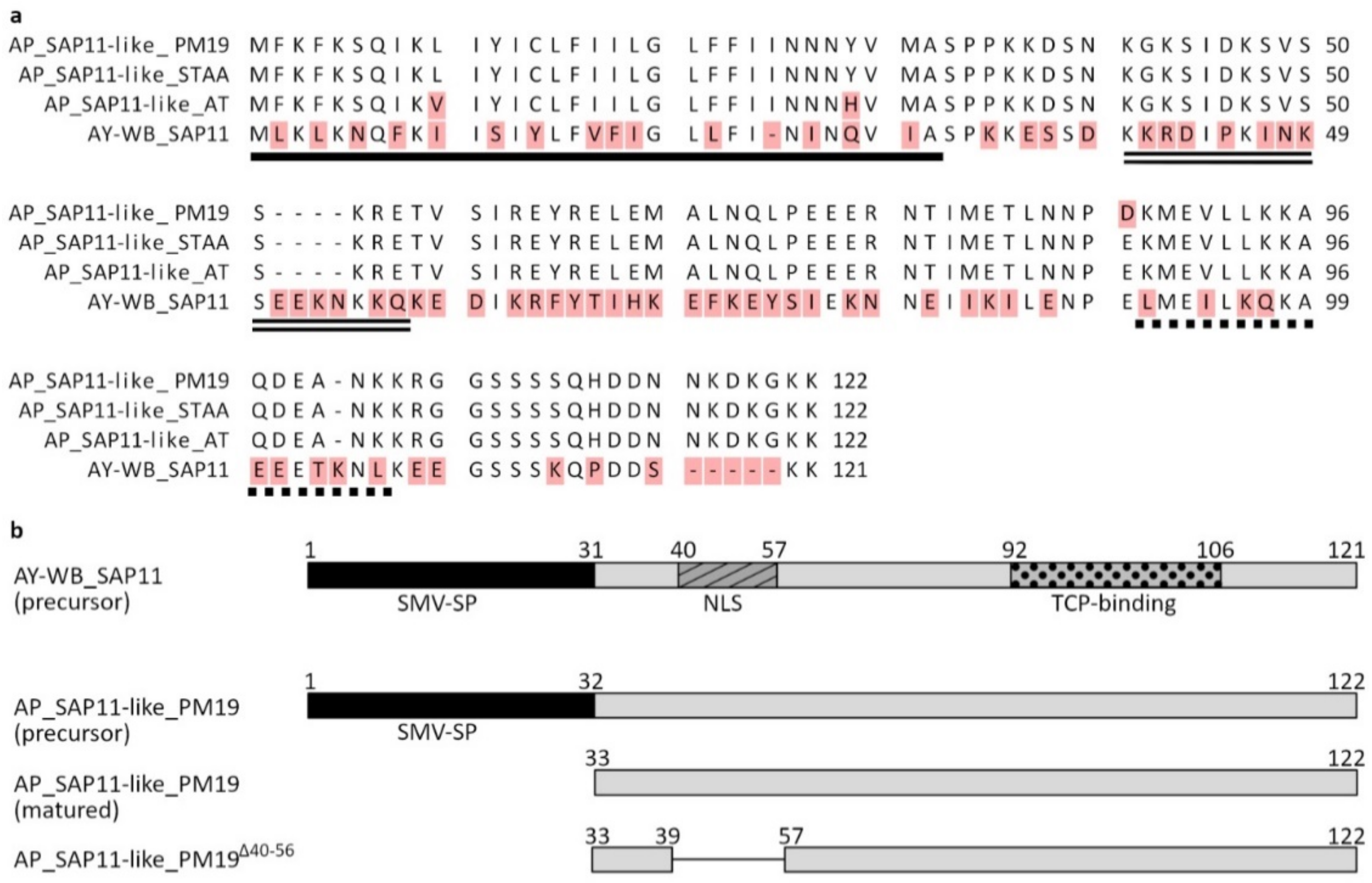
3.1. Sequence Analysis of SAP11-like Protein of ‘Candidatus Phytoplasma mali’ Strain PM19
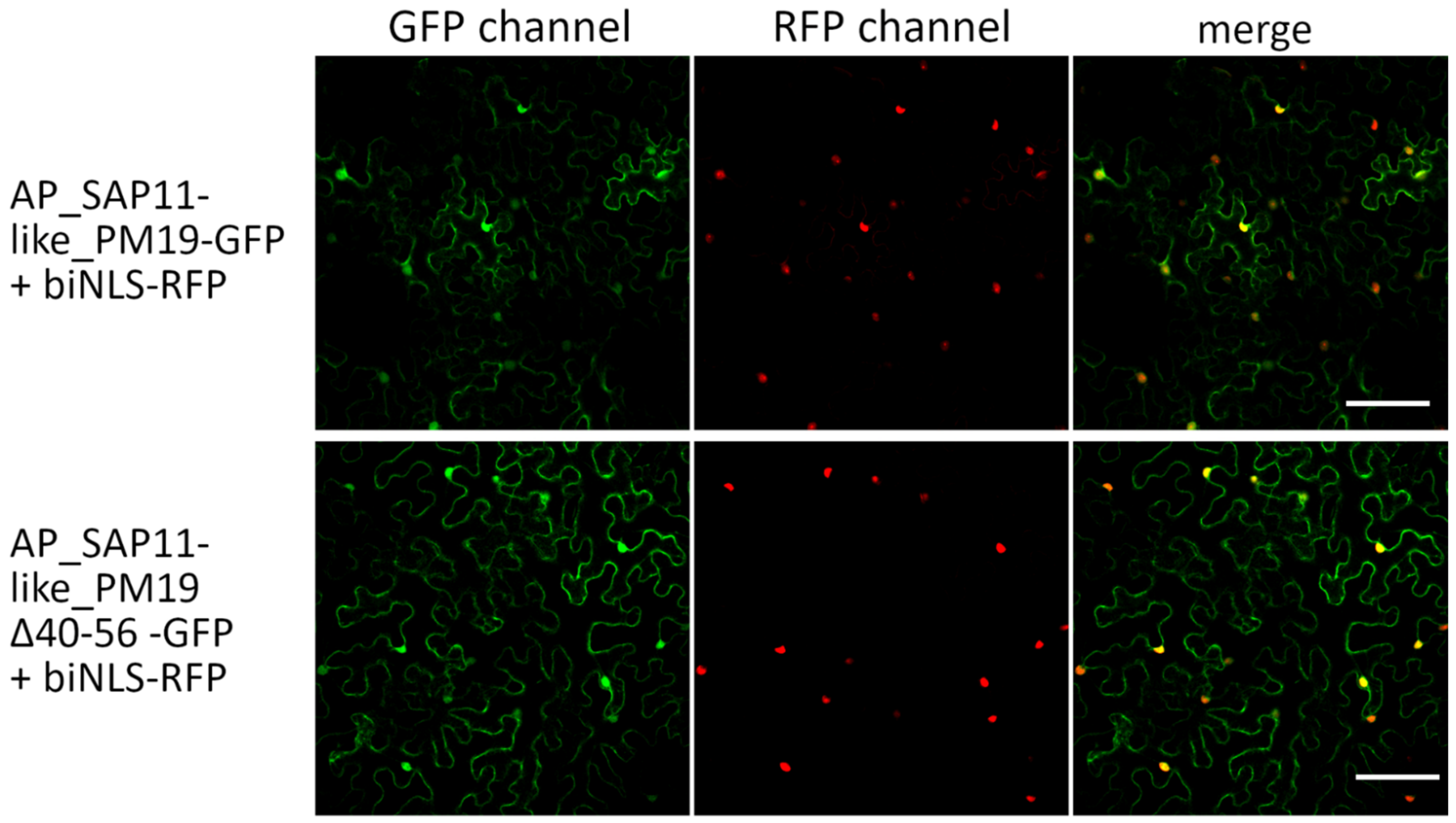
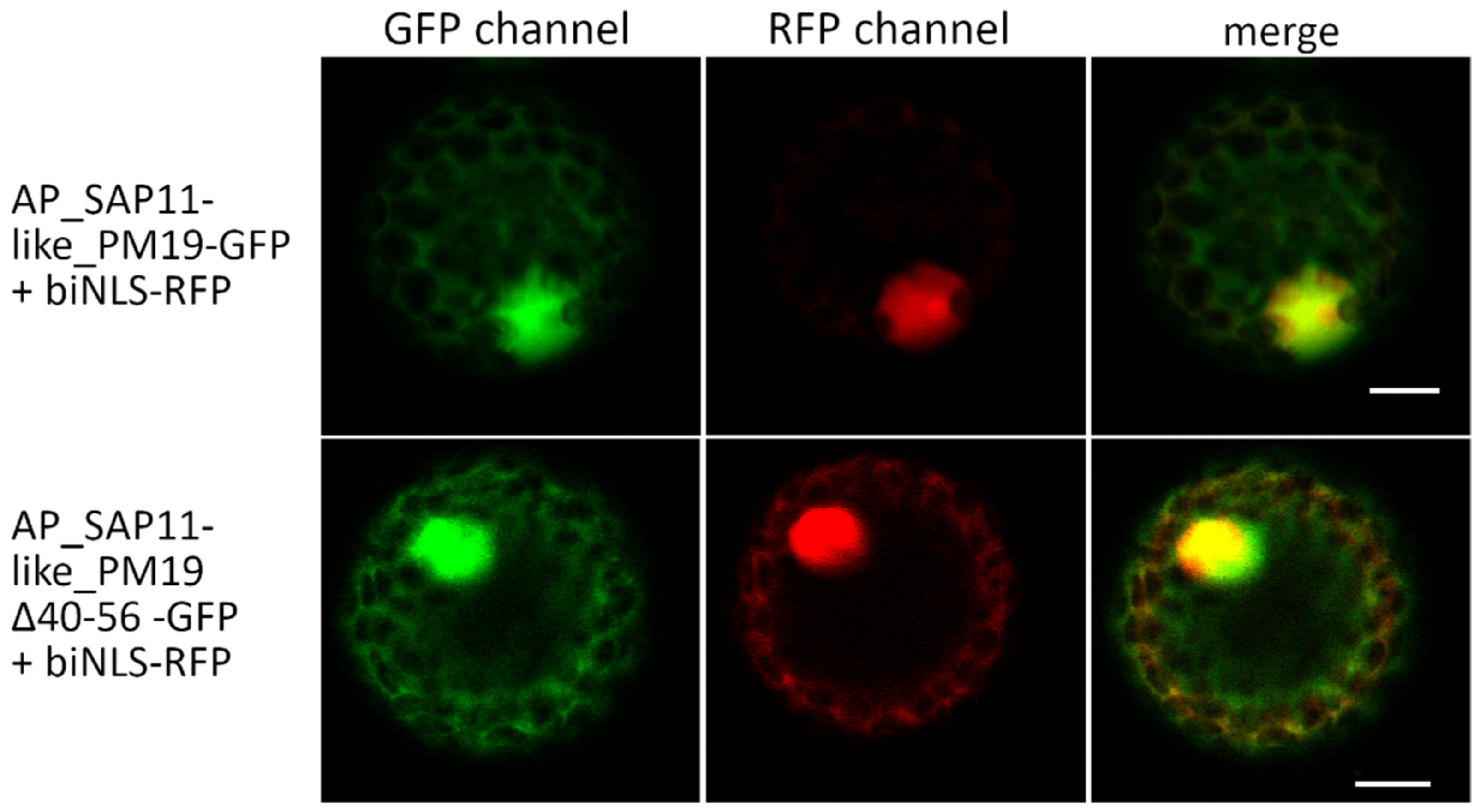
3.2. Amino Acids 40 to 56 of AP_SAP11-like_PM19 Are Not Necessary for Nuclear Localization of the Protein in N. benthamiana
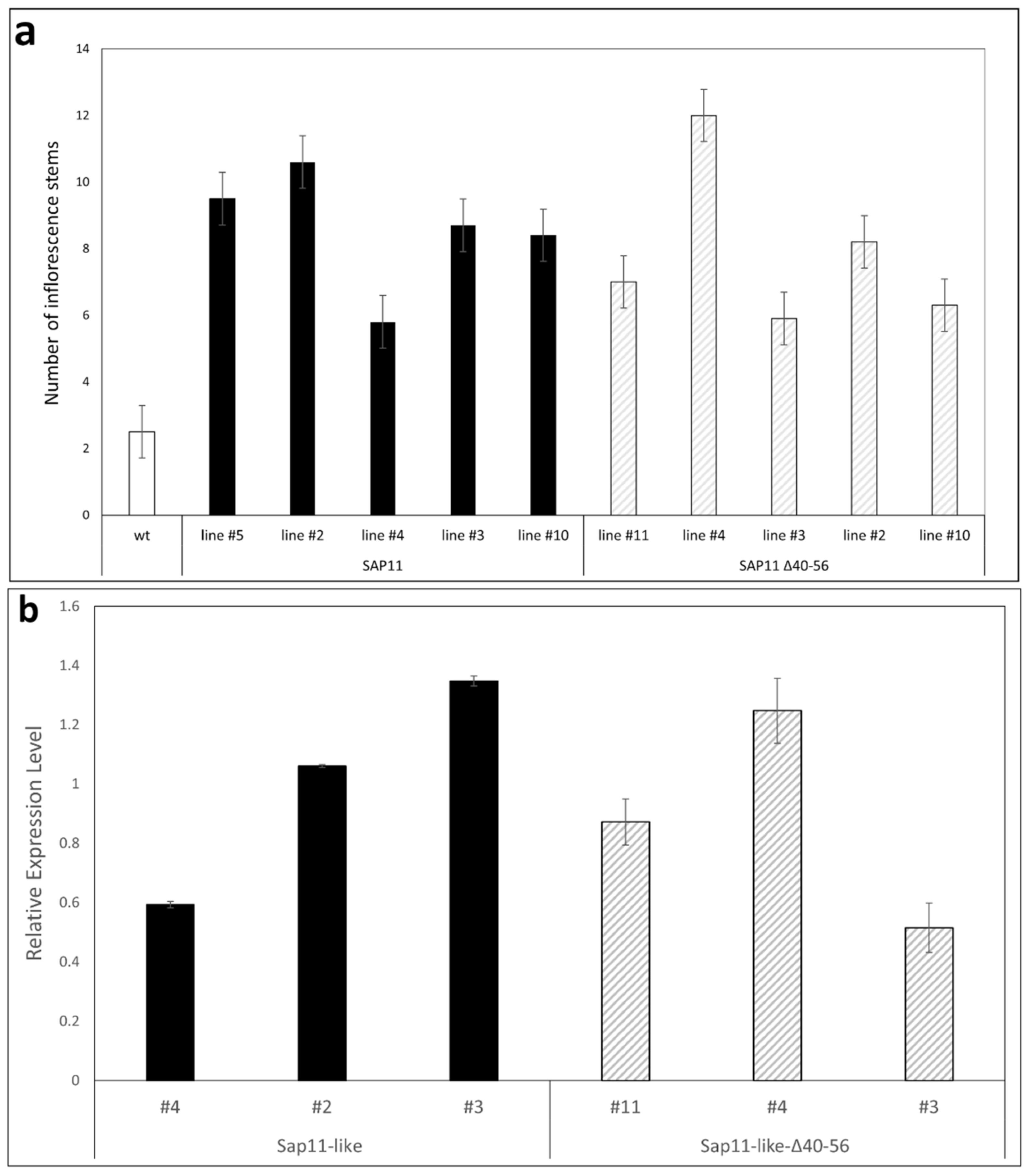
3.3. AP_SAP11-like_PM19 Induces Crinkled Leaves and Siliques and Witches’ Broom Symptoms in Arabidopsis
3.4. Amino Acids 40 to 56 of AP_ SAP11-like Protein Are Important for Symptom Development
3.5. Amino Acids 40 to 56 of AP_SAP11-like_PM19 Are Important for Binding to Some A. thaliana (At) TCPs in Yeast Two-Hybrid (Y2H) Analysis
3.6. The 40–56 Amino Acid Stretch of AP_SAP11-like_PM19 Is Not Required to Destabilize Some of the TCPs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogenhout, S.A.; van der Hoorn, R.A.L.; Terauchi, R.; Kamoun, S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009, 22, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Sugio, A.; MacLean, A.M.; Kingdom, H.N.; Grieve, V.M.; Manimekalai, R.; Hogenhout, S.A. Diverse targets of phytoplasma effectors: From plant development to defense against insects. Annu. Rev. Phytopathol. 2011, 49, 175–195. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Oshima, K.; Ammar, E.-D.; Kakizawa, S.; Kingdom, H.N.; Namba, S. Phytoplasmas: Bacteria that manipulate plants and insects. Mol. Plant Pathol. 2008, 9, 403–423. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso Radek, A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Correa, V.R.; Toruño, T.Y.; Ammar, E.-D.; Kamoun, S.; Hogenhout, S.A. AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant Microbe Interact. 2009, 22, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugio, A.; MacLean, A.M.; Hogenhout, S.A. The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef] [Green Version]
- Siewert, C.; Luge, T.; Duduk, B.; Seemüller, E.; Büttner, C.; Sauer, S.; Kube, M. Analysis of expressed genes of the bacterium ‘Candidatus phytoplasma Mali’ highlights key features of virulence and metabolism. PLoS ONE 2014, 9, e94391. [Google Scholar] [CrossRef] [Green Version]
- Jomantiene, R.; Zhao, Y.; Davis, R.E. Sequence-variable mosaics: Composites of recurrent transposition characterizing the genomes of phylogenetically diverse phytoplasmas. DNA Cell Biol. 2007, 26, 557–564. [Google Scholar] [CrossRef]
- Janik, K.; Mithöfer, A.; Raffeiner, M.; Stellmach, H.; Hause, B.; Schlink, K. An effector of apple proliferation phytoplasma targets TCP transcription factors—A generalized virulence strategy of phytoplasma? Mol. Plant Pathol. 2017, 18, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.J.; Vilcinskas, A.; Gross, J. Pathogen-induced release of plant allomone manipulates vector insect behavior. J. Chem. Ecol. 2008, 34, 1518–1522. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.J.; Vilcinskas, A.; Gross, J. Phytopathogen lures its insect vector by altering host plant odor. J. Chem. Ecol. 2008, 34, 1045–1049. [Google Scholar] [CrossRef]
- Tan, C.M.; Li, C.-H.; Tsao, N.-W.; Su, L.-W.; Lu, Y.-T.; Chang, S.H.; Lin, Y.Y.; Liou, J.-C.; Hsieh, L.-C.; Yu, J.-Z.; et al. Phytoplasma SAP11 alters 3-isobutyl-2-methoxypyrazine biosynthesis in Nicotiana benthamiana by suppressing NbOMT1. J. Exp. Bot. 2016, 67, 4415–4425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef]
- Jarausch, B.; Schwind, N.; Fuchs, A.; Jarausch, W. Characteristics of the spread of apple proliferation by its vector Cacopsylla picta. Phytopathology 2011, 101, 1471–1480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kube, M.; Schneider, B.; Kuhl, H.; Dandekar, T.; Heitmann, K.; Migdoll, A.M.; Reinhardt, R.; Seemüller, E. The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali’. BMC Genom. 2008, 9, 306. [Google Scholar] [CrossRef] [Green Version]
- Schöb, H.; Kunz, C.; Meins, F. Silencing of transgenes introduced into leaves by agroinfiltration: A simple, rapid method for investigating sequence requirements for gene silencing. Mol. Gen. Genet. 1997, 256, 581–585. [Google Scholar] [CrossRef]
- Hajdukiewicz, P.; Svab, Z.; Maliga, P. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 1994, 25, 989–994. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Chang, S.H.; Tan, C.M.; Wu, C.-T.; Lin, T.-H.; Jiang, S.-Y.; Liu, R.-C.; Tsai, M.-C.; Su, L.-W.; Yang, J.-Y. Alterations of plant architecture and phase transition by the phytoplasma virulence factor SAP11. J. Exp. Bot. 2018, 69, 5389–5401. [Google Scholar] [CrossRef] [Green Version]
- Beckwith, J. The Sec-dependent pathway. Res. Microbiol. 2013, 164, 497–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, Y.; Ohya, H.; Yamaguchi, Y.; Koizumi, N.; Sano, H. Preferential de novo methylation of cytosine residues in non-CpG sequences by a domains rearranged DNA methyltransferase from tobacco plants. J. Biol. Chem. 2003, 278, 42386–42393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–36. [Google Scholar] [CrossRef]
- Cokol, M.; Nair, R.; Rost, B. Finding nuclear localization signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Brameier, M.; Krings, A.; MacCallum, R.M. NucPred—Predicting nuclear localization of proteins. Bioinformatics 2007, 23, 1159–1160. [Google Scholar] [CrossRef]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.J.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef] [Green Version]
- Tsuda, K.; Qi, Y.; Le Nguyen, V.; Bethke, G.; Tsuda, Y.; Glazebrook, J.; Katagiri, F. An efficient Agrobacterium-mediated transient transformation of Arabidopsis. Plant J. 2012, 69, 713–719. [Google Scholar] [CrossRef]
- Wroblewski, T.; Tomczak, A.; Michelmore, R. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 2005, 3, 259–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Yang, Y. Transient expression assay by agroinfiltration of leaves. Methods Mol. Biol. 2006, 323, 225–229. [Google Scholar] [CrossRef]
- Li, J.-F.; Park, E.; von Arnim, A.G.; Nebenführ, A. The FAST technique: A simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 2009, 5, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar] [CrossRef] [Green Version]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [Green Version]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef] [Green Version]
- Finlayson, S.A. Arabidopsis Teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 2007, 48, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Poza-Carrión, C.; Aguilar-Martínez, J.A.; Cubas, P. Role of TCP Gene BRANCHED1 in the Control of Shoot Branching in Arabidopsis. Plant Signal. Behav. 2007, 2, 551–552. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Qin, G. The Regulation of CIN-like TCP Transcription Factors. Int. J. Mol. Sci. 2020, 21, 4498. [Google Scholar] [CrossRef] [PubMed]
- Efroni, I.; Han, S.-K.; Kim, H.J.; Wu, M.-F.; Steiner, E.; Birnbaum, K.D.; Hong, J.C.; Eshed, Y.; Wagner, D. Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 2013, 24, 438–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, Q.; Guo, D.; Wei, B.; Zhang, F.; Pang, C.; Jiang, H.; Zhang, J.; Wei, T.; Gu, H.; Qu, L.-J.; et al. The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 2013, 25, 421–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Nicolas, M.; Zhang, J.; Yu, H.; Guo, D.; Yuan, R.; Zhang, T.; Yang, J.; Cubas, P.; Qin, G. The TIE1 transcriptional repressor controls shoot branching by directly repressing BRANCHED1 in Arabidopsis. PLoS Genet. 2018, 14, e1007296. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Davis, R.E.; Nuss, D.L.; Zhao, Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. USA 2013, 110, 19149–19154. [Google Scholar] [CrossRef] [Green Version]
- Lenhard, M.; Jürgens, G.; Laux, T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 2002, 129, 3195–3206. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.S.; Chen, C.; Wagner, D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005, 19, 992–1003. [Google Scholar] [CrossRef] [Green Version]
- Scarpella, E.; Barkoulas, M.; Tsiantis, M. Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2010, 2, a001511. [Google Scholar] [CrossRef] [Green Version]

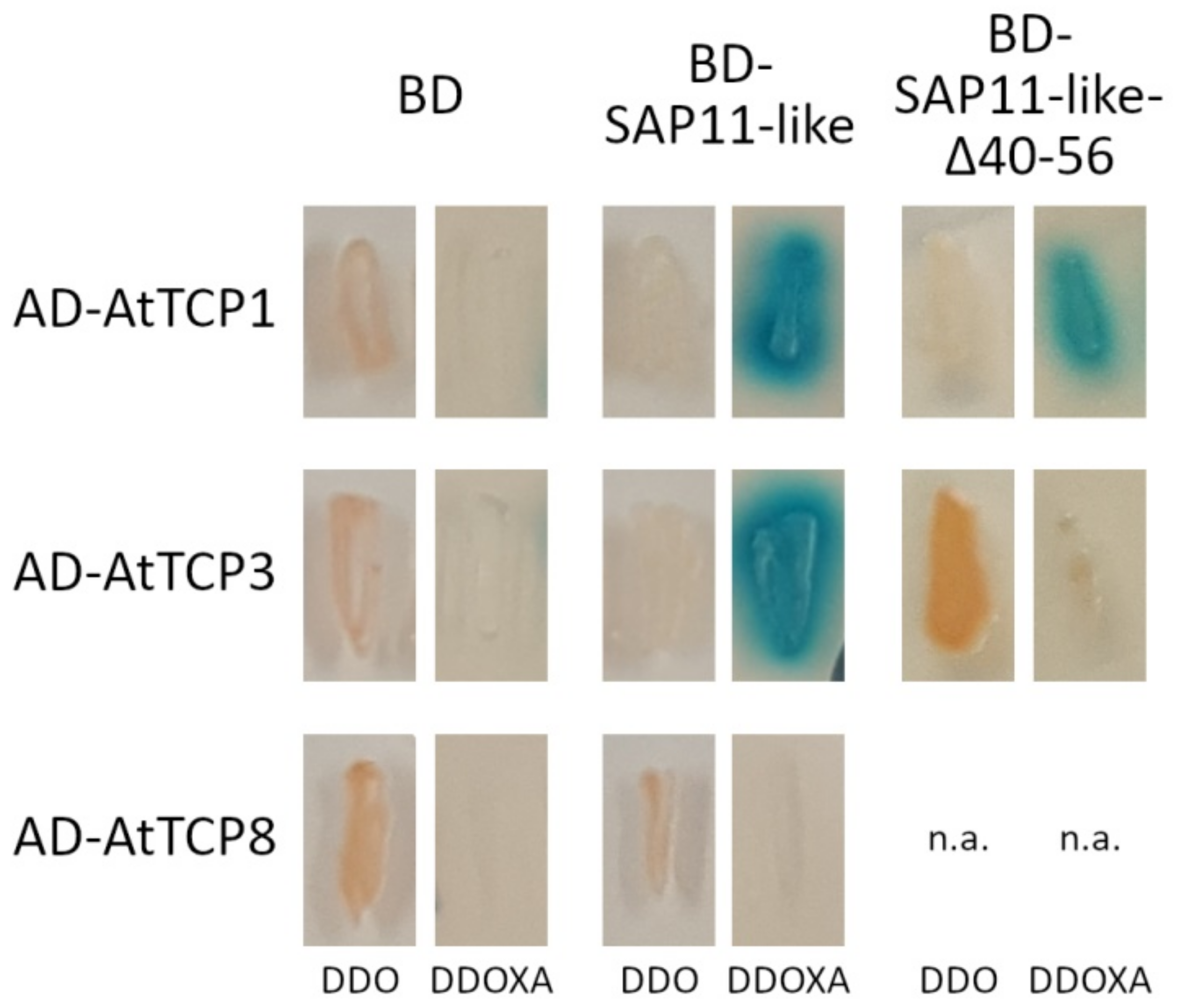
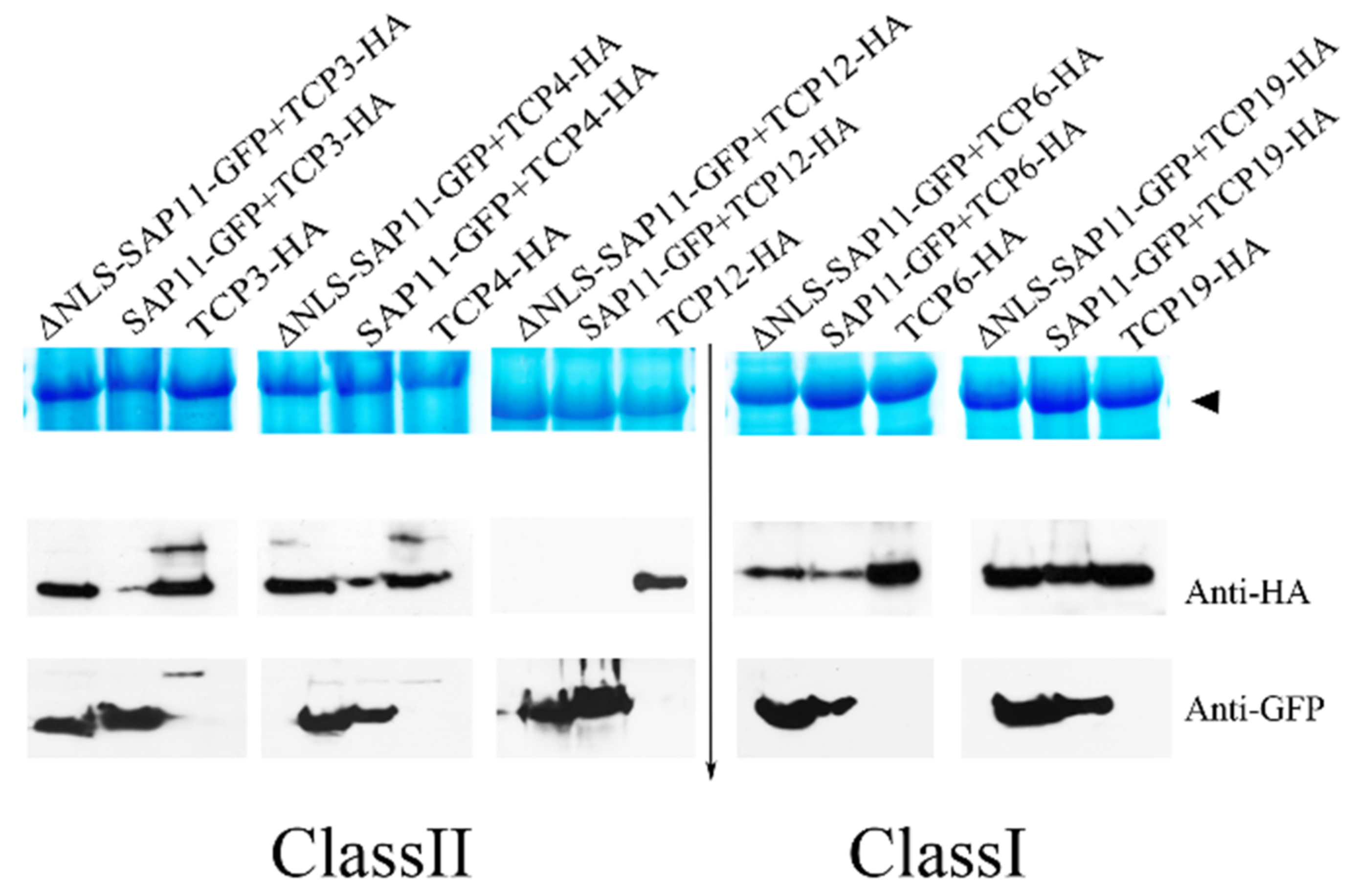
| AtTCP | Subclass | Interaction with AP_SAP11_like_ PM19 | Interaction with AP_SAP11_like_ PM19Δ40–56 |
|---|---|---|---|
| 1 | Class II | pos. | pos. |
| 2 | Class II | pos. | pos. |
| 3 | Class II | pos. | neg. |
| 4 | Class II | pos. | neg. |
| 5 | Class II | pos. | pos. |
| 6 | Class I | pos. | pos. |
| 7 | Class I | pos. | pos. |
| 8 | Class I | neg. | n.a. |
| 9 | Class I | pos. | pos. |
| 10 | Class II | pos. | neg. |
| 11 | Class I | neg. | n.a. |
| 12 | Class II | pos. | pos. |
| 13 | Class II | pos. | pos. |
| 14 | Class I | pos. | pos. |
| 15 | Class I | neg. | n.a. |
| 16 | Class I | neg. | n.a. |
| 17 | Class II | pos. | pos. |
| 18 | Class II | pos. | pos. |
| 19 | Class I | pos. | neg. |
| 20 | Class I | neg. | n.a. |
| 21 | Class I | pos. | pos. |
| 22 | Class I | neg. | n.a. |
| 23 | Class I | neg. | n.a. |
| 24 | Class II | pos. | pos. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strohmayer, A.; Schwarz, T.; Braun, M.; Krczal, G.; Boonrod, K. The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor. Microorganisms 2021, 9, 1756. https://doi.org/10.3390/microorganisms9081756
Strohmayer A, Schwarz T, Braun M, Krczal G, Boonrod K. The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor. Microorganisms. 2021; 9(8):1756. https://doi.org/10.3390/microorganisms9081756
Chicago/Turabian StyleStrohmayer, Alisa, Timothy Schwarz, Mario Braun, Gabi Krczal, and Kajohn Boonrod. 2021. "The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor" Microorganisms 9, no. 8: 1756. https://doi.org/10.3390/microorganisms9081756
APA StyleStrohmayer, A., Schwarz, T., Braun, M., Krczal, G., & Boonrod, K. (2021). The Effect of the Anticipated Nuclear Localization Sequence of ‘Candidatus Phytoplasma mali’ SAP11-like Protein on Localization of the Protein and Destabilization of TCP Transcription Factor. Microorganisms, 9(8), 1756. https://doi.org/10.3390/microorganisms9081756







