First Genome Description of Providencia vermicola Isolate Bearing NDM-1 from Blood Culture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation
2.2. Antimicrobial Susceptibility Testing
2.3. Molecular Mechanisms of Antibiotic Resistance and Whole Genome Sequencing
2.4. Bioinformatic Analysis
2.5. Conjugation Experiment
3. Results
3.1. Clinical Information and Phenotypic Characterisation of Isolates
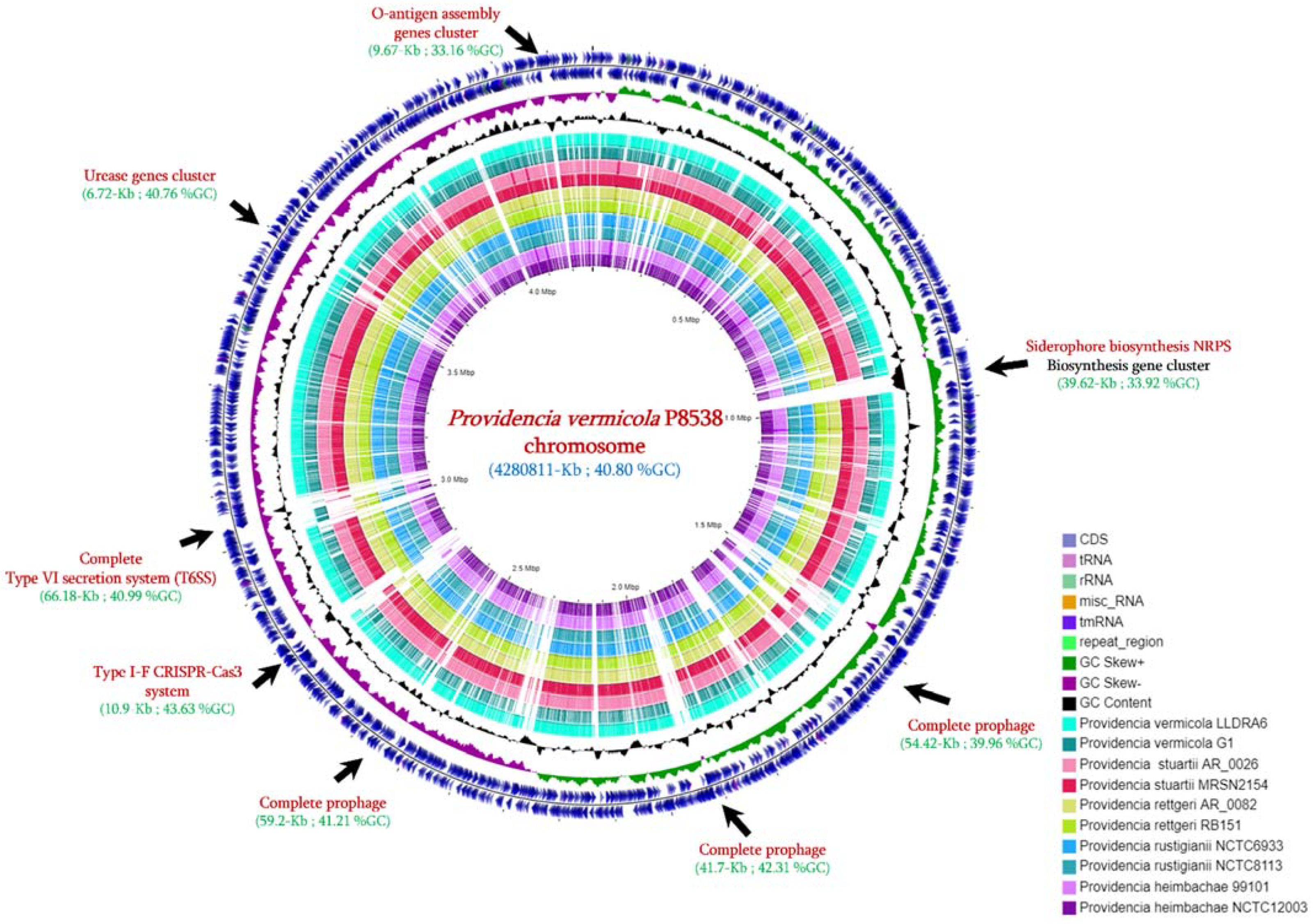
3.2. Genome Features
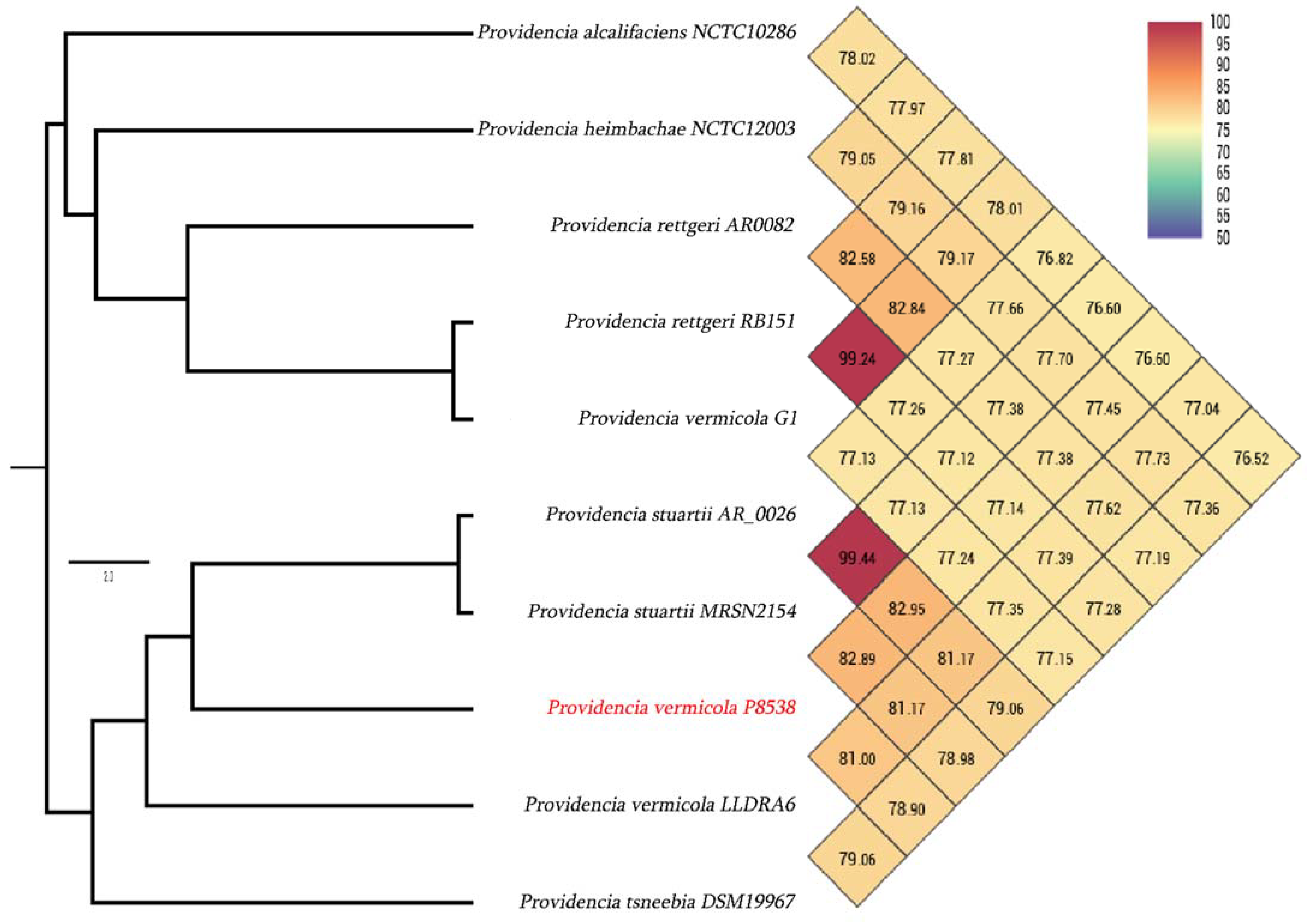
3.3. Genome Comparison with Closely Related Species
3.4. Resistome
3.5. Genomic Analysis of the E. coli P8540 Isolate
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Considerations
References
- Abdallah, M.; Balshi, A. First literature review of carbapenem-resistant Providencia. New Microbes New Infect. 2018, 25, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Sharma, P.; Soni, P. First case report of Providencia Rettgeri neonatal sepsis. BMC Res. Notes 2017, 10, 536. [Google Scholar] [CrossRef] [Green Version]
- Yuan, C.; Wei, Y.; Zhang, S.; Cheng, J.; Cheng, X.; Qian, C.; Wang, Y.; Zhang, Y.; Yin, Z.; Chen, H. Comparative Genomic Analysis Reveals Genetic Mechanisms of the Variety of Pathogenicity, Antibiotic Resistance, and Environmental Adaptation of Providencia Genus. Front. Microbiol. 2020, 11, 572642. [Google Scholar] [CrossRef]
- Mbelle, N.M.; Sekyere, J.O.; Amoako, D.G.; Maningi, N.; Modipane, L.; Essack, S.; Feldman, C. Genomic analysis of a multidrug-resistant clinical Providencia rettgeri (PR002) strain with the novel integron ln 1483 and an A/C plasmid replicon. Ann. N. Y. Acad. Sci. 2020, 1462, 92–103. [Google Scholar] [CrossRef]
- Guerfali, M.M.; Djobbi, W.; Charaabi, K.; Hamden, H.; Fadhl, S.; Marzouki, W.; Dhaouedi, F.; Chevrier, C. Evaluation of Providencia rettgeri pathogenicity against laboratory Mediterranean fruit fly strain (Ceratitis capitata). PLoS ONE 2018, 13, e0196343. [Google Scholar] [CrossRef]
- Ksentini, I.; Gharsallah, H.; Sahnoun, M.; Schuster, C.; Amri, S.H.; Gargouri, R.; Triki, M.A.; Ksantini, M.; Leclerque, A. Providencia entomophila sp. nov., a new bacterial species associated with major olive pests in Tunisia. PLoS ONE 2019, 14, e0223943. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, V.S.; Lang, E.; Sträubler, B.; Spröer, C.; Schumann, P.; Ganguly, S.; Saxena, A.K.; Stackebrandt, E. Providencia vermicola sp. nov., isolated from infective juveniles of the entomopathogenic nematode Steinernema thermophilum. Int. J. Syst. Evol. Microbiol. 2006, 56, 629–633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erajpara, N.; Kutar, B.M.R.N.S.; Esinha, R.; Enag, D.; Ekoley, H.; Eramamurthy, T.; Bhardwaj, A.E. Role of integrons, plasmids and SXT elements in multidrug resistance of Vibrio cholerae and Providencia vermicola obtained from a clinical isolate of diarrhea. Front. Microbiol. 2015, 6, 57. [Google Scholar] [CrossRef] [Green Version]
- Mnif, B.; Ktari, S.; Chaari, A.; Medhioub, F.; Rhimi, F.; Bouaziz, M.; Hammami, A. Nosocomial dissemination of Providencia stuartii isolates carrying blaOXA-48, blaPER-1, blaCMY-4 and qnrA6 in a Tunisian hospital. J. Antimicrob. Chemother. 2013, 68, 329–332. [Google Scholar] [CrossRef] [Green Version]
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: An updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Tritt, A.; Eisen, J.A.; Facciotti, M.T.; Darling, A.E. An Integrated Pipeline for de Novo Assembly of Microbial Genomes. PLoS ONE 2012, 7, e42304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Stothard, P.; Grant, J.R.; Van Domselaar, G. Visualizing and comparing circular genomes using the CGView family of tools. Briefings Bioinform. 2018, 20, 1576–1582. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Gupta, S.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Ramoul, A.; Loucif, L.; Bakour, S.; Amiri, S.; Dekhil, M.; Rolain, J.-M. Co-occurrence of bla NDM-1 with bla OXA-23 or bla OXA-58 in clinical multidrug-resistant Acinetobacter baumannii isolates in Algeria. J. Glob. Antimicrob. Resist. 2016, 6, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.; Diene, S.M.; Kempf, M.; Berrazeg, M.; Bakour, S.; Gupta, S.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: An epidemiological and molecular study. Int. J. Antimicrob. Agents 2014, 44, 500–507. [Google Scholar] [CrossRef] [Green Version]
- Diene, S.M.; Merhej, V.; Henry, M.; El Filali, A.; Roux, V.; Robert, C.; Azza, S.; Gavory, F.; Barbe, V.; La Scola, B.; et al. The Rhizome of the Multidrug-Resistant Enterobacter aerogenes Genome Reveals How New “Killer Bugs” Are Created because of a Sympatric Lifestyle. Mol. Biol. Evol. 2013, 30, 369–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, C.F.; Proudfoot, M.; Brown, G.; Korniyenko, Y.; Mori, H.; Savchenko, A.V.; Yakunin, A. Molecular Basis of Formaldehyde Detoxification. J. Biol. Chem. 2006, 281, 14514–14522. [Google Scholar] [CrossRef] [Green Version]
- Harms, N.; Ras, J.; Reijnders, W.N.; van Spanning, R.; Stouthamer, A.H. S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: A universal pathway for formaldehyde detoxification? J. Bacteriol. 1996, 178, 6296–6299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruns, H.; Crüsemann, M.; Letzel, A.-C.; Alanjary, M.; McInerney, J.O.; Jensen, P.; Schulz, S.; Moore, B.S.; Ziemert, N. Function-related replacement of bacterial siderophore pathways. ISME J. 2018, 12, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Garcia, E.C.; Brumbaugh, A.R.; Mobley, H.L.T. Redundancy and Specificity of Escherichia coli Iron Acquisition Systems during Urinary Tract Infection. Infect. Immun. 2011, 79, 1225–1235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weakland, D.R.; Smith, S.N.; Bell, B.; Tripathi, A.; Mobley, H.L.T. The Serratia marcescens Siderophore Serratiochelin Is Necessary for Full Virulence during Bloodstream Infection. Infect. Immun. 2020, 88. [Google Scholar] [CrossRef] [PubMed]
- Holden, V.; Breen, P.; Houle, S.; Dozois, C.M.; Bachman, M.A. Klebsiella pneumoniae Siderophores Induce Inflammation, Bacterial Dissemination, and HIF-1α Stabilization during Pneumonia. mBio 2016, 7, e01397-16. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Mekalanos, J.J. Type 6 Secretion Dynamics Within and Between Bacterial Cells. Science 2012, 337, 815. [Google Scholar] [CrossRef] [Green Version]
- Records, A.R. The Type VI Secretion System: A Multipurpose Delivery System with a Phage-Like Machinery. Mol. Plant Microbe Interact. 2011, 24, 751–757. [Google Scholar] [CrossRef] [Green Version]
- Leiman, P.; Basler, M.; Ramagopal, U.A.; Bonanno, J.B.; Sauder, J.M.; Pukatzki, S.; Burley, S.; Almo, S.C.; Mekalanos, J.J. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 2009, 106, 4154–4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.-W.; Xue, P.; Fu, Y.; Yang, L. T6SS Mediated Stress Responses for Bacterial Environmental Survival and Host Adaptation. Int. J. Mol. Sci. 2021, 22, 478. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Ma, A.; Sturtevant, D.; Krastins, B.; Sarracino, D.; Nelson, W.; Heidelberg, J.; Mekalanos, J.J. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 2006, 103, 1528–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, W.; Nie, L.; Baskaran, R.; Kumar, A.; Liu, Z. Characterization and improved properties of Glutamine synthetase from Providencia vermicola by site-directed mutagenesis. Sci. Rep. 2018, 8, 15640. [Google Scholar] [CrossRef] [PubMed]


| Strain Names | Sex | Age | Sample | Status | Service | Resistance Phenotype | MIC IPM (µg/mL) | Antimicrobial Resistance Genes | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β-lactams | Aminoglycosides | Sulfonamides and Trimethoprim | Phenicols | Quinolones and Imidazoles | Cyclines | ||||||||
| P. vermicola P8538 | M | 58 | Blood | Inpatient | ICU | AX, AMC, TZP, KF, CRO, ERT, IPM, AK, GEN, CIP, DO | >32 | blaCMY-6, blaNDM-1, H-NS | rmtC, aac(6′)-Ib3, aac(6′)-Ib10, aac(2)-Ia | sul1 | catA1 | aac(6′)-Ib-cr, acrB, msbA, mdtH, crp | tetA, tetB, tetD |
| E. coli P8540 | F | 26 | Urine | Inpatient | ICU | AX, AMC, KF, CRO, FEP, ERT, IPM, AK, GEN, CIP, DO | >32 | blaTEM-1B, blaSHV-12, blaAmpC1, blaCMY-42, evgS, evgA, blaCTX-M-88, blaNDM-1, blaCTX-M-15, ampH H-NS | aph(6)-Id, aph(3′’)-Ib, aadA2, aadA1, aac(3′)-IId, aac(6′)-Ib3, rmtC, aadA16, baeR, baeS, strB, strA | dfrA27, dfr12 | catA2, aadA2, mdtm, catII | qnrB6, qnrS1, aac(6′)-Ib-cr, qepA, emrR, emrA, emrB, mdtE, mdtH, mdtF, gadW, gadX, acrB, acrA, crp, acrE, acrF, aadA1-pm, | tet(D), tet(A), tetR, tetD, tet34, mdfA, emrK, emrY |
| Features | P. vermicola P8538 | E. coli P8540 |
|---|---|---|
| Genome size | 4,432,495-bp | 4,809,673-bp |
| % GC content | 41.1% | 50.9% |
| No. of contigs | 2 | 210 |
| N50 | 184,648-bp | 80,966-bp |
| No. of predicted genes | 4166 | 4951 |
| No. of CDS | 3991 | 4553 |
| No. of predicted tRNAs | 74 | 83 |
| No. of predicted rRNA | 11 | 13 |
| No. of predicted tmRNA | 1 | 1 |
| No. of predicted miscRNAs | 89 | 303 |
| No. of phage sequences | 5 | 2 |
| Sequence Type (ST) | - | 1421 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lupande-Mwenebitu, D.; Khedher, M.B.; Khabthani, S.; Rym, L.; Phoba, M.-F.; Nabti, L.Z.; Lunguya-Metila, O.; Pantel, A.; Lavigne, J.-P.; Rolain, J.-M.; et al. First Genome Description of Providencia vermicola Isolate Bearing NDM-1 from Blood Culture. Microorganisms 2021, 9, 1751. https://doi.org/10.3390/microorganisms9081751
Lupande-Mwenebitu D, Khedher MB, Khabthani S, Rym L, Phoba M-F, Nabti LZ, Lunguya-Metila O, Pantel A, Lavigne J-P, Rolain J-M, et al. First Genome Description of Providencia vermicola Isolate Bearing NDM-1 from Blood Culture. Microorganisms. 2021; 9(8):1751. https://doi.org/10.3390/microorganisms9081751
Chicago/Turabian StyleLupande-Mwenebitu, David, Mariem Ben Khedher, Sami Khabthani, Lalaoui Rym, Marie-France Phoba, Larbi Zakaria Nabti, Octavie Lunguya-Metila, Alix Pantel, Jean-Philippe Lavigne, Jean-Marc Rolain, and et al. 2021. "First Genome Description of Providencia vermicola Isolate Bearing NDM-1 from Blood Culture" Microorganisms 9, no. 8: 1751. https://doi.org/10.3390/microorganisms9081751
APA StyleLupande-Mwenebitu, D., Khedher, M. B., Khabthani, S., Rym, L., Phoba, M.-F., Nabti, L. Z., Lunguya-Metila, O., Pantel, A., Lavigne, J.-P., Rolain, J.-M., & Diene, S. M. (2021). First Genome Description of Providencia vermicola Isolate Bearing NDM-1 from Blood Culture. Microorganisms, 9(8), 1751. https://doi.org/10.3390/microorganisms9081751








