Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally
Abstract
:1. Introduction
2. Methodology
2.1. Epidemiology and Phylogeny of Environmental Strains of SARS-CoV-2
2.2. Mutational Analysis of SARS-CoV-2 Retrieved from the Environment
3. Results
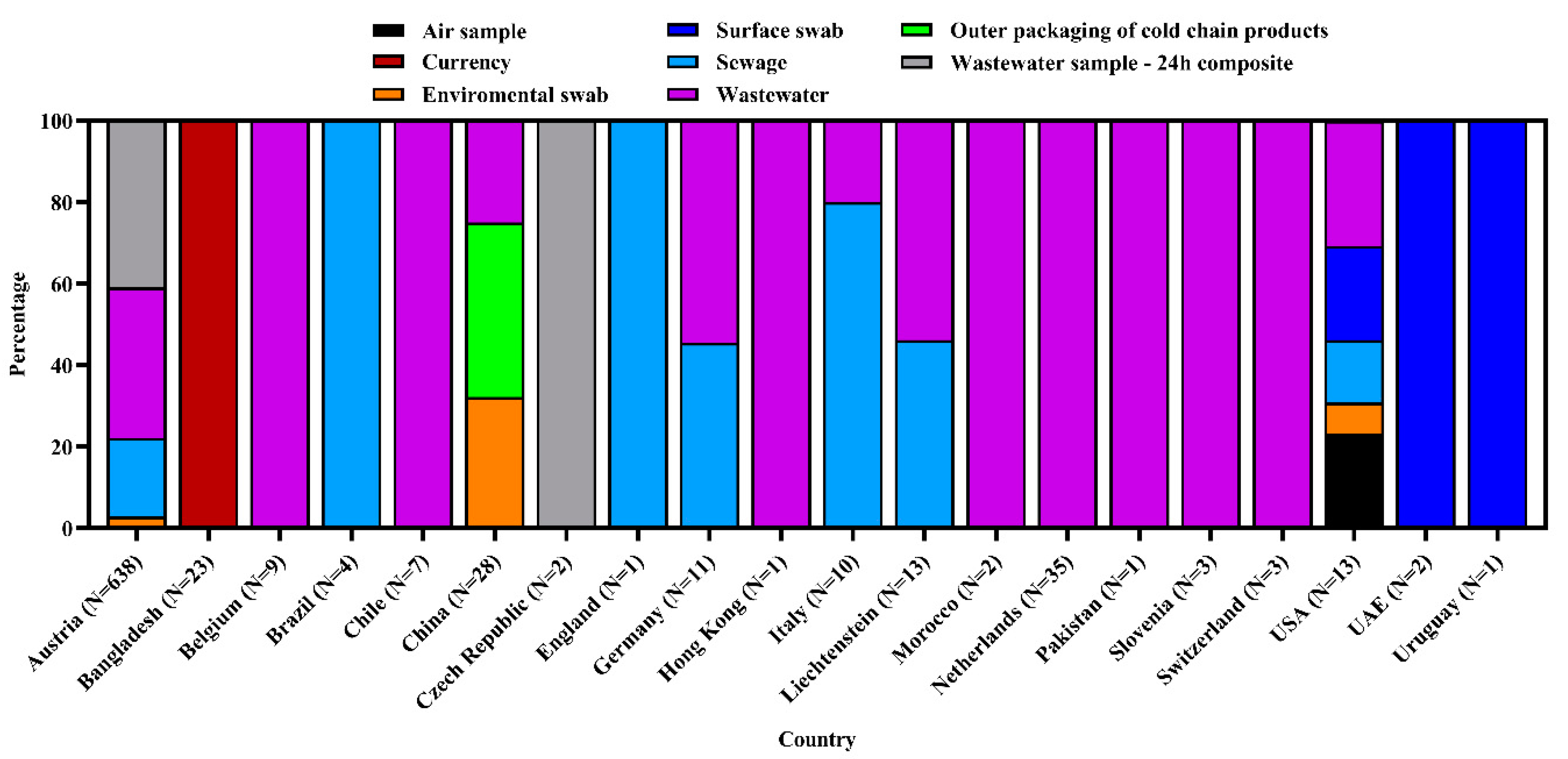
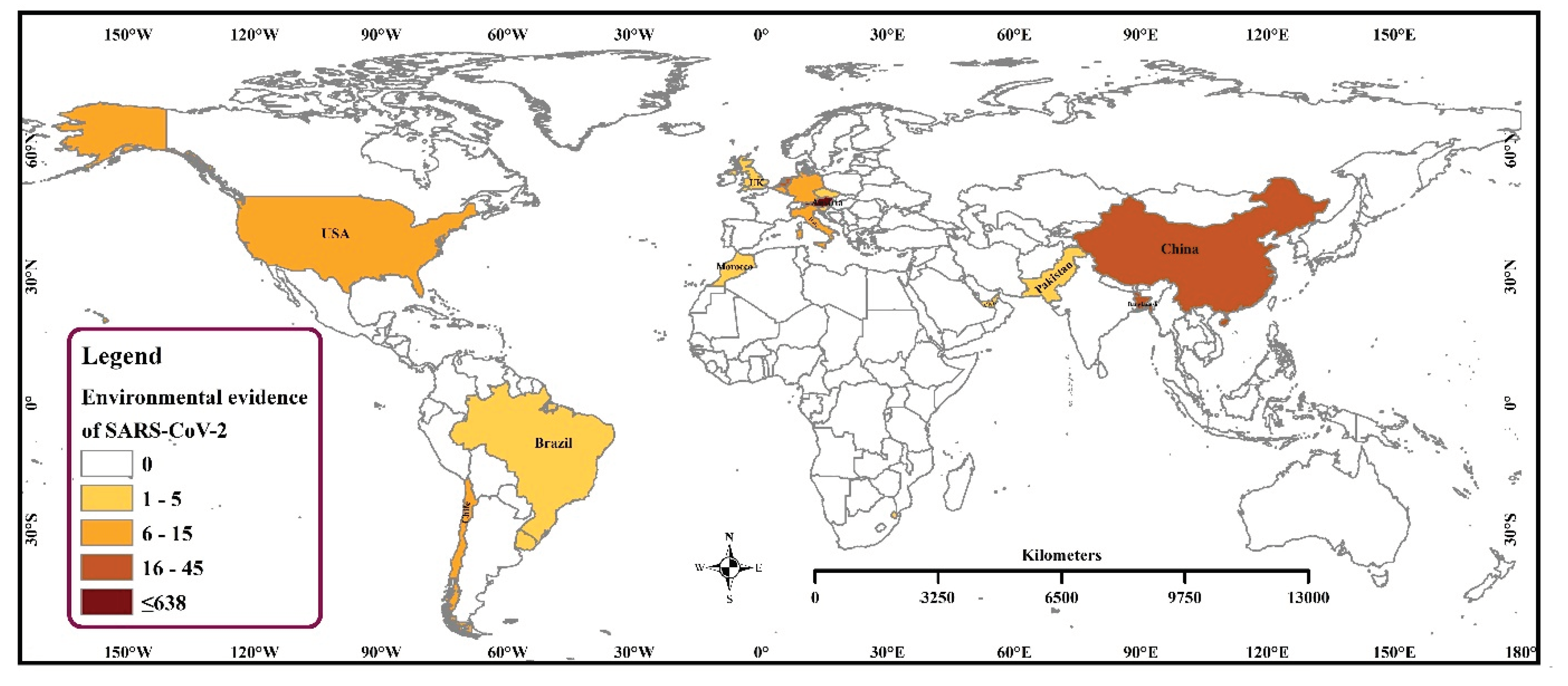
3.1. Frequency and Spatial Distribution of SARS-CoV-2 Sequences of Environmental Strains
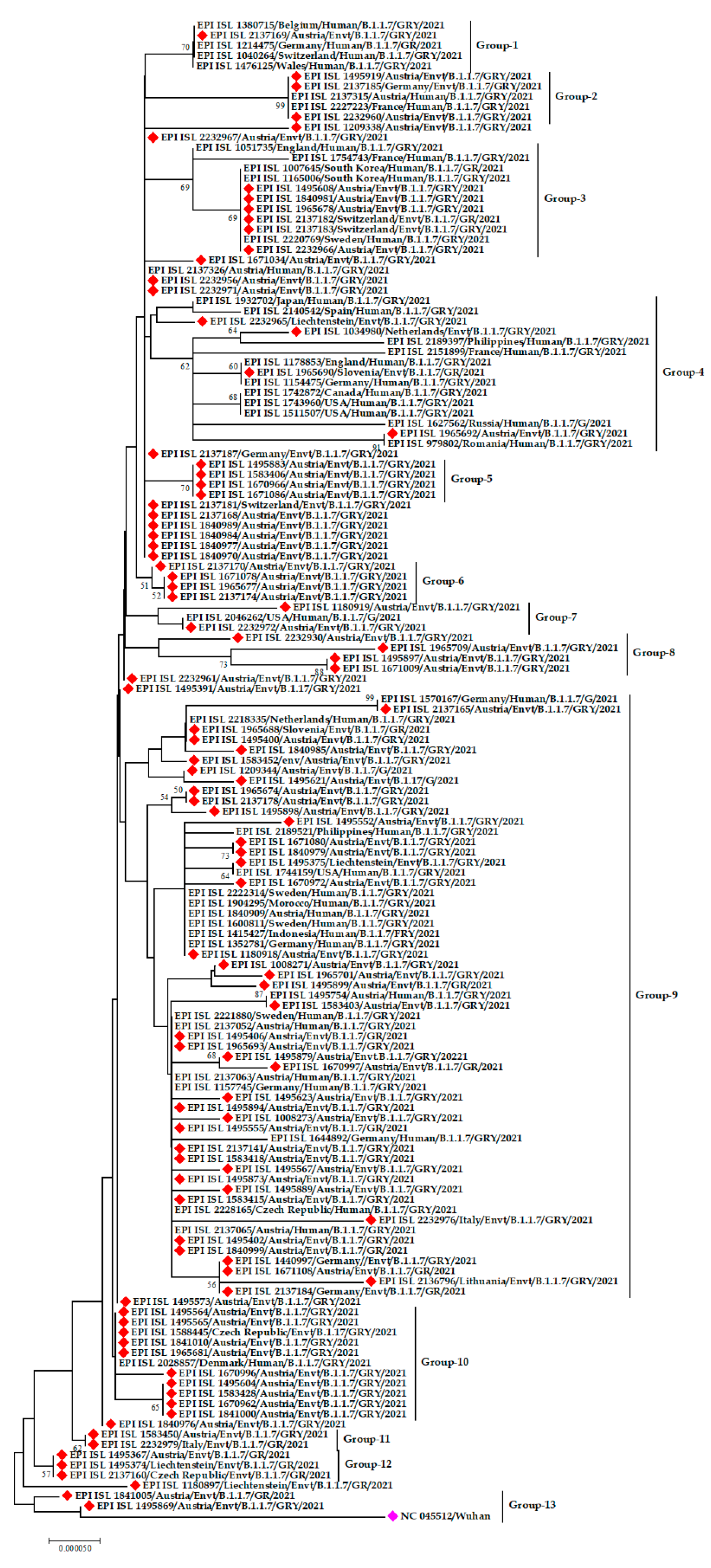
3.2. Phylogenetic Analysis of SARS-CoV-2 from the Environment
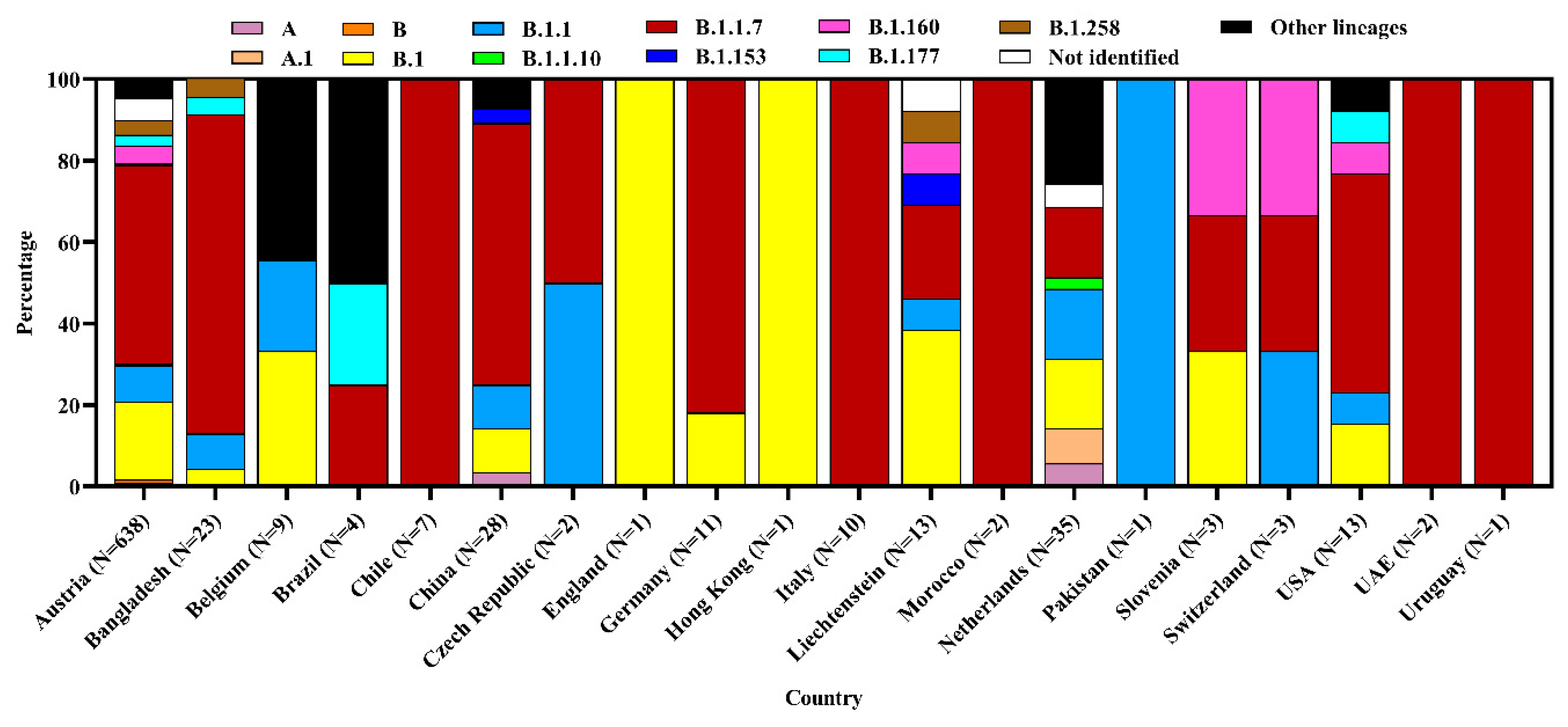
3.3. Clade and Lineage Diversity of SARS-CoV-2 Circulating in the Environment
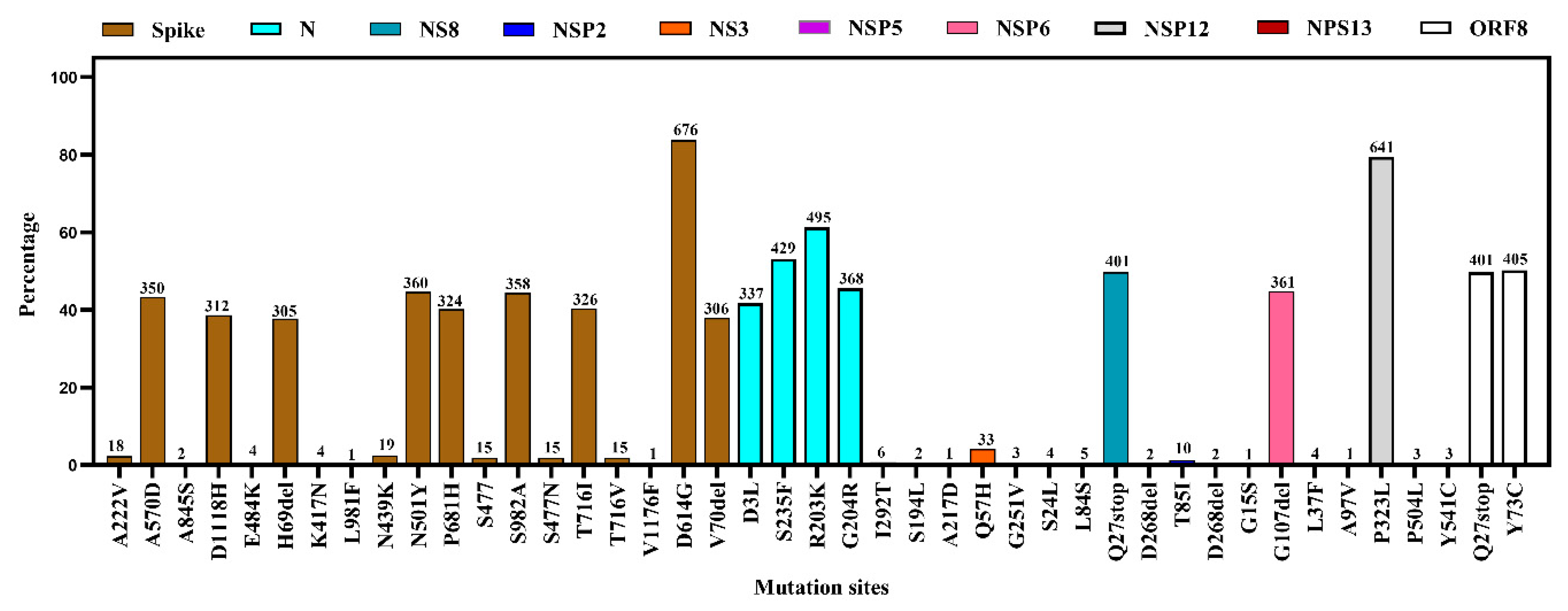
3.4. Mutational Diversity in SARS-CoV-2 from Environmental Samples
4. Discussion
5. Conclusions and Recommendations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hassan, M.M.; Kalam, M.; Shano, S.; Nayem, M.; Khan, R.; Rahman, M.; Khan, S.A.; Islam, A. Assessment of Epidemiological Determinants of COVID-19 Pandemic Related to Social and Economic Factors Globally. J. Risk Financ. Manag. 2020, 13, 194. [Google Scholar] [CrossRef]
- McKibbin, W.; Fernando, R. The global macroeconomic impacts of COVID-19: Seven scenarios. Asian Econ. Pap. 2020, 20, 1–55. [Google Scholar] [CrossRef]
- Holshue, M.L.; DeBolt, C.; Lindquist, S.; Lofy, K.H.; Wiesman, J.; Bruce, H.; Spitters, C.; Ericson, K.; Wilkerson, S.; Tural, A. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020, 382, 929–936. [Google Scholar] [CrossRef]
- Kitajima, M.; Ahmed, W.; Bibby, K.; Carducci, A.; Gerba, C.P.; Hamilton, K.A.; Haramoto, E.; Rose, J.B. SARS-CoV-2 in wastewater: State of the knowledge and research needs. Sci. Total Environ. 2020, 739, 139076. [Google Scholar] [CrossRef]
- Ling, Y.; Xu, S.-B.; Lin, Y.-X.; Tian, D.; Zhu, Z.-Q.; Dai, F.-H.; Wu, F.; Song, Z.-G.; Huang, W.; Chen, J. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin. Med. J. 2020, 133, 1039. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. WHO issues consensus document on the epidemiology of SARS. Wkly. Epidemiol. Rec. 2003, 78, 373–375. [Google Scholar]
- McKinney, K.R.; Gong, Y.Y.; Lewis, T.G. Environmental transmission of SARS at Amoy Gardens. J. Environ. Health 2006, 68, 26. [Google Scholar] [PubMed]
- WHO. Consensus Document on the Epidemiology of Severe Acute Respiratory Syndrome (SARS); World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Gormley, M.; Aspray, T.J.; Kelly, D.A. COVID-19: Mitigating transmission via wastewater plumbing systems. Lancet Glob. Health 2020, 8, e643. [Google Scholar] [CrossRef] [Green Version]
- Dharavath, B.; Yadav, N.; Desai, S.; Sunder, R.; Mishra, R.; Ketkar, M.; Bhanshe, P.; Gupta, A.; Redhu, A.K.; Patkar, N. A one-step, one-tube real-time RT-PCR based assay with an automated analysis for detection of SARS-CoV-2. Heliyon 2020, 6, e04405. [Google Scholar] [CrossRef]
- Lodder, W.; de Roda Husman, A.M. SARS-CoV-2 in wastewater: Potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020, 5, 533–534. [Google Scholar] [CrossRef]
- Lodder, W.; Buisman, A.; Rutjes, S.; Heijne, J.; Teunis, P.; de Roda Husman, A. Feasibility of quantitative environmental surveillance in poliovirus eradication strategies. Appl. Environ. Microbiol. 2012, 78, 3800–3805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef] [PubMed]
- Orive, G.; Lertxundi, U.; Barcelo, D. Early SARS-CoV-2 outbreak detection by sewage-based epidemiology. Sci. Total Environ. 2020, 732, 139298. [Google Scholar] [CrossRef] [PubMed]
- Lodder, W.J.; Rutjes, S.A.; Takumi, K.; de Roda Husman, A.M. Aichi virus in sewage and surface water, the Netherlands. Emerg. Infect. Dis. 2013, 19, 1222. [Google Scholar] [CrossRef] [PubMed]
- Kazama, S.; Miura, T.; Masago, Y.; Konta, Y.; Tohma, K.; Manaka, T.; Liu, X.; Nakayama, D.; Tanno, T.; Saito, M. Environmental surveillance of norovirus genogroups I and II for sensitive detection of epidemic variants. Appl. Environ. Microbiol. 2017, 83, e03406–e03416. [Google Scholar] [CrossRef] [Green Version]
- WHO. Status of Environmental Surveillance for SARS-CoV-2 Virus: Scientific Brief, 5 August 2020; World Health Organization: Geneva, Switzerland, 2020; Volume 5. [Google Scholar]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Wu, F.; Xiao, A.; Zhang, J.; Gu, X.; Lee, W.L.; Kauffman, K.; Hanage, W.; Matus, M.; Ghaeli, N.; Endo, N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems 2020, 5, e00614–e00620. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.-M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in Paris wastewaters. Sci. Total Environ. 2020, 778, 146270. [Google Scholar]
- Chen, C.; Gao, G.; Xu, Y.; Pu, L.; Wang, Q.; Wang, L.; Wang, W.; Song, Y.; Chen, M.; Wang, L. SARS-CoV-2–positive sputum and feces after conversion of pharyngeal samples in patients with COVID-19. Ann. Internal Med. 2020, 172, 832–834. [Google Scholar] [CrossRef]
- Hassan, M.M.; El Zowalaty, M.E.; Khan, S.A.; Islam, A.; Nayem, M.R.K.; Järhult, J.D. Role of Environmental Temperature on the Attack rate and Case fatality rate of Coronavirus Disease 2019 (COVID-19) Pandemic. Inf. Eco. Epid. 2020, 10, 1792620. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Ferdous, J.; Islam, S.; Hassan, M.M. Geospatial dynamics of COVID-19 clusters and hotspots in Bangladesh. In Transboundary and Emerging Diseases; Wiley: Hoboken, NJ, USA, 2021; pp. 1–15. [Google Scholar]
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Ferdous, J.; Shano, S.; Choudhury, S.D.; Hassan, M.M. Spatiotemporal patterns and trends of community transmission of the pandemic COVID-19 in South Asia: Bangladesh as a case study. Biosaf. Health 2020, 3, 39–49. [Google Scholar] [CrossRef]
- Zambrano-Monserrate, M.A.; Ruano, M.A.; Sanchez-Alcalde, L. Indirect effects of COVID-19 on the environment. Sci. Total Environ. 2020, 728, 138813. [Google Scholar] [CrossRef]
- Ul-Rahman, A.; Shabbir, M.A.B.; Aziz, M.W.; Yaqub, S.; Mehmood, A.; Raza, M.A.; Shabbir, M.Z. A comparative phylogenomic analysis of SARS-CoV-2 strains reported from non-human mammalian species and environmental samples. Mol. Biol. Rep. 2020, 47, 9207–9217. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiyotani, K.; Toyoshima, Y.; Nemoto, K.; Nakamura, Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J. Hum. Genet. 2020, 65, 569–575. [Google Scholar] [CrossRef]
- Toyoshima, Y.; Nemoto, K.; Matsumoto, S.; Nakamura, Y.; Kiyotani, K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J. Hum. Genet. 2020, 65, 1075–1082. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Anand, U.; Adelodun, B.; Pivato, A.; Suresh, S.; Indari, O.; Jakhmola, S.; Jha, H.C.; Jha, P.K.; Tripathi, V.; Di Maria, F. A review of the presence of SARS-CoV-2 RNA in wastewater and airborne particulates and its use for virus spreading surveillance. Environ. Res. 2021, 196, 110929. [Google Scholar] [CrossRef]
- Bhattacharya, P.; Kumar, M.; Islam, M.T.; Haque, R.; Chakraborty, S.; Ahmad, A.; Niazi, N.K.; Cetecioglu, Z.; Nilsson, D.; Ijumulana, J. Prevalence of SARS-CoV-2 in Communities Through Wastewater Surveillance—a Potential Approach for Estimation of Disease Burden. Curr. Pollut. Rep. 2021, 7, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Lara, R.W.I.; Elsinga, G.; Heijnen, L.; Munnink, B.B.O.; Schapendonk, C.M.; Nieuwenhuijse, D.; Kon, M.; Lu, L.; Aarestrup, F.M.; Lycett, S. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the netherlands and belgium. Emerg. Infec. Dis. 2020, 25, 1405. [Google Scholar]
- Ahmed, F.; Islam, M.A.; Kumar, M.; Hossain, M.; Bhattacharya, P.; Islam, M.T.; Hossen, F.; Hossain, M.S.; Islam, M.S.; Uddin, M.M. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation centre in Bangladesh: Variation along the sewer network. Sci. Total Environ. 2021, 776, 145724. [Google Scholar] [CrossRef]
- Akter, S.; Roy, P.C.; Ferdaus, A.; Ibnat, H.; Alam, A.R.U.; Nigar, S.; Jahid, I.K.; Hossain, M.A. Prevalence and stability of SARS-CoV-2 RNA on Bangladeshi banknotes. Sci. Total Environ. 2021, 779, 146133. [Google Scholar] [CrossRef]
- Behnam, B.; Oishi, S.N.; Uddin, S.M.N.; Rafa, N.; Nasiruddin, S.M.; Mollah, A.M.; Hongzhi, M. Inadequacies in Hospital Waste and Sewerage Management in Chattogram, Bangladesh: Exploring Environmental and Occupational Health Hazards. Sustainability 2020, 12, 9077. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020. [Google Scholar] [CrossRef]
- La Rosa, G.; Bonadonna, L.; Lucentini, L.; Kenmoe, S.; Suffredini, E. Coronavirus in water environments: Occurrence, persistence and concentration methods—A scoping review. Water Res. 2020, 115899. [Google Scholar] [CrossRef] [PubMed]
- Das, A.K.; Islam, M.N.; Billah, M.M.; Sarker, A. COVID-19 and municipal solid waste (MSW) management: A review. Environ. Sci. Pollut. Res. 2021, 1–16. [Google Scholar] [CrossRef]
- Kirby, T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021, 9, e20–e21. [Google Scholar] [CrossRef]
- Jungnick, S.; Hobmaier, B.; Mautner, L.; Hoyos, M.; Haase, M.; Baiker, A.; Lahne, H.; Eberle, U.; Wimmer, C.; Hepner, S. Detection of the new SARS-CoV-2 variants of concern B. 1.1. 7 and B. 1.351 in five SARS-CoV-2 rapid antigen tests (RATs), Germany, March 2021. Eurosurveillance 2021, 26, 2100413. [Google Scholar] [CrossRef]
- Brejová, B.; Hodorová, V.; Boršová, K.; Čabanová, V.; Reizigová, L.; Paul, E.; Čekan, P.; Klempa, B.; Nosek, J.; Vinař, T.B. 1.258$ Δ $, a SARS-CoV-2 variant with $ Δ $ H69/$ Δ $ V70 in the Spike protein circulating in the Czech Republic and Slovakia. arXiv 2021, arXiv:2102.04689. [Google Scholar]
- Smyth, D.S.; Trujillo, M.; Cheung, K.; Gao, A.; Hoxie, I.; Kannoly, S.; Kubota, N.; Markman, M.; San, K.; Sompanya, G. Detection of Mutations Associated with Variants of Concern Via High Throughput Sequencing of SARS-CoV-2 Isolated from NYC Wastewater. medRxiv 2021, 1, 1–12. [Google Scholar] [CrossRef]
- Washington, N.L.; Gangavarapu, K.; Zeller, M.; Bolze, A.; Cirulli, E.T.; Schiabor Barrett, K.M.; Larsen, B.B.; Anderson, C.; White, S.; Cassens, T.; et al. Genomic epidemiology identifies emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. medRxiv 2021, 1, 1–19. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Sayeed, M.A.; Rahman, M.K.; Zamil, S.; Abedin, J.; Saha, O.; Hassan, M.M. Assessment of basic reproduction number (R0), spatial and temporal epidemiological determinants, and genetic characterization of SARS-CoV-2 in Bangladesh. Infect. Genet. Evol. 2021, 92, 104884. [Google Scholar] [CrossRef]
- Napit, R.; Manandhar, P.; Chaudhary, A.; Shrestha, B.; Poudel, A.; Raut, R.; Pradhan, S.; Raut, S.; Mathema, S.; Rajbhandari, R. Rapid genomic surveillance of SARS-CoV-2 in a dense urban community using environmental (sewage) samples. medRxiv 2021, 1, 1–41. [Google Scholar] [CrossRef]
- Cortey, M.; Li, Y.; Diaz, I.; Clilverd, H.; Darwich, L.; Mateu, E. SARS-CoV-2 amino acid substitutions widely spread in the human population are mainly located in highly conserved segments of the structural proteins. bioRxiv 2020, 1, 1–25. [Google Scholar] [CrossRef]
- Crits-Christoph, A.; Kantor, R.S.; Olm, M.R.; Whitney, O.N.; Al-Shayeb, B.; Lou, Y.C.; Flamholz, A.; Kennedy, L.C.; Greenwald, H.; Hinkle, A. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. MBio 2021, 12, e02703–e02720. [Google Scholar] [CrossRef]
- Cristescu, M.E. Can environmental RNA revolutionize biodiversity science? Trends Ecol. Evol. 2019, 34, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Polo, D.; Lois, M.; Fernández-Núñez, M.T.; Romalde, J.L. Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments. Sci. Total Environ. 2021, 786, 147534. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.-Y.; Wang, W.-B.; Hao, Y.-G.; Zhang, H.-R.; Wang, Z.-C.; Chen, Y.-L.; Gao, R.-D. Stability and infectivity of coronaviruses in inanimate environments. World J. Clin. Cases 2020, 8, 1391. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, A.; Sayeed, M.A.; Kalam, M.A.; Ferdous, J.; Rahman, M.K.; Abedin, J.; Islam, S.; Shano, S.; Saha, O.; Shirin, T.; et al. Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally. Microorganisms 2021, 9, 1696. https://doi.org/10.3390/microorganisms9081696
Islam A, Sayeed MA, Kalam MA, Ferdous J, Rahman MK, Abedin J, Islam S, Shano S, Saha O, Shirin T, et al. Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally. Microorganisms. 2021; 9(8):1696. https://doi.org/10.3390/microorganisms9081696
Chicago/Turabian StyleIslam, Ariful, Md. Abu Sayeed, Md. Abul Kalam, Jinnat Ferdous, Md. Kaisar Rahman, Josefina Abedin, Shariful Islam, Shahanaj Shano, Otun Saha, Tahmina Shirin, and et al. 2021. "Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally" Microorganisms 9, no. 8: 1696. https://doi.org/10.3390/microorganisms9081696
APA StyleIslam, A., Sayeed, M. A., Kalam, M. A., Ferdous, J., Rahman, M. K., Abedin, J., Islam, S., Shano, S., Saha, O., Shirin, T., & Hassan, M. M. (2021). Molecular Epidemiology of SARS-CoV-2 in Diverse Environmental Samples Globally. Microorganisms, 9(8), 1696. https://doi.org/10.3390/microorganisms9081696








