Cross-Shore and Depth Zonations in Bacterial Diversity Are Linked to Age and Source of Dissolved Organic Matter across the Intertidal Area of a Sandy Beach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Sediment Sampling and Porewater
2.3. Bioinformatics and Statistical Analysis of Sequence Data
2.4. Functional Prediction Using Tax4Fun2
3. Results
3.1. Clusters in Community Composition
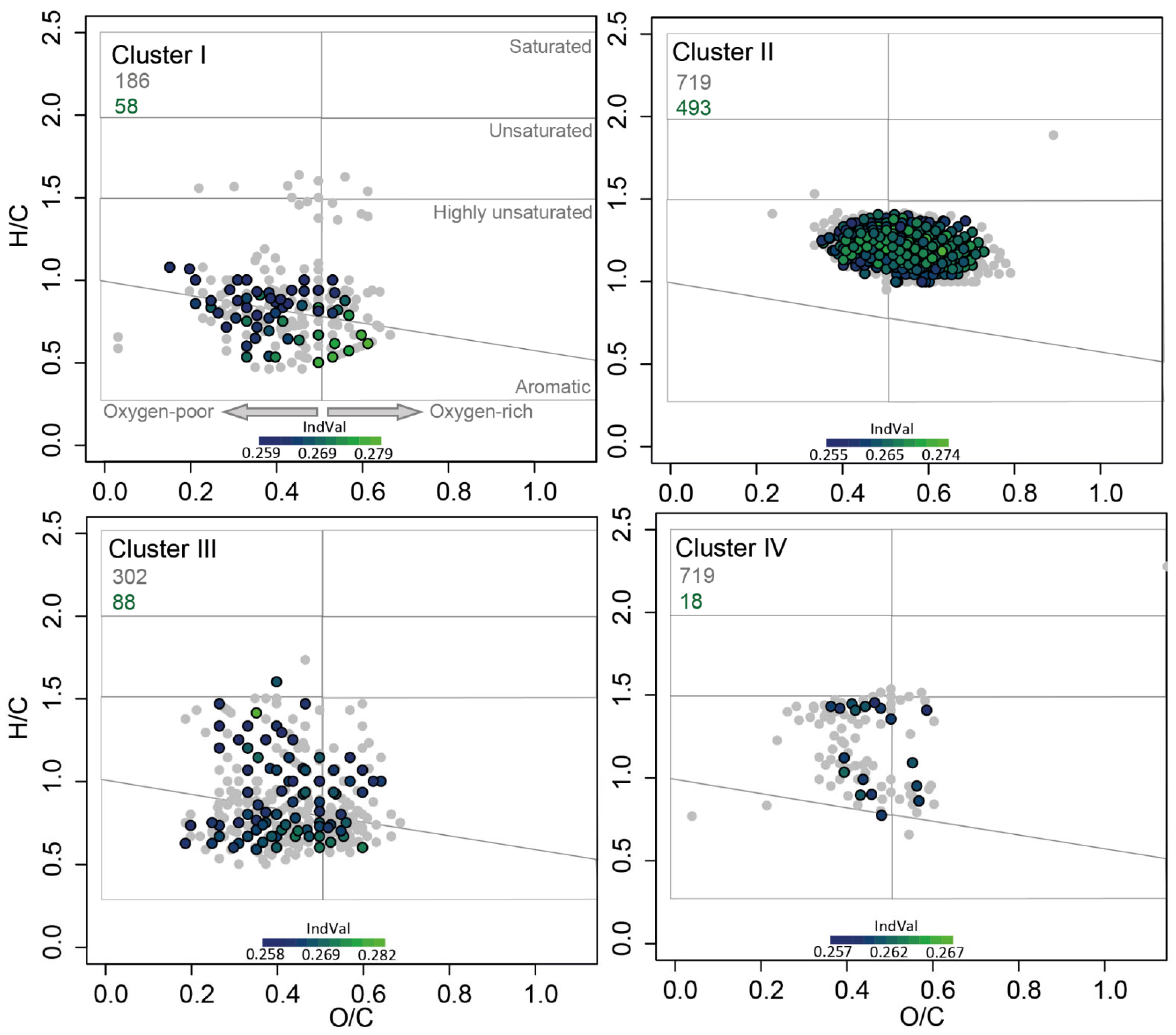
3.2. DOM Components Related to Clusters within the Bacterial Community
3.3. Predicted Functional Diversity and Its Influence on Cluster Formation
4. Discussion
4.1. Depth and Cross-Shore Distribution of Bacterial Clusters Is Governed by DOM Age and Source
4.2. Candidate Phyla and the Island of Stability
4.3. SGD Impacted Sediments Are Characterized by a Typical Deep Subsurface Community
4.4. Algal Polymer Degraders and the Lack of Algal Polymers
4.5. Predicted Functional Data Support the Hypothesis of a Subsurface Bloom of Denitrifiers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ridgwell, A.; Arndt, S. Why Dissolved Organics Matter. In Biogeochemistry of Marine Dissolved Organic Matter; Academic Press: Cambridge, MA, USA, 2015; pp. 1–20. [Google Scholar]
- Thornton, D.C.O. Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur. J. Phycol. 2014, 49, 20–46. [Google Scholar] [CrossRef] [Green Version]
- Azam, F.; Fenchel, T.; Field, J.G.; Gray, J.S.; Meyer-Reil, L.A.; Thingstad, F. The Ecological Role of Water-Column Microbes in the Sea. Mar. Ecol. Prog. Ser. 1983, 10, 257–263. [Google Scholar] [CrossRef]
- De Leeuw, J.W.; Largeau, C. A Review of Macromolecular Organic Compounds That Comprise Living Organisms and Their Role in Kerogen, Coal, and Petroleum Formation. In Organic Geochemistry. Topics in Geobiology; Springer: Boston, MA, USA, 1993. [Google Scholar]
- Avery, G.B.; Willey, J.D.; Kieber, R.J.; Shank, G.C.; Whitehead, R.F. Flux and bioavailability of Cape Fear River and rainwater dissolved organic carbon to Long Bay, southeastern United States. Glob. Biogeochem. Cycles 2003, 17. [Google Scholar] [CrossRef]
- Avery, G.B.; Kieber, R.J.; Taylor, K.J.; Dixon, J.L. Dissolved organic carbon release from surface sand of a high energy beach along the Southeastern Coast of North Carolina, USA. Mar. Chem. 2012, 132–133, 23–27. [Google Scholar] [CrossRef]
- Mahmoudi, N.; Beaupre, S.R.; Steen, A.D.; Pearson, A. Sequential bioavailability of sedimentary organic matter to heterotrophic bacteria. Environ. Microbiol. 2017, 19, 2629–2644. [Google Scholar] [CrossRef] [PubMed]
- Hansell, D.A.; Carlson, C.A.; Schlitzer, R. Net removal of major marine dissolved organic carbon fractions in the subsurface ocean. Glob. Biogeochem. Cycles 2012, 26. [Google Scholar] [CrossRef]
- Lechtenfeld, O.J.; Kattner, G.; Flerus, R.; McCallister, S.L.; Schmitt-Kopplin, P.; Koch, B.P. Molecular transformation and degradation of refractory dissolved organic matter in the Atlantic and Southern Ocean. Geochim. Cosmochim. Acta 2014, 126, 321–337. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Simpson, A.J.; Kujawinski, E.B.; Freitas, M.A.; Hatcher, P.G. High resolution electrospray ionization mass spectrometry and 2D solution NMR for the analysis of DOM extracted by C18 solid phase disk. Org. Geochem. 2003, 34, 1325–1335. [Google Scholar] [CrossRef]
- Koch, B.P.; Witt, M.; Engbrodt, R.; Dittmar, T.; Kattner, G. Molecular formulae of marine and terrigenous dissolved organic matter detected by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Geochim. Cosmochim. Acta 2005, 69, 3299–3308. [Google Scholar] [CrossRef]
- Tremblay, L.B.; Dittmar, T.; Marshall, A.G.; Cooper, W.J.; Cooper, W.T. Molecular characterization of dissolved organic matter in a North Brazilian mangrove porewater and mangrove-fringed estuaries by ultrahigh resolution Fourier Transform-Ion Cyclotron Resonance mass spectrometry and excitation/emission spectroscopy. Mar. Chem. 2007, 105, 15–29. [Google Scholar] [CrossRef]
- Sleighter, R.L.; Hatcher, P.G. Molecular characterization of dissolved organic matter (DOM) along a river to ocean transect of the lower Chesapeake Bay by ultrahigh resolution electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Mar. Chem. 2008, 110, 140–152. [Google Scholar] [CrossRef]
- Einsiedl, F.; Hertkorn, N.; Wolf, M.; Frommberger, M.; Schmitt-Kopplin, P.; Koch, B.P. Rapid biotic molecular transformation of fulvic acids in a karst aquifer. Geochim. Cosmochim. Acta 2007, 71, 5474–5482. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, F.; Elvert, M.; Koch, B.P.; Witt, M.; Hinrichs, K.-U. Molecular characterization of dissolved organic matter in pore water of continental shelf sediments. Geochim. Cosmochim. Acta 2009, 73, 3337–3358. [Google Scholar] [CrossRef]
- Longnecker, K.; Kujawinski, E.B. Composition of dissolved organic matter in groundwater. Geochim. Cosmochim. Acta 2011, 75, 2752–2761. [Google Scholar] [CrossRef] [Green Version]
- Santos, I.R.; Burnett, W.C.; Dittmar, T.; Suryaputra, I.G.N.A.; Chanton, J. Tidal pumping drives nutrient and dissolved organic matter dynamics in a Gulf of Mexico subterranean estuary. Geochim. Cosmochim. Acta 2009, 73, 1325–1339. [Google Scholar] [CrossRef]
- Reckhardt, A.; Beck, M.; Seidel, M.; Riedel, T.; Wehrmann, A.; Bartholomä, A.; Schnetger, B.; Dittmar, T.; Brumsack, H.-J. Carbon, nutrient and trace metal cycling in sandy sediments: A comparison of high-energy beaches and backbarrier tidal flats. Estuar. Coast. Shelf Sci. 2015, 159, 1–14. [Google Scholar] [CrossRef]
- Beck, M.; Reckhardt, A.; Amelsberg, J.; Bartholomä, A.; Brumsack, H.-J.; Cypionka, H.; Dittmar, T.; Engelen, B.; Greskowiak, J.; Hillebrand, H.; et al. The drivers of biogeochemistry in beach ecosystems: A cross-shore transect from the dunes to the low-water line. Mar. Chem. 2017, 190, 35–50. [Google Scholar] [CrossRef] [Green Version]
- Seidel, M.; Beck, M.; Riedel, T.; Waska, H.; Suryaputra, I.G.N.A.; Schnetger, B.; Niggemann, J.; Simon, M.; Dittmar, T. Biogeochemistry of dissolved organic matter in an anoxic intertidal creek bank. Geochim. Cosmochim. Acta 2014, 140, 418–434. [Google Scholar] [CrossRef]
- Seidel, M.; Beck, M.; Greskowiak, J.; Riedel, T.; Waska, H.; Suryaputra, I.G.N.A.; Schnetger, B.; Niggemann, J.; Simon, M.; Dittmar, T. Benthic-pelagic coupling of nutrients and dissolved organic matter composition in an intertidal sandy beach. Mar. Chem. 2015, 176, 150–163. [Google Scholar] [CrossRef]
- Jorgensen, B.B.; Boetius, A. Feast and famine--microbial life in the deep-sea bed. Nat. Rev. Microbiol. 2007, 5, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Biddle, J.F.; Lipp, J.S.; Leverd, M.A.; Lloydd, K.G.; Sørensen, K.B.; Andersonc, R.; Fredricks, H.F.; Elvertc, M.; Kelly, T.J.; Schragh, D.P.; et al. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. USA 2006, 103, 3846–3851. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, F.; Nunoura, T.; Nakagawa, S.; Teske, A.; Lever, M.; Lauer, A.; Suzuki, M.; Takai, K.; Delwiche, M.; Colwell, F.S.; et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 2006, 103, 2815–2820. [Google Scholar] [CrossRef] [Green Version]
- Gan, S.; Schmidt, F.; Heuer, V.B.; Goldhammer, T.; Witt, M.; Hinrichs, K.-U. Impacts of redox conditions on dissolved organic matter (DOM) quality in marine sediments off the River Rhône, Western Mediterranean Sea. Geochim. Cosmochim. Acta 2020, 276, 151–169. [Google Scholar] [CrossRef]
- Wakeham, S.G.; Canuel, E.A. Degradation and Preservation of Organic Matter in Marine Sediments. In Marine Organic Matter: Biomarkers, Isotopes and DNA; Springer: Berlin/Heidelberg, Germany, 2006; pp. 295–321. [Google Scholar]
- LaRowe, D.E.; Arndt, S.; Bradley, J.A.; Estes, E.R.; Hoarfrost, A.; Lang, S.Q.; Lloyd, K.G.; Mahmoudi, N.; Orsi, W.D.; Shah Walter, S.R.; et al. The fate of organic carbon in marine sediments—New insights from recent data and analysis. Earth-Sci. Rev. 2020, 204, 103146. [Google Scholar] [CrossRef]
- Arnosti, C. Microbial extracellular enzymes and the marine carbon cycle. Ann. Rev. Mar. Sci. 2011, 3, 401–425. [Google Scholar] [CrossRef]
- Kamalanathan, M.; Doyle, S.M.; Xu, C.; Achberger, A.M.; Wade, T.L.; Schwehr, K.; Santschi, P.H.; Sylvan, J.B.; Quigg, A. Exoenzymes as a Signature of Microbial Response to MarineEnvironmental Conditions. Appl. Environ. Sci. 2020, 5, e00290-20. [Google Scholar] [CrossRef]
- Bernardet, J.-F.; Bowman, J.P. The Genus Flavobacterium. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006; pp. 481–531. [Google Scholar]
- Dyksma, S.; Lenk, S.; Sawicka, J.E.; Mussmann, M. Uncultured Gammaproteobacteria and Desulfobacteraceae Account for Major Acetate Assimilation in a Coastal Marine Sediment. Front. Microbiol. 2018, 9, 3124. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.L.; Pelikan, C.; de Rezende, J.R.; Wasmund, K.; Putz, M.; Glombitza, C.; Kjeldsen, K.U.; Jorgensen, B.B.; Loy, A. Bacterial interactions during sequential degradation of cyanobacterial necromass in a sulfidic arctic marine sediment. Environ. Microbiol. 2018, 20, 2927–2940. [Google Scholar] [CrossRef] [Green Version]
- Glöckner, F.O.; Kube, M.; Bauer, M.; Teeling, H.; Lombardot, T.; Ludwig, W.; Gade, D.; Beck, A.; Borzym, K.; Heitmann, K.; et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 2003, 100, 8298–8303. [Google Scholar] [CrossRef] [Green Version]
- Linz, A.M.; Crary, B.C.; Shade, A.; Owens, S.; Gilbert, J.A.; Knight, R.; McMahon, K.D. Bacterial Community Composition and Dynamics Spanning Five Years in Freshwater Bog Lakes. mSphere 2017, 2, 00169-17. [Google Scholar] [CrossRef] [Green Version]
- Dombrowski, N.; Teske, A.P.; Baker, B.J. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat. Commun. 2018, 9, 4999. [Google Scholar] [CrossRef]
- Tully, B.J.; Graham, E.D.; Heidelberg, J.F. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci. Data 2018, 5, 170203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Kantor, R.S.; Wrighton, K.C.; Handley, K.M.; Sharon, I.; Hug, L.A.; Castelle, C.J.; Thomas, B.C.; Banfield, J.F. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 2013, 4, e00708–e00713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.Y.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef]
- Vigneron, A.; Cruaud, P.; Langlois, V.; Lovejoy, C.; Culley, A.I.; Vincent, W.F. Ultra-small and abundant: Candidate phyla radiation bacteria are potential catalysts of carbon transformation in a thermokarst lake ecosystem. Limnol. Oceanogr. Lett. 2019, 5, 212–220. [Google Scholar] [CrossRef] [Green Version]
- Guenet, B.; Danger, M.; Abbadie, L.; Lacroix, G. Priming effect: Bridging the gap between terrestrial and aquatic ecology. Ecology 2010, 91, 2850–2861. [Google Scholar] [CrossRef] [PubMed]
- Osterholz, H.; Singer, G.; Wemheuer, B.; Daniel, R.; Simon, M.; Niggemann, J.; Dittmar, T. Deciphering associations between dissolved organic molecules and bacterial communities in a pelagic marine system. ISME J. 2016, 10, 1717–1730. [Google Scholar] [CrossRef]
- Osterholz, H.; Niggemann, J.; Giebel, H.A.; Simon, M.; Dittmar, T. Inefficient microbial production of refractory dissolved organic matter in the ocean. Nat. Commun. 2015, 6, 7422. [Google Scholar] [CrossRef]
- Oni, O.E.; Schmidt, F.; Miyatake, T.; Kasten, S.; Witt, M.; Hinrichs, K.U.; Friedrich, M.W. Microbial Communities and Organic Matter Composition in Surface and Subsurface Sediments of the Helgoland Mud Area, North Sea. Front. Microbiol. 2015, 6, 1290. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, J.; Beck, M.; Marchant, H.K.; Ahmerkamp, S.; Schnetger, B.; Brumsack, H.J. Seasonality of Organic Matter Degradation Regulates Nutrient and Metal Net Fluxes in a High Energy Sandy Beach. J. Geophys. Res. Biogeosci. 2020, 125, e2019JG005399. [Google Scholar] [CrossRef]
- Waska, H.; Simon, H.; Ahmerkamp, S.; Greskowiak, J.; Ahrens, J.; Seibert, S.L.; Schwalfenberg, K.; Zielinski, O.; Dittmar, T. Molecular Traits of Dissolved Organic Matter in the Subterranean Estuary of a High-Energy Beach: Indications of Sources and Sinks. Front. Mar. Sci. 2021, 8, 54. [Google Scholar] [CrossRef]
- Degenhardt, J.; Dlugosch, L.; Ahrens, J.; Beck, M.; Waska, H.; Engelen, B. Seasonal Dynamics of Microbial Diversity at a Sandy High Energy Beach Reveal a Resilient Core Community. Front. Mar. Sci. 2020, 7, 869. [Google Scholar] [CrossRef]
- Degenhardt, J.; Khodami, S.; Milke, F.; Waska, H.; Engelen, B.; Martinez Arbizu, P. The Three Domains of Life Within the Discharge Area of a Shallow Subterranean Estuary at a High Energy Beach. Front. Environ. Sci. 2021, 9, 642098. [Google Scholar] [CrossRef]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Grünenbaum, N.; Ahrens, J.; Beck, M.; Gilfedder, B.S.; Greskowiak, J.; Kossack, M.; Massmann, G. A Multi-Method Approach for Quantification of In- and Exfiltration Rates from the Subterranean Estuary of a High Energy Beach. Front. Earth Sci. 2020, 8, 571310. [Google Scholar] [CrossRef]
- Grünenbaum, N.; Greskowiak, J.; Sültenfuß, J.; Massmann, G. Groundwater flow and residence times below a meso-tidal high-energy beach: A model-based analyses of salinity patterns and 3H-3He groundwater ages. J. Hydrol. 2020, 587, 124948. [Google Scholar] [CrossRef]
- Waska, H.; Greskowiak, J.; Ahrens, J.; Beck, M.; Ahmerkamp, S.; Böning, P.; Brumsack, H.J.; Degenhardt, J.; Ehlert, C.; Engelen, B.; et al. Spatial and Temporal Patterns of Pore Water Chemistry in the Inter-Tidal Zone of a High Energy Beach. Front. Mar. Sci. 2019, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Itaya, K.; Ui, M. A New Micromethod For The Colorimetric Determination of Inorganic Phosphate. Clin. Chim. Acta 1966, 14, 361–366. [Google Scholar] [CrossRef]
- Laskov, C.; Herzog, C.; Lewandowski, J.; Hupfe, M. Miniaturized photometrical methods for the rapid analysis of phosphate, ammonium, ferrous iron, and sulfate in pore water of freshwater sediments. Limnol. Oceanogr. Methods 2007, 4, 63–71. [Google Scholar] [CrossRef]
- Merder, J.; Freund, J.A.; Feudel, U.; Hansen, C.T.; Hawkes, J.A.; Jacob, B.; Klaproth, K.; Niggemann, J.; Noriega-Ortega, B.E.; Osterholz, H.; et al. ICBM-OCEAN: Processing Ultrahigh-Resolution Mass Spectrometry Data of Complex Molecular Mixtures. Anal. Chem. 2020, 92, 6832–6838. [Google Scholar] [CrossRef]
- Lueders, T.; Manefield, M.; Friedrich, M.W. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 2004, 6, 73–78. [Google Scholar] [CrossRef]
- Gabor, E.M.; Vries, E.J.; Janssen, D.B. Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol. Ecol. 2003, 44, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012; Volume 24, p. 1006. [Google Scholar]
- Hahs-Vaughn, D.L. Applied Multivariate Statistical Concepts; Taylor & Francis: Abingdon, UK, 2016. [Google Scholar]
- Dufrene, M.; Legendre, P. Species Assemblages and Indicator Species: The Need for a Flexible Asymmetrical Approach. Ecol. Monogr. 1997, 67, 345–366. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hothorn, T.; Hornik, K.; Zeileis, A. Unbiased Recursive Partitioning: A Conditional Inference Framework. J. Comput. Graph. Stat. 2006, 15, 651–674. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Auguie, B.; Antonov, A.; Auguie, M.B. Package ‘gridextra’, Miscellaneous Functions for “Grid” Graphics. 2017. Available online: https://cran.rproject.org/web/packages/gridExtra/gridExtra.pdf (accessed on 10 October 2020).
- Collingro, A.; Toenshoff, E.R.; Taylor, M.W.; Fritsche, T.R.; Wagner, M.; Horn, M. ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 2005, 55, 1863–1866. [Google Scholar] [CrossRef] [PubMed]
- Flerus, R.; Lechtenfeld, O.J.; Koch, B.P.; McCallister, S.L.; Schmitt-Kopplin, P.; Benner, R.; Kaiser, K.; Kattner, G. A molecular perspective on the ageing of marine dissolved organic matter. Biogeosciences 2012, 9, 1935–1955. [Google Scholar] [CrossRef] [Green Version]
- D’Andrilli, J.; Cooper, W.T.; Foreman, C.M.; Marshall, A.G. An ultrahigh-resolution mass spectrometry index to estimate natural organic matter lability. Rapid Commun. Mass Spectrom. 2015, 29, 2385–2401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medeiros, P.M.; Seidel, M.; Niggemann, J.; Spencer, R.G.M.; Hernes, P.J.; Yager, P.L.; Miller, W.L.; Dittmar, T.; Hansell, D.A. A novel molecular approach for tracing terrigenous dissolved organic matter into the deep ocean. Glob. Biogeochem. Cycles 2016, 30, 689–699. [Google Scholar] [CrossRef]
- D’Andrilli, J.; Junker, J.R.; Smith, H.J.; Scholl, E.A.; Foreman, C.M. DOM composition alters ecosystem function during microbial processing of isolated sources. Biogeochemistry 2019, 142, 281–298. [Google Scholar] [CrossRef] [Green Version]
- Linkhorst, A.; Dittmar, T.; Waska, H. Molecular Fractionation of Dissolved Organic Matter in a Shallow Subterranean Estuary: The Role of the Iron Curtain. Environ. Sci. Technol. 2017, 51, 1312–1320. [Google Scholar] [CrossRef]
- Riedel, T.; Zark, M.; Vähätalo, A.V.; Niggemann, J.; Spencer, R.G.M.; Hernes, P.J.; Dittmar, T. Molecular Signatures of Biogeochemical Transformations in Dissolved Organic Matter from Ten World Rivers. Front. Earth Sci. 2016, 4, 85. [Google Scholar] [CrossRef] [Green Version]
- Mostovaya, A.; Hawkes, J.A.; Dittmar, T.; Tranvik, L.J. Molecular Determinants of Dissolved Organic Matter Reactivity in Lake Water. Front. Earth Sci. 2017, 5, 106. [Google Scholar] [CrossRef]
- Solden, L.; Lloyd, K.; Wrighton, K. The bright side of microbial dark matter: Lessons learned from the uncultivated majority. Curr. Opin. Microbiol. 2016, 31, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danczak, R.E.; Johnston, M.D.; Kenah, C.; Slattery, M.; Wrighton, K.C.; Wilkins, M.J. Members of the Candidate Phyla Radiation are functionally differentiated by carbon- and nitrogen-cycling capabilities. Microbiome 2017, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Shinohara, A.; Fukui, M. Sulfurifustis variabilis gen. nov., sp. nov., a sulfur oxidizer isolated from a lake, and proposal of Acidiferrobacteraceae fam. nov. and Acidiferrobacterales ord. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 3709–3713. [Google Scholar] [CrossRef] [PubMed]
- Kolinko, S.; Richter, M.; Glockner, F.O.; Brachmann, A.; Schuler, D. Single-cell genomics of uncultivated deep-branching magnetotactic bacteria reveals a conserved set of magnetosome genes. Environ. Microbiol. 2016, 18, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Dharamshi, J.E.; Tamarit, D.; Eme, L.; Stairs, C.W.; Martijn, J.; Homa, F.; Jorgensen, S.L.; Spang, A.; Ettema, T.J.G. Marine Sediments Illuminate Chlamydiae Diversity and Evolution. Curr. Biol. 2020, 30, 1032–1048.e1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedel, T.; Lettmann, K.; Schnetger, B.; Beck, M.; Brumsack, H.-J. Rates of trace metal and nutrient diagenesis in an intertidal creek bank. Geochim. Cosmochim. Acta 2011, 75, 134–147. [Google Scholar] [CrossRef]
- Waska, H.; Brumsack, H.-J.; Massmann, G.; Koschinsky, A.; Schnetger, B.; Simon, H.; Dittmar, T. Inorganic and organic iron and copper species of the subterranean estuary: Origins and fate. Geochim. Cosmochim. Acta 2019, 259, 211–232. [Google Scholar] [CrossRef]
- Krzmarzick, M.J.; Crary, B.B.; Harding, J.J.; Oyerinde, O.O.; Leri, A.C.; Myneni, S.C.; Novak, P.J. Natural niche for organohalide-respiring Chloroflexi. Appl. Environ. Microbiol. 2012, 78, 393–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adrian, L.; Löffler, F.E. Organohalide—Respiring Bacteria; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Kaster, A.K.; Mayer-Blackwell, K.; Pasarelli, B.; Spormann, A.M. Single cell genomic study of Dehalococcoidetes species from deep-sea sediments of the Peruvian Margin. ISME J. 2014, 8, 1831–1842. [Google Scholar] [CrossRef]
- Biddle, J.F.; Fitz-Gibbon, S.; Schuster, S.C.; Brenchley, J.E.; House, C.H. Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. USA 2008, 105, 10583–10588. [Google Scholar] [CrossRef] [Green Version]
- Wasmund, K.; Schreiber, L.; Lloyd, K.G.; Petersen, D.G.; Schramm, A.; Stepanauskas, R.; Jorgensen, B.B.; Adrian, L. Genome sequencing of a single cell of the widely distributed marine subsurface Dehalococcoidia, phylum Chloroflexi. ISME J. 2014, 8, 383–397. [Google Scholar] [CrossRef] [Green Version]
- Löffler, F.E.; Yan, J.; Ritalahti, K.M.; Adrian, L.; Edwards, E.A.; Konstantinidis, K.T.; Müller, J.A.; Fullerton, H.; Zinder, S.H.; Spormann, A.M. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 2013, 63, 625–635. [Google Scholar] [CrossRef] [Green Version]
- Wilms, R.; Sass, H.; Kopke, B.; Koster, J.; Cypionka, H.; Engelen, B. Specific bacterial, archaeal, and eukaryotic communities in tidal-flat sediments along a vertical profile of several meters. Appl. Environ. Microbiol. 2006, 72, 2756–2764. [Google Scholar] [CrossRef] [Green Version]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; Gonzalez, J.M. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 2014, 12, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.T.; et al. Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, Y.; Kurahashi, M.; Sakiyama, Y.; Ohuchi, M.; Yokota, A.; Harayama, S. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 2009, 55, 267–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostgaard, K. Enzymatic microassay for the determination and characterization of alginates. Carbohydr. Polym. 1992, 19, 51–59. [Google Scholar] [CrossRef]
- Raeke, J.; Lechtenfeld, O.J.; Wagner, M.; Herzsprung, P.; Reemtsma, T. Selectivity of solid phase extraction of freshwater dissolved organic matter and its effect on ultrahigh resolution mass spectra. Environ. Sci. Process. Impacts 2016, 18, 918–927. [Google Scholar] [CrossRef] [PubMed]



| Cluster I | Cluster II | Cluster III | Cluster IV | |
|---|---|---|---|---|
| Salinity | 29.86 ± 3.59 | 30.21 ± 2.83 | 27.43 ± 6.65 | 29.18 ± 2.49 |
| O2 (µM) | 20.63 ± 40.73 | 101.32 ± 122.48 | 13.11 ± 26.96 | 46.01 ± 53.33 |
| NH4 (µM) | 16.81 ± 15.46 | 3.17 ± 3.85 | 24.73 ± 22.25 | 11.03 ± 11.97 |
| NO3 (µM) | 6.77 ± 13.61 | 13.71 ± 15.8 | 6.41 ± 17.54 | 9.99 ± 16.66 |
| Si (µM) | 49.81 ± 27.35 | 34.05 ± 16.01 | 92.38 ± 57.31 | 49.1 ± 18.65 |
| Fe (µM) | 29.59 ± 40.18 | 0.16 ± 0.19 | 36.96 ± 35.19 | 18.28 ± 30.73 |
| Mn (µM) | 10.69 ± 15.54 | 0.74 ± 1.82 | 5.59 ± 6.06 | 3.91 ± 3.89 |
| DOC (µM) | 136.08 ± 29.95 | 105.49 ± 20.66 | 128.5 ± 19.02 | 131.25 ± 21.08 |
| FDOM (ppb QSE) | 41.47 ± 10.65 | 29.65 ± 10.71 | 50.51 ± 10.26 | 38.37 ± 10.99 |
| Homologous series | 8326 ± 251 | 13039 ± 81 | 12925 ± 55 | 11835 ± 98 |
| H/C ratio | 0.82 ± 0.01 | 1.21 ± 0.003 | 0.99 ± 0.01 | 1.24 ± 0.02 |
| O/C ratio | 0.41 ± 0.01 | 0.53 ± 0.003 | 0.41 ± 0.003 | 0.46 ± 0.003 |
| N | 1.23 ± 0.06 | 0.21 ± 0.02 | 0.02 ± 0.002 | 0.17 ± 0.01 |
| S | 0 | 0.005 ± 0.0004 | 0.13 ± 0.01 | 0.19 ± 0.01 |
| CHO | 10 (17) | 314 (68) | 59 (67) | 9 (50) |
| CHON | 48 (83) | 141 (30) | 4 (5) | 4 (22) |
| CHOS | 0 (0) | 9 (2) | 25 (28) | 5 (28) |
| Mass (Da) | 294.25 ± 1.85 | 464.37 ± 1.88 | 315.01 ± 2.01 | 488.55 ± 1.25 |
| AI.mod | 0.58 ± 0.01 | 0.23 ± 0.002 | 0.45 ± 0.01 | 0.24 ± 0.01 |
| DBE | 10.01 ± 0.06 | 9.48 ± 0.04 | 9.18 ± 0.16 | 9.92 ± 0.2 |
| Aromatic | 72.45 ± 1.71 | 0 | 37.34 ± 2.63 | 4.11 ± 0.76 |
| Highly Unsaturated | 27.55 ± 1.71 | 100 ± 0 | 62.08 ± 2.60 | 95.89 ± 0.76 |
| Unsaturated | 0 | 0 | 0.58 ± 0.09 | 0 |
| Unsaturated with N | 0 | 0 | 0 | 0 |
| Saturated | 0 | 0 | 0 | 0 |
| * Ideg | 0.70 ± 0.07 | 0.79 ± 0.09 | 0.70 ± 0.05 | 0.72 ± 0.07 |
| * MLB_l | 9.22 ± 1.66 | 8.63 ± 2.28 | 8.73 ± 1.47 | 9.18 ± 1.63 |
| * ITerr | 0.21 ± 0.02 | 0.17 ± 0.02 | 0.22 ± 0.02 | 0.20 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degenhardt, J.; Merder, J.; Heyerhoff, B.; Simon, H.; Engelen, B.; Waska, H. Cross-Shore and Depth Zonations in Bacterial Diversity Are Linked to Age and Source of Dissolved Organic Matter across the Intertidal Area of a Sandy Beach. Microorganisms 2021, 9, 1720. https://doi.org/10.3390/microorganisms9081720
Degenhardt J, Merder J, Heyerhoff B, Simon H, Engelen B, Waska H. Cross-Shore and Depth Zonations in Bacterial Diversity Are Linked to Age and Source of Dissolved Organic Matter across the Intertidal Area of a Sandy Beach. Microorganisms. 2021; 9(8):1720. https://doi.org/10.3390/microorganisms9081720
Chicago/Turabian StyleDegenhardt, Julius, Julian Merder, Benedikt Heyerhoff, Heike Simon, Bert Engelen, and Hannelore Waska. 2021. "Cross-Shore and Depth Zonations in Bacterial Diversity Are Linked to Age and Source of Dissolved Organic Matter across the Intertidal Area of a Sandy Beach" Microorganisms 9, no. 8: 1720. https://doi.org/10.3390/microorganisms9081720
APA StyleDegenhardt, J., Merder, J., Heyerhoff, B., Simon, H., Engelen, B., & Waska, H. (2021). Cross-Shore and Depth Zonations in Bacterial Diversity Are Linked to Age and Source of Dissolved Organic Matter across the Intertidal Area of a Sandy Beach. Microorganisms, 9(8), 1720. https://doi.org/10.3390/microorganisms9081720






