Abstract
Gut microbiota appears to be involved in the pathogenesis of primary sclerosing cholangitis (PSC). The protein tyrosine phosphatase nonreceptor 2 (PTPN2) gene risk variant rs1893217 is associated with gut dysbiosis in inflammatory bowel disease (IBD), and PTPN2 was mentioned as a possible risk gene for PSC. This study assessed the microbial profile of ulcerative colitis (UC) patients with PSC and without PSC (non-PSC). Additionally, effects of the PTPN2 risk variant were assessed. In total, 216 mucosal samples from ileum, colon, and rectum were collected from 7 PSC and 42 non-PSC patients, as well as 28 control subjects (non-IBD). The microbial composition was derived from 16S rRNA sequencing data. Overall, bacterial richness was highest in PSC patients, who also had a higher relative abundance of the genus Roseburia compared to non-PSC, as well as Haemophilus, Fusobacterium, Bifidobacterium, and Actinobacillus compared to non-IBD, as well as a lower relative abundance of Bacteroides compared to non-PSC and non-IBD, respectively. After exclusion of patients with the PTPN2 risk variant, Brachyspira was higher in PSC compared to non-PSC, while, solely in colon samples, Eubacterium and Tepidimonas were higher in PSC vs. non-IBD. In conclusion, this study underlines the presence of gut mucosa-associated microbiome changes in PSC patients and rather weakens the role of PTPN2 as a PSC risk gene.
1. Introduction
Primary sclerosing cholangitis (PSC) is a rare cholestatic liver disease causing intra- and extrahepatic bile duct strictures and fibrosis, which ultimately leads to liver cirrhosis [1,2]. PSC is often associated with ulcerative colitis (UC), affecting roughly 4% of all UC patients [3,4,5]. While the exact pathogenesis of PSC remains largely unknown, UC is the main risk factor, with 60–80% of PSC patients suffering from concurrent UC [4,6,7,8].
In recent years, changes in the microbiome, metabolome, and intestinal barrier function were extensively studied to unravel the pathogenesis of PSC [9]. This system is often referred to as “gut–liver axis”, which symbolizes the interaction of gut microbiota and the liver: Intestinal dysbiosis, i.e., the change in the microbiota linked to disease, may weaken the intestinal barrier and lead to translocation of harmful bacterial particles, which in the end leads to liver inflammation and fibrosis. It is thought that intestinal dysbiosis in PSC patients with UC leads to translocation of pathogenic microbes or their products, which not only cause liver inflammation, but also biliary strictures [10,11,12,13]. Studies investigating changes in the fecal microbiota showed a distinctly different bacterial composition in PSC patients compared to healthy controls and UC patients [14,15,16], with certain bacterial taxa such as Fusobacterium associated with intestinal inflammation, and taxa such as Enterococcus rather associated with cholangitis [17]. This suggests a strong link between the composition of the gut microbiome and the pathogenesis of UC and PSC, respectively. Only few studies with inconsistent findings investigated the mucosa-associated microbiome in PSC [18]. Furthermore, changes in the gut microbiota profile resulting in altered bile acid homeostasis and increased intestinal inflammation contribute to the pathogenesis of PSC [19,20].
A dysfunction of protein tyrosine phosphatase nonreceptor 2 (PTPN2), caused by the single nucleotide polymorphism (SNP) rs1893217, is associated with intestinal dysbiosis and a more severe disease course in inflammatory bowel disease (IBD) patients [21,22,23]. While the homozygous mutation of PTPN2 occurs in roughly 1.5% of the normal population, it is present in 3.8% of IBD patients [24]. PTPN2 has also been discussed as a risk gene for PSC [25]. However, it remains elusive whether this SNP is also associated with intestinal dysbiosis in PSC patients as reported in IBD patients.
We hypothesized that the presence of PTPN2 SNP rs1893217 is associated with intestinal dysbiosis in PSC patients. To this aim, the study compared the mucosa-associated microbiota of UC patients with and without PSC according to the presence of the PTPN2 SNP, in reference to a control group, by analyzing mucosal biopsies from multiple intestinal locations from UC patients of the Swiss IBD Cohort (SIBDCS) and a (local) Bern cohort.
2. Materials and Methods
Sample dataset: This study analyzed biopsies from the terminal ileum, the right colon, the left colon, and the rectum collected during ileocolonoscopies, as published previously [26]. The SIBDCS comprises primarily IBD patients, who may simultaneously suffer from PSC, which is a rare liver disease. Therefore, data from seven UC patients with PSC (PSC) could be retrieved from the SIBDCS. A total of 42 UC patients without PSC (non-PSC) and 28 controls (non-IBD) were selected based on matching age, gender, and Modified Truelove and Witts activity index (MTWAI) values of the PSC patients, if applicable [26]. The asymptomatic control subjects underwent a screening ileocolonoscopy for colorectal carcinoma, were negative for all other biochemical and hematological tests, and presented without any macroscopic or microscopic abnormalities [26].
Ethics statement: This study is a sub-study of the Swiss IBD Cohort Study. Patient biopsy samples and clinical data collection from patients of the SIBDCS were approved by the Ethics Committee of the Canton Zürich (KEK-ZH-Nr. 2013-0284). Control data were retrieved from the Bern Human Intestinal Community project, approved by the Ethics Commission of the Canton Bern (KEK-BE-Nr 251/14 and 336/14). Informed consent for data collection and analysis was obtained from all patients of both study cohorts.
DNA extraction: Biopsies were collected in 2 mL microcentrifuge tubes (Sigma-Aldrich, St. Louis, MO, USA) and stored at −80 °C until DNA extraction, as published previously [21]. Total DNA was isolated using AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In brief, 600 μL of RLT lysis buffer (Qiagen), plus β-mercaptoethanol and a metal bead were added into each tube. Samples were then homogenized using the Retsch Tissue Lyser (Qiagen) at 30/s for 3 min and 3 min centrifugation at 13,000× g. Total supernatants were transferred into a Prep DNA mini spin column (Eppendorf, Hamburg, Germany) and centrifuged at 9000× g for 30 s. DNA attached to spin columns was washed and desalted using 500 μL of Buffer AW1 and Buffer AW2 (Qiagen). Total DNA was eluted in 30 μL RNase-free water into 1.5 mL microfuge tubes. The concentration and purity of the isolated DNA was analyzed using NanoDrop® (Thermo Fisher Scientific, Waltham, MA, USA).
16S rRNA sequencing: Amounts of 500 to 1500 ng of DNA per sample were used to amplify the V5/V6 region of the 16S rRNA gene. The expected product length was ~350 bp including adaptors and barcodes. Bacteria-specific primers (forward 5′ CCATCTCATCCCTGCGTGTCTCCGACTCAGC barcode ATTAGATACCCYGGTAGTCC 3′ and reverse 5′ CCTCTCTATGGGCAGTCGGTGATACGAGCTGACGACARCCATG-3′) were used [27]. PCR conditions consisted of an initial 5 min at 94 °C denaturation step, followed by 35 cycles of 1 min denaturation at 94 °C, 20 s annealing cycle at 46 °C, and 30 s extension cycle at 72 °C, with a final extension for 7 min at 72 °C. Samples were kept at 4 °C until loading onto a 1% agarose gel. Amplicons were then purified using the Gel Extraction Kit (Qiagen), and the pooled amplicon library at 26 pM concentration was used for sequencing, which was performed within the Ion PGMTM System (Thermo Fisher) using an Ion PGMTM Sequencing 400 Kit and an Ion 316TM Chip V2 [28].
Computation analysis of the 16S rRNA microbial data: Combined FASTQ sequencing files were first processed in QIIME 1.9.1 pipeline, as described [29] using custom analysis scripts for analysis on the UBELIX Linux cluster of the University of Bern [26]. Samples with more than 5000 high-quality reads were then used for downstream analysis in QIIME and R (R Foundation for Statistical Computing, Vienna, Austria). Operational taxonomic units were picked using UCLUST with a 97% sequence identity threshold and followed by taxonomy assignment using the latest Greengenes database (version gg_13_5; greengenes.secondgenome.com (accessed on 1 February 2021)). The operational taxonomic unit (OTU) abundance biome file and mapping file were used for statistical analyses and data was visualized with the phyloseq R package [30]. Species richness within samples was calculated using the α-diversity indices Simpson and Shannon. β-diversity between samples was calculated using Bray–Curtis genus-level community dissimilarities. Mann–Whitney U tests were performed for α diversity and Adonis (PERMANOVA) for β diversity was performed as a statistical test to confirm the strength and statistical significance of groups in the same distance metrics in the QIIME pipeline and phyloseq in R [30,31]. Taxonomy profile of samples was performed using multivariate analysis by linear models R package [32] to find associations between tested groups. The q-value package was implemented in MaAsLin2 to correct for multiple testing (Benjamini–Hochberg false discovery rate correction; a false discovery rate (FDR), q-value of 0.2). Taxa present in at least 30% of the samples and that had more than 0.0001% of total abundance were set as cut-off values for further analysis. After correction for a false discovery rate, q < 0.05 was considered significant. Plots were generated with ggplot2 using phyloseq object.
3. Results
A total of 216 biopsy samples taken from the terminal ileum, right colon, left colon, as well as rectum, were analyzed by 16S rRNA sequencing. All included subjects were male. Table 1 summarizes the demographics of the included patients.

Table 1.
Demographics of study subjects. BMI: body mass index; PSC: primary sclerosing cholangitis; IBD: inflammatory bowel disease; MTWAI: Modified Truelove and Witts activity index; PTPN2: protein tyrosine phosphatase nonreceptor type 2.
3.1. Differences in Bacterial Diversity
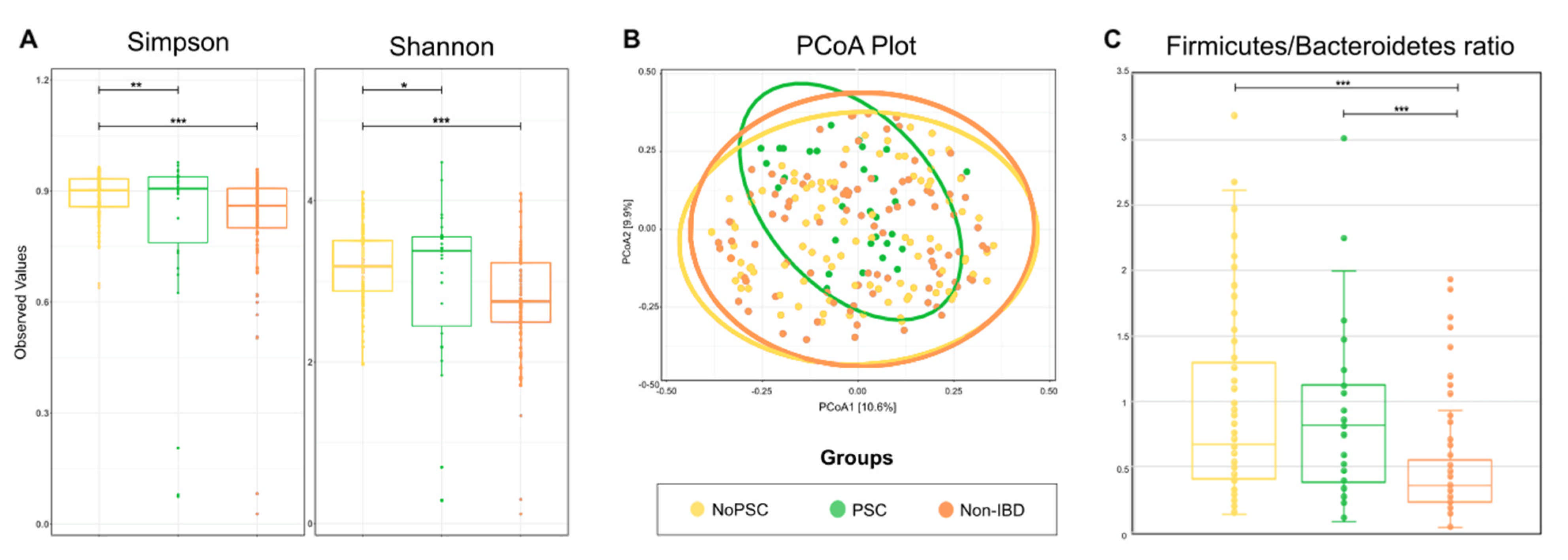
To obtain an overall impression of the bacterial diversity between groups, samples of the different locations were pooled and analyzed per patient. Results are given in Figure 1. α-diversity was higher in PSC patients compared to non-PSC patients and the control group (Figure 1A). The bacterial community in non-PSC patients and controls clustered broader than the microbiome of PSC patients (p < 0.001) (Figure 1B).

Figure 1.
Comparison of the microbial composition between groups. (A) α-diversity was compared using the Simpson and Shannon indices in the indicated groups. (B) β-diversity was compared by a principal coordinates analysis (PCoA) of the Bray-Curtis dissimilarity between groups. Ellipsoids in PCoA plots represent the 95% confidence interval surrounding each group. (C) Firmicutes/Bacteroides ratio in the indicated groups. The boxplots represent the median and the interquartile range. Differences were determined using a one-way ANOVA and marked as follows: * p < 0.05; ** p < 0.01; *** p < 0.001; PSC: primary sclerosing cholangitis; IBD: inflammatory bowel disease. PSC: n = 30, non-PSC: n = 94, non-IBD: n = 92.
Firmicutes and Bacteroides are the most abundant bacterial phyla within the gut, and the ratio of these phyla is a sign of health status. Therefore, the Firmicutes/Bacteroidetes (F/B) ratio can be used as a measure for general microbiome changes based on the patients’ health status [33]. The F/B ratio was significantly higher in non-PSC and PSC compared to non-IBD (p < 0.001) (Figure 1C). Specific changes on the genus level are given below.
3.2. Gut Microbial Signature in UC Patients with PSC
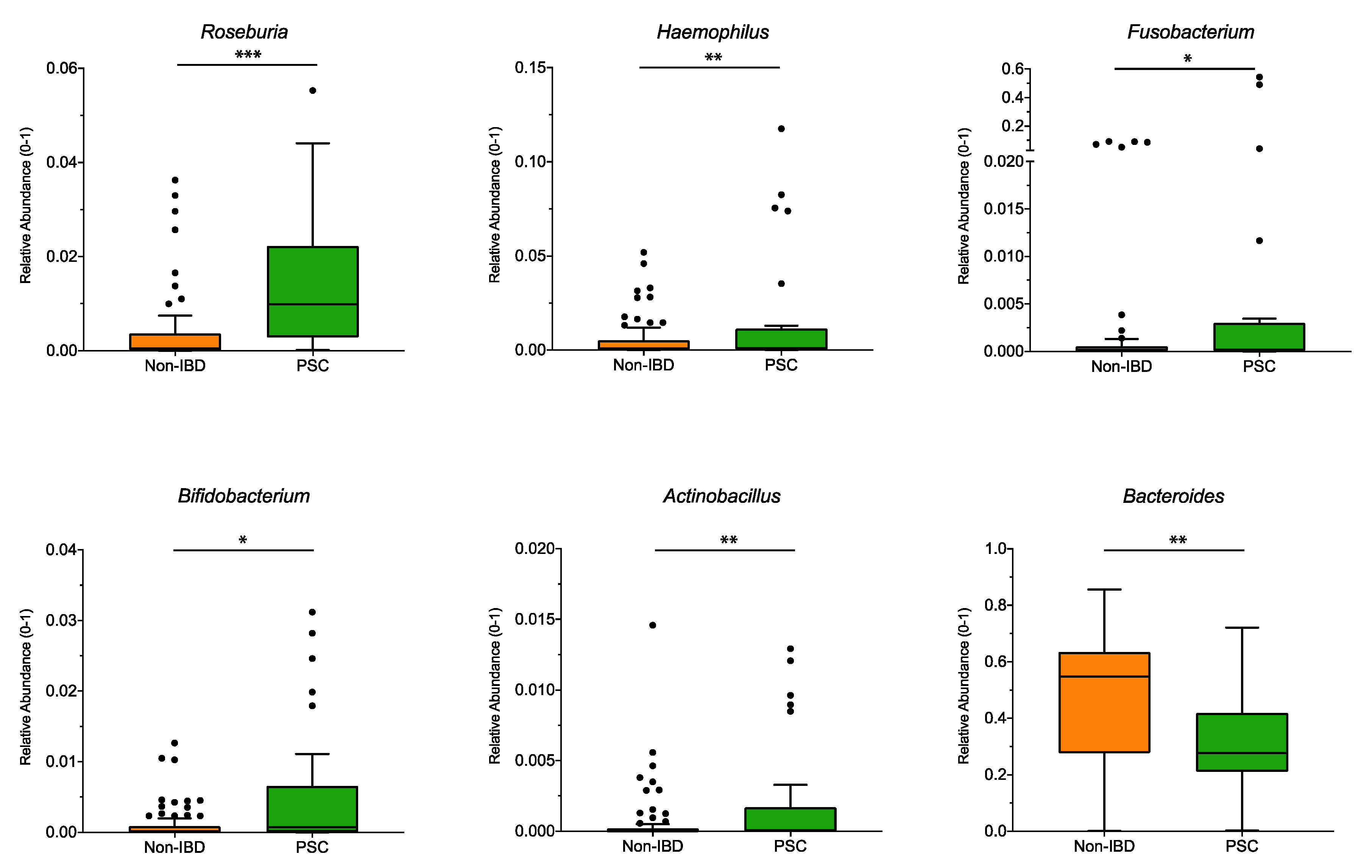
Significant differences at the genus level for the pooled sampling sites (Figure 2) were observed for Roseburia, Haemophilus, Fusobacterium, Bifidobacterium, and Actinobacillus, and were higher in PSC patients compared to non-IBD, while the genus Bacteroides was significantly lower in PSC patients. However, when analyzed for individual sampling site, no significant differences at the genus level were detected for PSC vs. non-IBD at the ileum, right colon, left colon, or the rectum.
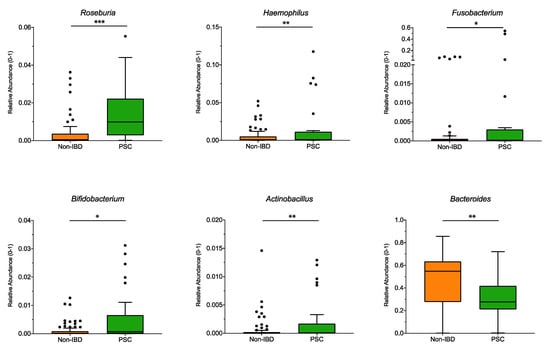
Figure 2.
Genus differences between PSC and non-IBD: The relative abundance of significantly different genera (p < 0.05; q < 0.05) is shown for pooled samples for PSC and non-IBD as indicated. The boxplots represent the median and the interquartile range. The adj-p value was reported with *** adj-p < 0.001; ** adj-p < 0.01; * adj-p < 0.05.; PSC: primary sclerosing cholangitis; IBD: inflammatory bowel disease. PSC: n = 30, non-IBD: n = 92.
3.3. Gut Microbial Signature in UC Patients without PSC
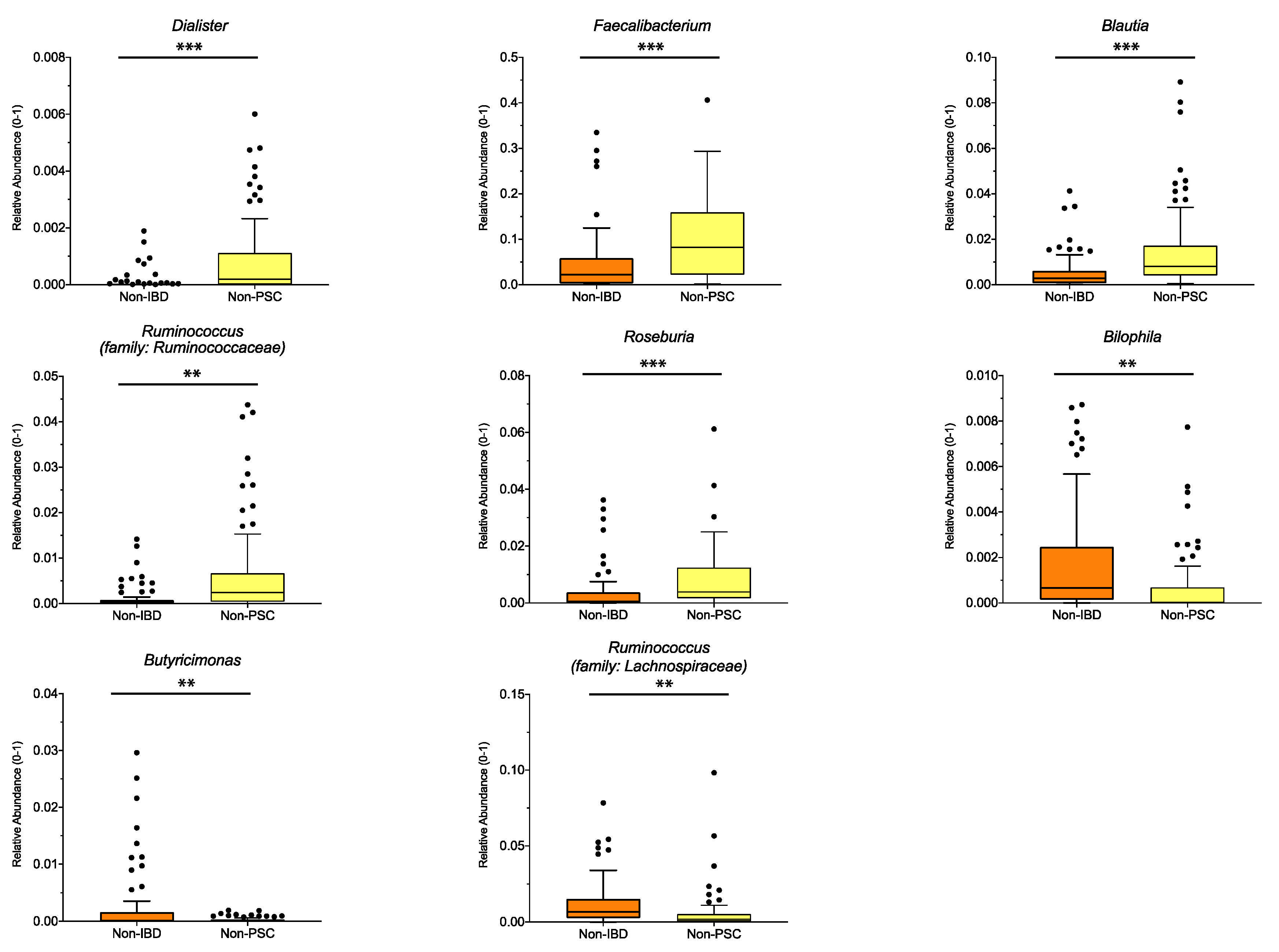
To distinguish which microbiome differences are mainly connected to UC, UC patients without PSC (non-PSC) were compared to controls (non-IBD). Significant differences for the pooled sampling sites are shown in Figure 3, with higher relative abundance of the genus Dialister, Faecalibacterium, Blautia, Ruminococcus (family: Ruminococcaceae) and Roseburia (all belonging to the phylum Firmicutes), and lower relative abundance of Bilophila, Butyricimonas and Ruminococcus (family: Lachnospiraceae) in non-PSC compared to non-IBD. Analyzed for individual sampling sites, Ruminococcus (family: Ruminococcaceae), Blautia and Dialister were higher at the left colon and Roseburia was higher at the ileum in non-PSC compared to non-IBD, while no significant differences at the genus level were detected at the right colon and rectum.
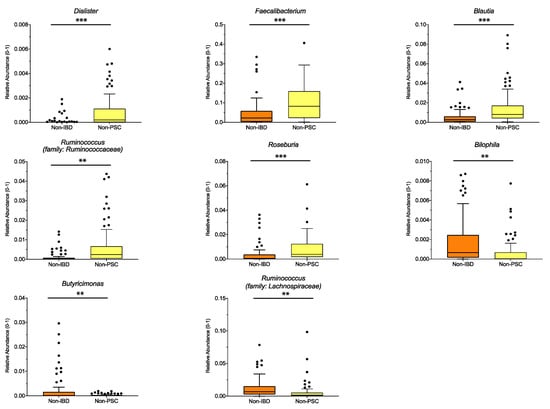
Figure 3.
Genus differences between non-PSC and non-IBD: The relative abundance of significantly different genera is shown for pooled samples in non-PSC and non-IBD as indicated. The boxplots represent the median and the interquartile range. The adj-p value was reported with *** adj-p < 0.001; ** adj-p < 0.01; PSC: primary sclerosing cholangitis; IBD: inflammatory bowel disease. Non-PSC: n = 94, non-IBD: n = 92.
3.4. PSC Status in UC Results in Minor Taxonomic Changes
All investigated PSC patients suffered from concurrent UC. Hence, to detect microbiome changes associated solely with PSC, the PSC population was directly compared to UC patients without PSC (non-PSC).
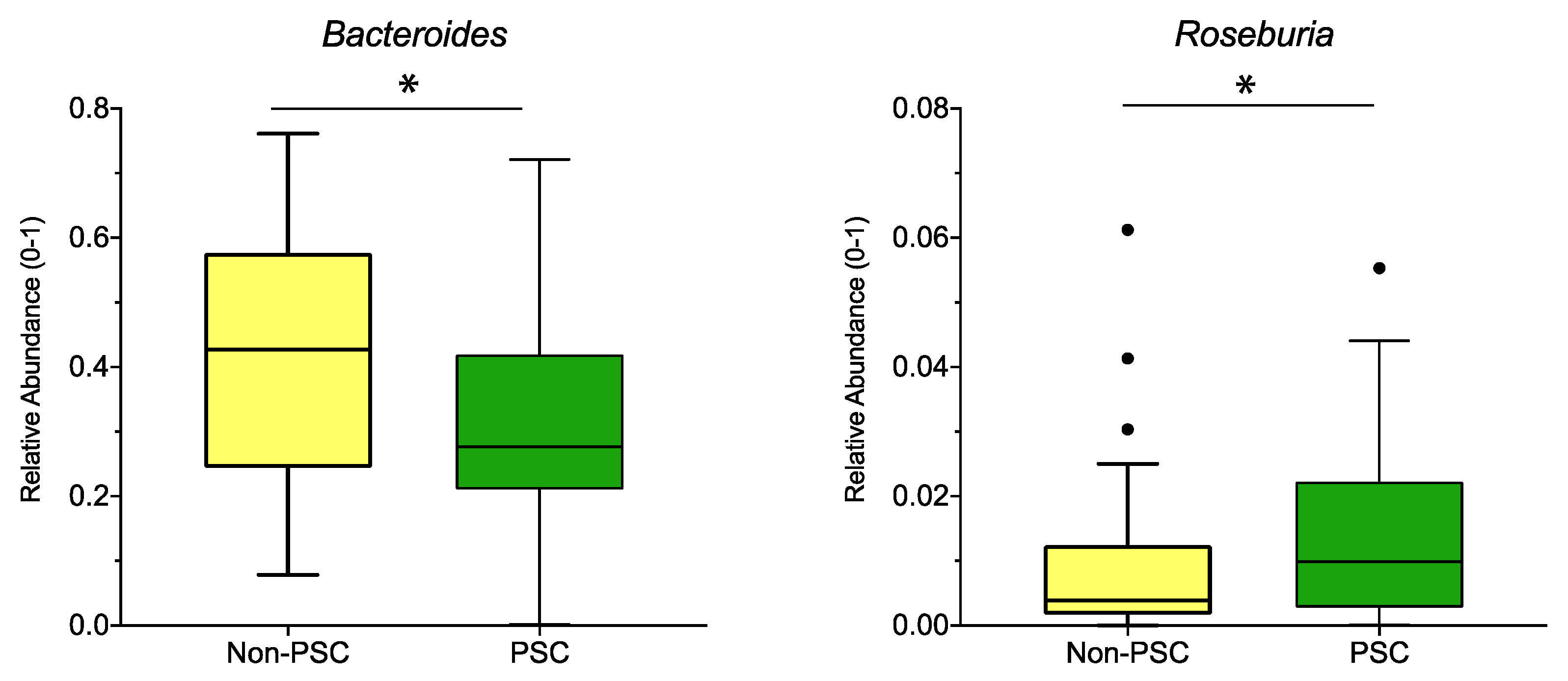
When pooling the sampling sites, relative abundance of the genus Bacteroides was lower, while the abundance of Roseburia was higher in PSC compared to non-PSC (Figure 4). According to sampling sites, no significant differences at the genus level were detected for PSC vs. non-PSC at the ileum, right colon, left colon, or rectum.
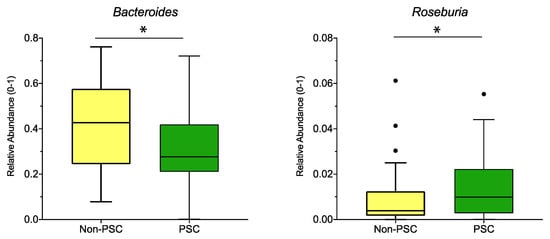
Figure 4.
Genus differences between PSC and non-PSC: The relative abundance of significantly different genera (p < 0.05; q < 0.05) is shown for PSC, non-PSC, and non-IBD as indicated. The boxplots represent the median and the interquartile range. The adj-p value was reported with * adj-p < 0.05.; PSC: primary sclerosing cholangitis; IBD: inflammatory bowel disease. PSC: n = 30, non-PSC: n = 94.
3.5. Results According to Genetic Variation of PTPN2
PTPN2 has been discussed as risk gene for PSC [25]. UC patients carrying the C-allele of the PTPN2 SNP rs1893217 are more likely to have a more severe disease course, including gallstones—which suggests altered bile acid homeostasis [34]. As the C-allele is rather rare and was detected in only seven of the investigated patients (see Table 1), microbial differences between those patients and the wild-type carriers were not detected. However, after exclusion of the patients carrying the C-allele, the genus Brachyspira was significantly higher in PSC compared to non-PSC for the pooled sampling sites (p < 0.05; q < 0.05).
When analyzing the data in patients without the PTPN2 C-allele according to the origin of the biopsy, in the right colon the genus Eubacterium and Tepidimonas were increased in PSC compared to non-IBD, and Dialister was increased in non-PSC compared to non-IBD patients. Comparing PSC and non-PSC patients without the C-allele, no differences at the genus level were detected at the ileum, right colon, left colon, or the rectum.
4. Discussion
This study analyzed the mucosa-associated microbiome of 7 UC patients with PSC in comparison with 42 UC patients without PSC and 28 controls. PSC patients had an increased bacterial richness and a shift in the overall microbial composition. This became apparent in several differences at the genus level detected between groups. By exclusion of patients with a potentially disease-aggravating genetic variation of the PTPN2 gene, a significant increase of Brachyspira was detected in biopsies from PSC patients.
PSC patients had a higher Firmicutes/Bacteroidetes (F/B) ratio than UC patients without PSC and healthy controls (Figure 1). The F/B ratio describes the microbiome on the phyla level [35]. As those two phyla represent 90% of the gut microbiota, changes are considered a sign of dysbiosis [36]. An increase in the F/B ratio was associated with obesity [37]. In IBD and PSC patients, the F/B ratio was reduced in earlier studies [38,39]. The higher ratio in PSC patients might be related to the increase in Roseburia, a genus belonging to the phylum Firmicutes, detected in the presented study. In addition, earlier studies found very low rates of Bacteroides [40], which is in line with our findings, and might add to the increase in the F/B ratio. Bacteroides are producers of sphingolipids, which are crucial for intestinal homeostasis. A decrease in Bacteroides and therefore sphingolipid levels resulted in intestinal inflammation [41]. Another genus involved in intestinal pathologies is Fusobacterium. In our study, Fusobacterium was significantly higher in PSC compared to non-IBD. Previous studies support this finding as Fusobacterium was overrepresented in stool samples of PSC patients [17,42]. Furthermore, it is thought that Fusobacterium leads to a proinflammatory microenvironment and is ultimately associated with colorectal cancer [43]. Patients with UC have a high risk of developing these tumors, and this risk is increased by 30% in patients simultaneously suffering from PSC [44]. Together with the reduction of Bacteroides (and presumably sphingolipids), the elevation of Fusobacterium might contribute to this increased cancer risk.
Gut bacteria are involved in bile acid homeostasis [45,46,47]. Their enzyme bile salt hydrolase (BSH) deconjugates primary bile acids in the gut [20] and represents one of their resistance mechanisms against bile acid toxicity [48,49]. A recent study by Quraishi et al. proved that mucosa-attaching bacterial genera expressing BSH are more abundant in PSC compared to UC patients without PSC [20]. The presented study detected a significant increase in the BSH producing genera Roseburia, Haemophilus, and Fusobacterium in PSC compared to non-IBD patients, Roseburia in PSC compared to UC patients, and Blautia, Ruminococcus, and Roseburia in UC compared to non-IBD patients. An increase of Roseburia and Haemophilus in PSC patients was also reported in earlier studies [20]. However, the BSH-producing genus Bacteroides is significantly lower in PSC compared to UC and non-IBD patients, which is a known finding in PSC, as mentioned above [40]. Therefore, the increase in the BSH producers might be a compensatory mechanism for the decrease in Bacteroides.
We have previously demonstrated that the SNP rs1893217 of the PTPN2 gene (which introduces a C-allele in the gene) promotes intestinal inflammation as well as a more severe disease course in IBD [50,51]. Furthermore, in the presence of the PTPN2 variant, distinct alterations of the mucosa-associated gut microbiome were detected in IBD patients, suggesting an interplay of genetic risk factors, intestinal microbiota, and disease course [21]. After the exclusion of patients with a C-allele in the PTPN2 gene, Brachyspira was elevated in PSC versus UC patients in our study. Increased mucosal Brachyspira colonization has also been detected in patients with irritable bowel syndrome (IBS), especially in IBS with predominant diarrhea [52]. In IBS, Brachyspira was associated with the induction of inflammatory mediators in the intestinal mucosa as well as mast cell activation and the induction of the actin related protein (ARP) 2/3 complex, a protein facilitating bacterial adhesion and invasion [52]. The ARP2/3 complex is involved in actin nucleation, a crucial process for cell migration and cytoskeletal motion, which is one of the mechanisms involved in cancer cell migration [53]. Thus, the subunit 4 (ARPC4) is associated with reduced survival in patients with hepatocellular carcinoma [54,55]. In addition, the expression of ARP3 is higher in inflamed intestinal samples of UC patients compared to healthy controls and it is related to epithelial apoptosis in vitro [56]. In contrast, the inhibition of the ARP2/3 complex activated the NF-κB pathway, causing a hypersensitive reaction to osmotic stress, which is associated with IBD [57]. Whether activation of ARP2/3 is protective or disease-aggravating in PSC, and if Brachyspira contributes to this process, remains to be elucidated. Analyzing the gut microbiome changes according to sampling site, the genera Eubacterium and Tepidimonas were significantly higher in the right colon of PSC patients without the C allele vs. control patients. Eubacterium are butyrate and BSH-producing bacteria that are associated with beneficial effects on gut and liver [49,58]. This is in line with the assumption that the absence of the PTPN2 risk allele is associated with a less-severe disease course in IBD [50]. Studies with Tepidimonas are scarce; however, a recent study detected Tepidimonas in extracellular vesicles of pancreatic tumors [59], demonstrating a mechanism by which the gut microbiome might influence extraintestinal organs. Whether this mechanism also applies to PSC, and if it might play a protective, or rather harmful, role in its pathogenesis is another unanswered question.
Despite the detected microbial differences that can be linked to intestinal and/or bile duct and liver inflammation, results need to be interpreted with caution, as the number of PSC patients in this study was rather small. Moreover, the investigated groups slightly differed in smoking status and BMI range. However, as published previously, smoking status did not influence the microbiome in UC patients [26].
5. Conclusions
In conclusion, this study underlines the presence of mucosa-associated microbiome changes in the gut of patients with PSC. These changes might be indirectly caused by altered bile acid synthesis or even directly be associated with mucosal inflammation and periductular fibrosis in PSC. These findings may lay the base for precision medicine studies investigating the impact of specific bacterial strains in the pathogenesis of PSC. The presence of a potentially pathogenic gut bacterium in patients without a genetic variation in PTPN2 rather weakens its role as a risk gene for PSC.
Author Contributions
Conceptualization, G.R., M.S., B.Y. and S.B.; methodology, B.Y.; resources, P.J., G.R. and B.Y.; formal analysis, L.D. and B.Y.; investigation, P.J. and B.Y.; writing—original draft, L.D. and S.B.; writing—review and editing, L.D., P.J., A.E.K., G.R., M.S., B.Y. and S.B.; visualization, L.D. and B.Y.; supervision, M.S. and S.B.; funding acquisition, G.R., M.S., B.Y. and S.B. All authors have read and agreed to the published version of the manuscript.
Funding
S.B. was supported by fellowships from University of Zurich (Walter and Gertrud Siegenthaler Fellowship, Hartmann Mueller Foundation and Fonds zur Foerderung des Akademischen Nachwuchses). B.Y. received funding from the Swiss National Science Foundation Ambizione grant PZ00P3_185880 and Novartis Foundation for Medical-Biological Research (#19A013). M.S. was supported by a grant from the Stiftung Experimentelle Biomedizin, a joint grant from the German Research Foundation/Swiss National Science Foundation (320030E_190969), and grants from the Swiss National Science Foundation (320030_184753 and 314730_166381). The SIBDCS is supported by a grant of the Swiss National Science Foundation to GR [Grant No. 33CS30-148422].
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the Canton Zürich (KEK-ZH-Nr. 2013-0284) and the Ethics Commission of the Canton Bern (KEK-BE-Nr 251/14 and 336/14).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
16S sequencing data of the SIBDCS can be found at https://doi.org/10.6084/m9.figshare.7335068.v1 (accessed on 19 December 2018). Metadata of PSC patients reported in this study was uploaded to https://figshare.com/articles/dataset/PSC/14923059 (accessed on 7 July 2021).
Acknowledgments
Thank you to all members of the Swiss IBD Cohort Study (SIBDCS): Members of the SIBDCS: Claudia Anderegg; Peter Bauerfeind; Christoph Beglinger; Stefan Begré; Dominique Belli; José M. Bengoa; Luc Biedermann; Beat Bigler; Janek Binek; Mirjam Blattmann; Stephan Boehm; Jan Borovicka; Christian P. Braegger; Nora Brunner; Patrick Bühr; Bernard Burnand; Emanuel Burri; Sophie Buyse; Matthias Cremer; Dominique H. Criblez; Philippe de Saussure; Lukas Degen; Joakim Delarive; Christopher Doerig; Barbara Dora; Gian Dorta; Mara Egger; Tobias Ehmann; Ali El-Wafa; Matthias Engelmann; Jessica Ezri; Christian Felley; Markus Fliegner; Nicolas Fournier; Montserrat Fraga; Pascal Frei; Remus Frei; Michael Fried; Florian Froehlich; Christian Funk; Raoul Ivano Furlano; Suzanne Gallot-Lavallée; Martin Geyer; Marc Girardin; Delphine Golay; Tanja Grandinetti; Beat Gysi; Horst Haack; Johannes Haarer; Beat Helbling; Peter Hengstler; Denise Herzog; Cyrill Hess; Klaas Heyland; Thomas Hinterleitner; Philippe Hiroz; Claudia Hirschi; Petr Hruz; Rika Iwata; Res Jost; Pascal Juillerat; Vera Kessler Brondolo; Christina Knellwolf; Christoph Knoblauch; Henrik Köhler; Rebekka Koller; Claudia Krieger-Grübel; Gerd Kullak-Ublick; Patrizia Künzler; Markus Landolt; Rupprecht Lange; Frank Serge Lehmann; Andrew Macpherson; Philippe Maerten; Michel H. Maillard; Christine Manser; Michael Manz; Urs Marbet; George Marx; Christoph Matter; Valérie McLin; Rémy Meier; Martina Mendanova; Christa Meyenberger; Pierre Michetti; Benjamin Misselwitz; Darius Moradpour; Bernhard Morell; Patrick Mosler; Christian Mottet; Christoph Müller; Pascal Müller; Beat Müllhaupt; Claudia Münger-Beyeler; Leilla Musso; Andreas Nagy; Michaela Neagu; Cristina Nichita; Jan Niess; Natacha Noël; Andreas Nydegger; Nicole Obialo; Carl Oneta; Cassandra Oropesa; Ueli Peter; Daniel Peternac; Laetitia Marie Petit; Franziska Piccoli-Gfeller; Julia Beatrice Pilz; Valérie Pittet; Nadia Raschle; Ronald Rentsch; Sophie Restellini; Jean-Pierre Richterich; Sylvia Rihs; Marc Alain Ritz; Jocelyn Roduit; Daniela Rogler; Gerhard Rogler; Jean-Benoît Rossel; Markus Sagmeister; Gaby Saner; Bernhard Sauter; Mikael Sawatzki; Michela Schäppi; Michael Scharl; Martin Schelling; Susanne Schibli; Hugo Schlauri; Sybille Schmid Uebelhart; Jean-François Schnegg; Alain Schoepfer; Frank Seibold; Mariam Seirafi; Gian-Marco Semadeni; David Semela; Arne Senning; Marc Sidler; Christiane Sokollik; Johannes Spalinger; Holger Spangenberger; Philippe Stadler; Michael Steuerwald; Alex Straumann; Bigna Straumann-Funk; Michael Sulz; Joël Thorens; Radu Tutuian; Stephan Vavricka; Francesco Viani; Jürg Vögtlin; Roland Von Känel; Alain Vonlaufen; Dominique Vouillamoz; Rachel Vulliamy; Jürg Wermuth; Helene Werner; Paul Wiesel; Reiner Wiest; Tina Wylie; Jonas Zeitz; Dorothee Zimmermann.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chapman, R.W.; Arborgh, B.A.; Rhodes, J.M.; Summerfield, J.A.; Dick, R.; Scheuer, P.J.; Sherlock, S. Primary sclerosing cholangitis: A review of its clinical features, cholangiography, and hepatic histology. Gut 1980, 21, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, T.H.; Franke, A.; Melum, E.; Kaser, A.; Hov, J.R.; Balschun, T.; Lie, B.A.; Bergquist, A.; Schramm, C.; Weismüller, T.J.; et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology 2010, 138, 1102–1111. [Google Scholar] [CrossRef]
- Conrad, K.; Roggenbuck, D.; Laass, M.W. Diagnosis and classification of ulcerative colitis. Autoimmun. Rev. 2014, 13, 463–466. [Google Scholar] [CrossRef]
- Olsson, R.; Danielsson, A.; Järnerot, G.; Lindström, E.; Lööf, L.; Rolny, P.; Rydén, B.O.; Tysk, C.; Wallerstedt, S. Prevalence of primary sclerosing cholangitis in patients with ulcerative colitis. Gastroenterology 1991, 100 Pt 1, 1319–1323. [Google Scholar] [CrossRef]
- Loftus, E.V.; Harewood, G.C.; Loftus, C.G.; Tremaine, W.J.; Harmsen, W.S.; Zinsmeister, A.R.; Jewell, D.A.; Sandborn, W.J. PSC-IBD: A unique form of inflammatory bowel disease associated with primary sclerosing cholangitis. Gut 2005, 54, 91–96. [Google Scholar] [CrossRef]
- Eaton, J.E.; Talwalkar, J.A.; Lazaridis, K.N.; Gores, G.J.; Lindor, K.D. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology 2013, 145, 521–536. [Google Scholar] [CrossRef] [Green Version]
- Chapman, R.; Fevery, J.; Kalloo, A.; Nagorney, D.M.; Boberg, K.M.; Shneider, B.; Gores, G.J. Diagnosis and management of primary sclerosing cholangitis. Hepatology 2010, 51, 660–678. [Google Scholar] [CrossRef]
- Bambha, K.; Kim, W.R.; Talwalkar, J.; Torgerson, H.; Benson, J.T.; Therneau, T.M.; Loftus, E.V.; Yawn, B.P.; Dickson, E.R.; Melton, L.J. Incidence, clinical spectrum, and outcomes of primary sclerosing cholangitis in a United States community. Gastroenterology 2003, 125, 1364–1369. [Google Scholar] [CrossRef]
- Little, R.; Wine, E.; Kamath, B.M.; Griffiths, A.M.; Ricciuto, A. Gut microbiome in primary sclerosing cholangitis: A review. World J. Gastroenterol. 2020, 26, 2768–2780. [Google Scholar] [CrossRef]
- Lichtman, S.N.; Keku, J.; Clark, R.L.; Schwab, J.H.; Sartor, R.B. Biliary tract disease in rats with experimental small bowel bacterial overgrowth. Hepatology 1991, 13, 766–772. [Google Scholar] [CrossRef]
- Lichtman, S.N.; Sartor, R.B.; Keku, J.; Schwab, J.H. Hepatic inflammation in rats with experimental small intestinal bacterial overgrowth. Gastroenterology 1990, 98, 414–423. [Google Scholar] [CrossRef]
- Tedesco, D.; Thapa, M.; Chin, C.Y.; Ge, Y.; Gong, M.; Li, J.; Gumber, S.; Speck, P.; Elrod, E.J.; Burd, E.M.; et al. Alterations in Intestinal Microbiota Lead to Production of Interleukin 17 by Intrahepatic γδ T-Cell Receptor-Positive Cells and Pathogenesis of Cholestatic Liver Disease. Gastroenterology 2018, 154, 2178–2193. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, N.; Sasaki, N.; Aoki, R.; Miyamoto, K.; Suda, W.; Teratani, T.; Suzuki, T.; Koda, Y.; Chu, P.S.; Taniki, N.; et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat. Microbiol. 2019, 4, 492–503. [Google Scholar] [CrossRef]
- Rühlemann, M.; Liwinski, T.; Heinsen, F.A.; Bang, C.; Zenouzi, R.; Kummen, M.; Thingholm, L.; Tempel, M.; Lieb, W.; Karlsen, T.; et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment. Pharmacol. Ther. 2019, 50, 580–589. [Google Scholar] [CrossRef] [Green Version]
- Kummen, M.; Holm, K.; Anmarkrud, J.A.; Nygård, S.; Vesterhus, M.; Høivik, M.L.; Trøseid, M.; Marschall, H.U.; Schrumpf, E.; Moum, B.; et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017, 66, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Lemoinne, S.; Kemgang, A.; Belkacem, K.B.; Straube, M.; Jegou, S.; Corpechot, C.; Chazouillères, O.; Housset, C.; Sokol, H.; Network, S.-A.I. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020, 69, 92–102. [Google Scholar] [CrossRef] [Green Version]
- Vieira-Silva, S.; Sabino, J.; Valles-Colomer, M.; Falony, G.; Kathagen, G.; Caenepeel, C.; Cleynen, I.; van der Merwe, S.; Vermeire, S.; Raes, J. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat. Microbiol. 2019, 4, 1826–1831. [Google Scholar] [CrossRef]
- Torres, J.; Bao, X.; Goel, A.; Colombel, J.F.; Pekow, J.; Jabri, B.; Williams, K.M.; Castillo, A.; Odin, J.A.; Meckel, K.; et al. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment. Pharmacol. Ther. 2016, 43, 790–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, A.; Macdonald, G.A.; Morrison, M.; Holtmann, G. Targeting the Gut Microbiome as a Treatment for Primary Sclerosing Cholangitis: A Conceptional Framework. Am. J. Gastroenterol. 2020, 115, 814–822. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Acharjee, A.; Beggs, A.D.; Horniblow, R.; Tselepis, C.; Gkoutos, G.; Ghosh, S.; Rossiter, A.E.; Loman, N.; van Schaik, W.; et al. A Pilot Integrative Analysis of Colonic Gene Expression, Gut Microbiota, and Immune Infiltration in Primary Sclerosing Cholangitis-Inflammatory Bowel Disease: Association of Disease with Bile Acid Pathways. J. Crohns Colitis 2020, 14, 935–947. [Google Scholar] [CrossRef] [Green Version]
- Yilmaz, B.; Spalinger, M.R.; Biedermann, L.; Franc, Y.; Fournier, N.; Rossel, J.B.; Juillerat, P.; Rogler, G.; Macpherson, A.J.; Scharl, M. The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients. PLoS ONE 2018, 13, e0199664. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; He, J.H.; Wang, J.; Song, J.; Lei, H.B.; Dong, W.G. Associations between PTPN2 polymorphisms and susceptibility to ulcerative colitis and Crohn’s disease: A meta-analysis. Inflamm. Res. 2014, 63, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Todd, J.A.; Walker, N.M.; Cooper, J.D.; Smyth, D.J.; Downes, K.; Plagnol, V.; Bailey, R.; Nejentsev, S.; Field, S.F.; Payne, F.; et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 2007, 39, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Scharl, M.; Mwinyi, J.; Fischbeck, A.; Leucht, K.; Eloranta, J.J.; Arikkat, J.; Pesch, T.; Kellermeier, S.; Mair, A.; Kullak-Ublick, G.A.; et al. Crohn’s disease-associated polymorphism within the PTPN2 gene affects muramyl-dipeptide-induced cytokine secretion and autophagy. Inflamm. Bowel Dis. 2012, 18, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Karlsen, T.H. Genetics of primary sclerosing cholangitis and pathophysiological implications. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Juillerat, P.; Øyås, O.; Ramon, C.; Bravo, F.D.; Franc, Y.; Fournier, N.; Michetti, P.; Mueller, C.; Geuking, M.; et al. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat. Med. 2019, 25, 323–336. [Google Scholar] [CrossRef]
- Sundquist, A.; Bigdeli, S.; Jalili, R.; Druzin, M.L.; Waller, S.; Pullen, K.M.; El-Sayed, Y.Y.; Taslimi, M.M.; Batzoglou, S.; Ronaghi, M. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC Microbiol. 2007, 7, 108. [Google Scholar] [CrossRef] [Green Version]
- Whiteley, A.S.; Jenkins, S.; Waite, I.; Kresoje, N.; Payne, H.; Mullan, B.; Allcock, R.; O’Donnell, A. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) Platform. J. Microbiol. Methods 2012, 91, 80–88. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor Workflow for Microbiome Data Analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Hov, J.R.; Karlsen, T.H. The Microbiome in Primary Sclerosing Cholangitis: Current Evidence and Potential Concepts. Semin Liver Dis. 2017, 37, 314–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spalinger, M.R.; Voegelin, M.; Biedermann, L.; Zeitz, J.; Rossel, J.B.; Sulz, M.C.; Frei, P.; Scharl, S.; Vavricka, S.R.; Fried, M.; et al. The Clinical Relevance of the IBD-Associated Variation within the Risk Gene Locus Encoding Protein Tyrosine Phosphatase Non-Receptor Type 2 in Patients of the Swiss IBD Cohort. Digestion 2016, 93, 182–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms 2020, 8, 1175. [Google Scholar] [CrossRef] [PubMed]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Aiello, V.; Romano, B.; De Lorenzo, A.; Izzo, A.A.; et al. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Z.H.; Zhu, C.X.; Quan, Y.S.; Yang, Z.Y.; Wu, S.; Luo, W.W.; Tan, B.; Wang, X.Y. Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018, 24, 5–14. [Google Scholar] [CrossRef]
- Cortez, R.V.; Moreira, L.N.; Padilha, M.; Bibas, M.D.; Toma, R.K.; Porta, G.; Taddei, C.R. Gut Microbiome of Children and Adolescents with Primary Sclerosing Cholangitis in Association with Ulcerative Colitis. Front. Immunol. 2020, 11, 598152. [Google Scholar] [CrossRef]
- Quraishi, M.N.; Sergeant, M.; Kay, G.; Iqbal, T.; Chan, J.; Constantinidou, C.; Trivedi, P.; Ferguson, J.; Adams, D.H.; Pallen, M.; et al. The gut-adherent microbiota of PSC-IBD is distinct to that of IBD. Gut 2017, 66, 386–388. [Google Scholar] [CrossRef]
- Brown, E.M.; Ke, X.; Hitchcock, D.; Jeanfavre, S.; Avila-Pacheco, J.; Nakata, T.; Arthur, T.D.; Fornelos, N.; Heim, C.; Franzosa, E.A.; et al. Bacteroides-Derived Sphingolipids Are Critical for Maintaining Intestinal Homeostasis and Symbiosis. Cell Host Microbe 2019, 25, 668–680. [Google Scholar] [CrossRef]
- Sabino, J.; Vieira-Silva, S.; Machiels, K.; Joossens, M.; Falony, G.; Ballet, V.; Ferrante, M.; Van Assche, G.; Van der Merwe, S.; Vermeire, S.; et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016, 65, 1681–1689. [Google Scholar] [CrossRef] [Green Version]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Leong, R.W. Primary sclerosing cholangitis as an independent risk factor for colorectal cancer in the context of inflammatory bowel disease: A review of the literature. World J. Gastroenterol. 2014, 20, 8783–8789. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Bajaj, J.S. The human gut sterolbiome: Bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm. Sin. B 2015, 5, 99–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Begley, M.; Gahan, C.G.; Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 2005, 29, 625–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gérard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Nat. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Cai, Y.; Lao, X.; Wang, X.; Lin, X.; Cui, Y.; Kalavagunta, P.K.; Liao, J.; Jin, L.; Shang, J.; et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 2019, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Scharl, M.; Wojtal, K.A.; Becker, H.M.; Fischbeck, A.; Frei, P.; Arikkat, J.; Pesch, T.; Kellermeier, S.; Boone, D.L.; Weber, A.; et al. Protein tyrosine phosphatase nonreceptor type 2 regulates autophagosome formation in human intestinal cells. Inflamm. Bowel Dis. 2012, 18, 1287–1302. [Google Scholar] [CrossRef] [PubMed]
- Spalinger, M.R.; Kasper, S.; Chassard, C.; Raselli, T.; Frey-Wagner, I.; Gottier, C.; Lang, S.; Atrott, K.; Vavricka, S.R.; Mair, F.; et al. PTPN2 controls differentiation of CD4⁺ T cells and limits intestinal inflammation and intestinal dysbiosis. Mucosal Immunol. 2015, 8, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Jabbar, K.S.; Dolan, B.; Eklund, L.; Wising, C.; Ermund, A.; Johansson, Å.; Törnblom, H.; Simren, M.; Hansson, G.C. Association between Brachyspira and irritable bowel syndrome with diarrhoea. Gut 2020, 70, 1117–1129. [Google Scholar] [CrossRef]
- Goley, E.D.; Welch, M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006, 7, 713–726. [Google Scholar] [CrossRef]
- Qin, G.; Dang, M.; Gao, H.; Wang, H.; Luo, F.; Chen, R. Deciphering the protein-protein interaction network regulating hepatocellular carcinoma metastasis. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Liu, J.; Xiao, M.; Xu, H.; Xu, C.; Zhang, X.; Tang, L.; Jiang, F.; Zhou, Y.; Zhang, Z.; et al. ARP3 promotes tumor metastasis and predicts a poor prognosis in hepatocellular carcinoma. Pathol. Res. Pract. 2018, 214, 1356–1361. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Ni, W.K.; Xiao, M.B.; Jiang, F.; Lu, C.H.; Wang, R.H.; Ni, R.Z. Actin related protein 3 (ARP3) promotes apoptosis of intestinal epithelial cells in ulcerative colitis. Pathol. Res. Pract. 2019, 215, 235–242. [Google Scholar] [CrossRef]
- Wu, C.; Haynes, E.M.; Asokan, S.B.; Simon, J.M.; Sharpless, N.E.; Baldwin, A.S.; Davis, I.J.; Johnson, G.L.; Bear, J.E. Loss of Arp2/3 induces an NF-κB-dependent, nonautonomous effect on chemotactic signaling. J. Cell Biol. 2013, 203, 907–916. [Google Scholar] [CrossRef]
- Mukherjee, A.; Lordan, C.; Ross, R.P.; Cotter, P.D. Gut microbes from the phylogenetically diverse genus. Gut Microbes 2020, 12, 1802866. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Kim, T.B.; Kim, J.; Choi, H.W.; Kim, E.J.; Yoo, H.J.; Lee, S.; Jun, H.R.; Yoo, W.; Kim, S.; et al. Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer. Cancers 2020, 12, 2346. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).