An Outbreak of Highly Pathogenic Avian Influenza (H7N7) in Australia and the Potential for Novel Influenza A Viruses to Emerge
Abstract
:1. Introduction
2. An Outbreak of Multiple Avian Influenza Virus Subtypes Simultaneously Emerge across Three Australian Farms
3. Amino Acid Substitutions in the Avian Influenza A (H7N7) Genome Can Increase Virulence and Pathogenicity in Humans
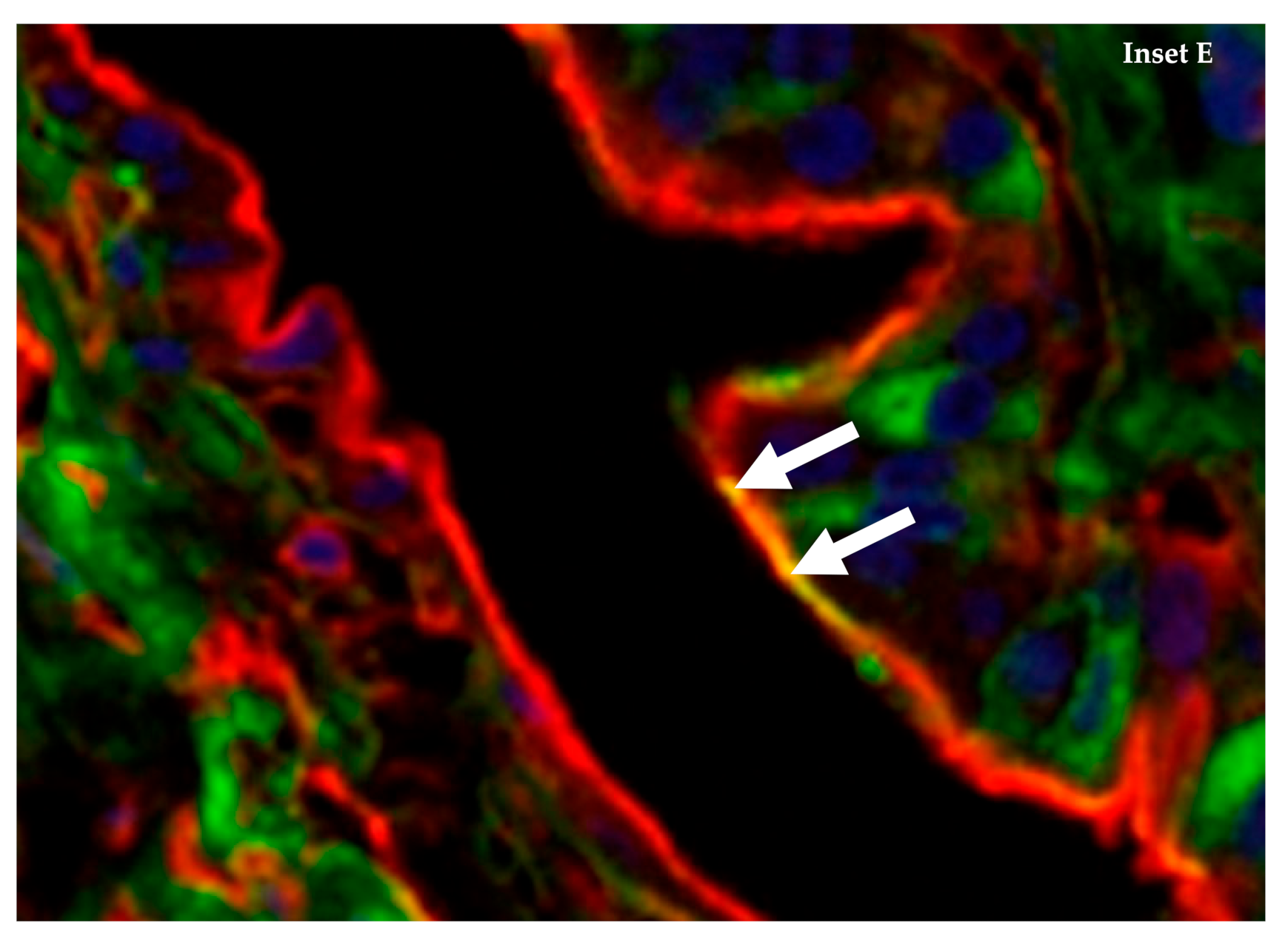
4. The Emu (Dromaius novaehollandiae) Has Abundant Co-Expression of α-2,3 and α-2,6 Sialic Acid Receptors on Internal Tissue and Organs
5. Dromaius novaehollandiae Is Susceptible to A Variety of Avian and Human Influenza A Viral Subtypes, Including the Pandemic H1N1
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Allen, T.; Murray, K.A.; Zambrana-Torrelio, C.; Morse, S.S.; Rondinini, C.; Di Marco, M.; Breit, N.; Olival, K.J.; Daszak, P. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Murray, K.; Selleck, P.; Hooper, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Westbury, H.; Hiley, L.; Selvey, L.; Rodwell, B. A morbillivirus that caused fatal disease in horses and humans. Science 1995, 268, 94–97. [Google Scholar] [CrossRef]
- Mahalingam, S.; Herrero, L.J.; Playford, E.G.; Spann, K.; Herring, B.; Rolph, M.S.; Middleton, D.; McCall, B.; Field, H.; Wang, L.-F. Hendra virus: An emerging paramyxovirus in Australia. Lancet Infect. Dis. 2012, 12, 799–807. [Google Scholar] [CrossRef]
- Halpin, K.; Young, P.L.; Field, H.; Mackenzie, J. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef] [PubMed]
- Selvey, L.A.; Wells, R.M.; McCormack, J.G.; Ansford, A.J.; Murray, K.; Rogers, R.J.; Lavercombe, P.S.; Selleck, P.; Sheridan, J.W. Infection of humans and horses by a newly described morbillivirus. Med. J. Aust. 1995, 162, 642–644. [Google Scholar] [CrossRef]
- Lindsay, S.A.; Gray, R. A novel presentation of tuberculosis with intestinal perforation in a free-ranging Australian sea lion (Neophoca cinerea). J. Wildl. Dis. 2021, 57, 220–224. [Google Scholar] [CrossRef]
- Wille, M.; Lisovski, S.; Risely, A.; Ferenczi, M.; Roshier, D.; Wong, F.Y.; Breed, A.C.; Klaassen, M.; Hurt, A.C. Serologic evidence of exposure to highly pathogenic avian influenza H5 viruses in migratory shorebirds, Australia. Emerg. Infect. Dis. 2019, 25, 1903–1910. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Deng, Y.-M.; Su, Y.C.; Fourment, M.; Iannello, P.; Arzey, G.G.; Hansbro, P.M.; Arzey, K.E.; Kirkland, P.D.; Warner, S. The recent establishment of North American H10 lineage influenza viruses in Australian wild waterfowl and the evolution of Australian avian influenza viruses. J. Virol. 2013, 87, 10182–10189. [Google Scholar] [CrossRef] [Green Version]
- Kishida, N.; Sakoda, Y.; Shiromoto, M.; Bai, G.-R.; Isoda, N.; Takada, A.; Laver, G.; Kida, H. H2N5 influenza virus isolates from terns in Australia: Genetic reassortants between those of the Eurasian and American lineages. Virus Genes 2008, 37, 16–21. [Google Scholar] [CrossRef]
- Spackman, E. Animal Influenza Virus: Methods and Protocols; Erica, S., Ed.; Springer: New York, NY, USA, 2020. [Google Scholar] [CrossRef]
- Webster, R.G.; Bean, W.J.; Gorman, O.T.; Chambers, T.M.; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev. 1992, 56, 152–179. [Google Scholar] [CrossRef]
- Yoon, S.-W.; Webby, R.J.; Webster, R.G. Evolution and ecology of influenza A viruses. In Influenza Pathogenesis and Control-Volume I; Compans, R., Oldstone, M., Eds.; Springer: Cham, Switzerland, 2014; Volume 385, pp. 359–375. [Google Scholar]
- More, S.; Bicout, D.; Bøtner, A.; Butterworth, A.; Calistri, P.; Depner, K.; Edwards, S.; Garin-Bastuji, B.; Good, M.; et al.; EFSA Panel on Animal Health and Welfare (AHAW) Avian influenza. EFSA J. 2017, 15, e04991. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Mostafa, A.; Abdelwhab, E.M.; Mettenleiter, T.C.; Pleschka, S. Zoonotic potential of influenza A viruses: A comprehensive overview. Viruses 2018, 10, 497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OIE World Animal Health Information System. Available online: https://wahis.oie.int/#/home (accessed on 2 March 2021).
- Scott, A.; Hernandez-Jover, M.; Groves, P.; Toribio, J.-A. An overview of avian influenza in the context of the Australian commercial poultry industry. One Health 2020, 10, 100139. [Google Scholar] [CrossRef]
- Commonwealth of Australia, Avian Influenza in Victoria. Available online: https://www.outbreak.gov.au/current-responses-to-outbreaks/avian-influenza (accessed on 2 March 2021).
- Forman, A.; Parsonson, I.; Doughty, W. The pathogenicity of an avian influenza virus isolated in Victoria. Aust. Vet. J. 1986, 63, 294–296. [Google Scholar] [CrossRef]
- Selleck, P.; Arzey, G.; Kirkland, P.; Reece, R.; Gould, A.; Daniels, P.; Westbury, H. An outbreak of highly pathogenic avian influenza in Australia in 1997 caused by an H7N4 virus. Avian Dis. 2003, 47, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, L.M.; Biggs, J.S.; Sheppeard, V.; Oakman, T.L. An evaluation of the use of short message service during an avian influenza outbreak on a poultry farm in Young. Commun. Dis. Intell. Q. Rep. 2016, 40, E195–E201. [Google Scholar]
- Long, J.S.; Mistry, B.; Haslam, S.M.; Barclay, W.S. Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 2019, 17, 67–81. [Google Scholar] [CrossRef]
- Koopmans, M.; Wilbrink, B.; Conyn, M.; Natrop, G.; van der Nat, H.; Vennema, H.; Meijer, A.; van Steenbergen, J.; Fouchier, R.; Osterhaus, A. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 2004, 363, 587–593. [Google Scholar] [CrossRef]
- De Jong, M.; Stegeman, J.; van der Goot, J.; Koch, G. Intra-and interspecies transmission of H7N7 highly pathogenic avian influenza virus during the avian influenza epidemic in The Netherlands in 2003. Revue Sci. Tech. 2009, 28, 333–340. [Google Scholar] [CrossRef] [Green Version]
- de Wit, E.; Munster, V.J.; van Riel, D.; Beyer, W.E.; Rimmelzwaan, G.F.; Kuiken, T.; Osterhaus, A.D.; Fouchier, R.A. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J. Virol. 2010, 84, 1597–1606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.-H.; Bertran, K.; Kwon, J.-H.; Swayne, D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3. 4.4. J. Vet. Sci. 2017, 18, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Laleye, A.T.; Abolnik, C. Emergence of highly pathogenic H5N2 and H7N1 influenza A viruses from low pathogenic precursors by serial passage in ovo. PLoS ONE 2020, 15, e0240290. [Google Scholar] [CrossRef]
- Kim, H.; Webster, R.G.; Webby, R.J. Influenza virus: Dealing with a drifting and shifting pathogen. Viral Immunol. 2018, 31, 174–183. [Google Scholar] [CrossRef]
- Steinhauer, D.A.; Domingo, E.; Holland, J.J. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 1992, 122, 281–288. [Google Scholar] [CrossRef]
- Tharakaraman, K.; Jayaraman, A.; Raman, R.; Viswanathan, K.; Stebbins, N.W.; Johnson, D.; Shriver, Z.; Sasisekharan, V.; Sasisekharan, R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell 2013, 153, 1486–1493. [Google Scholar] [CrossRef] [Green Version]
- de Vries, R.P.; Peng, W.; Grant, O.C.; Thompson, A.J.; Zhu, X.; Bouwman, K.M.; de la Pena, A.T.T.; van Breemen, M.J.; Wickramasinghe, I.N.A.; de Haan, C.A.; et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog. 2017, 13, e1006390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mok, C.K.P.; Lee, H.H.Y.; Lestra, M.; Nicholls, J.M.; Chan, C.W.M.; Sia, S.F.; Zhu, H.; Poon, L.L.M.; Guan, Y.; Peiris, J.M.S. Amino acid substitutions in polymerase basic protein 2 gene contributes to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J. Virol. 2014, 88, 3568–3576. [Google Scholar] [CrossRef] [Green Version]
- Kemink, S.; Fouchier, R.; Rozendaal, F.; Broekman, J.; Koopmans, M.; Osterhaus, A.; Schneeberger, P. A fatal infection due to avian influenza-A (H7N7) virus and adjustment of the preventive measures. Ned. Tijdschr. Geneesk. 2004, 148, 2190–2194. [Google Scholar]
- Fouchier, R.A.; Schneeberger, P.M.; Rozendaal, F.W.; Broekman, J.M.; Kemink, S.A.; Munster, V.; Kuiken, T.; Rimmelzwaan, G.F.; Schutten, M.; Van Doornum, G.J. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 1356–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonges, M.; Bataille, A.; Enserink, R.; Meijer, A.; Fouchier, R.A.; Stegeman, A.; Koch, G.; Koopmans, M. Comparative analysis of avian influenza virus diversity in poultry and humans during a highly pathogenic avian influenza A (H7N7) virus outbreak. J. Virol. 2011, 85, 10598–10604. [Google Scholar] [CrossRef] [Green Version]
- Munster, V.J.; de Wit, E.; van Riel, D.; Beyer, W.E.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Kuiken, T.; Fouchier, R.A. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J. Infect. Dis. 2007, 196, 258–265. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, S.; Kumlin, U.; Dimock, K.; Arnberg, N. Avian influenza and sialic acid receptors: More than meets the eye? Lancet Infect. Dis. 2005, 5, 184–188. [Google Scholar] [CrossRef]
- Belser, J.A.; Rota, P.A.; Tumpey, T.M. Ocular tropism of respiratory viruses. Microbiol. Mol. Biol. Rev. 2013, 77, 144–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belser, J.A.; Lash, R.R.; Garg, S.; Tumpey, T.M.; Maines, T.R. The eyes have it: Influenza virus infection beyond the respiratory tract. Lancet Infect. Dis. 2018, 18, e220–e227. [Google Scholar] [CrossRef]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef] [Green Version]
- Steel, J.; Lowen, A.C.; Mubareka, S.; Palese, P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 2009, 5, e1000252. [Google Scholar] [CrossRef]
- Butler, J.; Middleton, D.; Haining, J.; Layton, R.; Rockman, S.; Brown, L.E.; Sapats, S. Insights into the acquisition of virulence of avian influenza viruses during a single passage in ferrets. Viruses 2019, 11, 915. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Li, X.; Guo, J.; Li, L.; Chang, C.; Li, Y.; Bian, C.; Xu, K.; Chen, H.; Sun, B. The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J. Gen. Virol. 2014, 95, 779–786. [Google Scholar] [CrossRef]
- Xu, G.; Wang, F.; Li, Q.; Bing, G.; Xie, S.; Sun, S.; Bian, Z.; Sun, H.; Feng, Y.; Peng, X. Mutations in PB2 and HA enhanced pathogenicity of H4N6 avian influenza virus in mice. J. Gen. Virol. 2020, 101, 910–920. [Google Scholar] [CrossRef] [PubMed]
- Sang, X.; Wang, A.; Chai, T.; He, X.; Ding, J.; Gao, X.; Li, Y.; Zhang, K.; Ren, Z.; Li, L. Rapid emergence of a PB2-E627K substitution confers a virulent phenotype to an H9N2 avian influenza virus during adaption in mice. Arch. Virol. 2015, 160, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Peng, X.; Lu, R.; Xu, L.; Liu, F.; Cheng, L.; Lu, X.; Yao, H.; Wu, N. Virulence of an H5N8 highly pathogenic avian influenza is enhanced by the amino acid substitutions PB2 E627K and HA A149V. Infect. Genet. Evol. 2017, 54, 347–354. [Google Scholar] [CrossRef]
- Jonges, M.; Welkers, M.R.; Jeeninga, R.E.; Meijer, A.; Schneeberger, P.; Fouchier, R.A.; de Jong, M.D.; Koopmans, M. Emergence of the virulence-associated PB2 E627K substitution in a fatal human case of highly pathogenic avian influenza virus A (H7N7) infection as determined by Illumina ultra-deep sequencing. J. Virol. 2014, 88, 1694–1702. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Carney, P.J.; Donis, R.O.; Stevens, J. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J. Virol. 2012, 86, 8645–8652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Yu, Z.; Sun, W.; Li, X.; Chai, H.; Gao, X.; Guo, J.; Zhang, K.; Feng, N.; Zheng, X. Adaptive amino acid substitutions enhance the virulence of an H7N7 avian influenza virus isolated from wild waterfowl in mice. Vet. Microbiol. 2015, 177, 18–24. [Google Scholar] [CrossRef]
- Hu, M.; Chu, H.; Zhang, K.; Singh, K.; Li, C.; Yuan, S.; Chow, B.K.; Song, W.; Zhou, J.; Zheng, B.-J. Amino acid substitutions V63I or A37S/I61T/V63I/V100A in the PA N-terminal domain increase the virulence of H7N7 influenza A virus. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Scheibner, D.; Ulrich, R.; Fatola, O.I.; Graaf, A.; Gischke, M.; Salaheldin, A.H.; Harder, T.C.; Veits, J.; Mettenleiter, T.C.; Abdelwhab, E.M. Variable impact of the hemagglutinin polybasic cleavage site on virulence and pathogenesis of avian influenza H7N7 virus in chickens, turkeys and ducks. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Le, K.T.; Okamatsu, M.; Nguyen, L.T.; Matsuno, K.; Chu, D.-H.; Tien, T.N.; Le, T.T.; Kida, H.; Sakoda, Y. Genetic and antigenic characterization of the first H7N7 low pathogenic avian influenza viruses isolated in Vietnam. Infect. Genet. Evol. 2020, 78, 104117. [Google Scholar] [CrossRef]
- Wu, H.; Liu, F.; Yang, F.; Xiao, Y.; Yao, H.; Wu, N. Amino acid substitutions involved in the adaptation of a novel H7N7 avian influenza virus in mice. Res. Vet. Sci. 2020, 130, 203–206. [Google Scholar] [CrossRef]
- Puzelli, S.; Rossini, G.; Facchini, M.; Vaccari, G.; Di Trani, L.; Di Martino, A.; Gaibani, P.; Vocale, C.; Cattoli, G.; Bennett, M. Human infection with highly pathogenic A (H7N7) avian influenza virus, Italy, 2013. Emerg. Infect. Dis. 2014, 20, 1745. [Google Scholar] [CrossRef] [Green Version]
- Dietze, K.; Graaf, A.; Homeier-Bachmann, T.; Grund, C.; Forth, L.; Pohlmann, A.; Jeske, C.; Wintermann, M.; Beer, M.; Conraths, F.J. From low to high pathogenicity—characterization of H7N7 avian influenza viruses in two epidemiologically linked outbreaks. Transbound. Emerg. Dis. 2018, 65, 1576–1587. [Google Scholar] [CrossRef]
- Reid, S.M.; Núñez, A.; Seekings, A.H.; Thomas, S.S.; Slomka, M.J.; Mahmood, S.; Clark, J.R.; Banks, J.; Brookes, S.M.; Brown, I.H. Two single incursions of H7N7 and H5N1 low pathogenicity avian influenza in UK broiler breeders during 2015 and 2016. Avian Dis. 2019, 63, 181–192. [Google Scholar] [CrossRef]
- Byrne, A.M.; Reid, S.M.; Seekings, A.H.; Núñez, A.; Obeso Prieto, A.B.; Ridout, S.; Warren, C.J.; Puranik, A.; Ceeraz, V.; Essen, S. H7N7 avian influenza virus mutation from low to high pathogenicity on a layer chicken farm in the UK. Viruses 2021, 13, 259. [Google Scholar] [CrossRef]
- Dugan, V.G.; Chen, R.; Spiro, D.J.; Sengamalay, N.; Zaborsky, J.; Ghedin, E.; Nolting, J.; Swayne, D.E.; Runstadler, J.A.; Happ, G.M. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008, 4, e1000076. [Google Scholar] [CrossRef] [Green Version]
- Gujjar, N.; Chothe, S.K.; Gawai, S.; Nissly, R.; Bhushan, G.; Kanagaraj, V.; Jayarao, B.M.; Kathaperumal, K.; Subbiah, M.; Kuchipudi, S.V. Co-expression of sialic acid receptors compatible with avian and human influenza virus binding in emus (Dromaius novaehollandiae). Virology 2017, 500, 114–121. [Google Scholar] [CrossRef]
- Leigh Perkins, L.E.; Swayne, D.E. Pathogenicity of a Hong Kong–origin H5N1 highly pathogenic avian influenza virus for emus, geese, ducks, and pigeons. Avian Dis. 2002, 46, 53–63. [Google Scholar] [CrossRef]
- Panigrahy, B.; Senne, D.; Pearson, J. Presence of avian influenza virus (AIV) subtypes H5N2 and H7N1 in emus (Dromaius novaehollandiae) and rheas (Rhea americana): Virus isolation and serologic findings. Avian Dis. 1995, 39, 64–67. [Google Scholar] [CrossRef]
- Hassan, M.M.; El Zowalaty, M.E.; Islam, A.; Rahman, M.M.; Chowdhury, M.N.; Nine, H.S.; Rahman, M.K.; Järhult, J.D.; Hoque, M.A. Serological evidence of avian influenza in captive wild birds in a zoo and two safari parks in Bangladesh. Vet. Sci. 2020, 7, 122. [Google Scholar] [CrossRef]
- Rogers, G.N.; Paulson, J.C. Receptor determinants of human and animal influenza virus isolates: Differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983, 127, 361–373. [Google Scholar] [CrossRef]
- Woolcock, P.; Shivaprasad, H.; De Rosa, M. Isolation of avian influenza virus (H10N7) from an emu (Dromaius novaehollandiae) with conjunctivitis and respiratory disease. Avian Dis. 2000, 44, 737–744. [Google Scholar] [CrossRef]
- Amnon, I.; Shkoda, I.; Lapin, E.; Raibstein, I.; Rosenbluth, E.; Nagar, S.; Perk, S.; Bellaiche, M.; Davidson, I. Isolation and identification of highly pathogenic avian influenza virus subtype H5N1 from emus from the Ein Gedi Oasis by the Dead Sea. Avian Dis. 2011, 55, 499–502. [Google Scholar] [CrossRef]
- Kang, W.; Pang, W.; Hao, J.; Zhao, D. Isolation of avian influenza virus (H9N2) from emu in China. Ir. Vet. J. 2006, 59, 1–5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Avian Influenza Virus | Detected in | Location (Farm Numbers) | Date of Last Reported Occurrence |
|---|---|---|---|
| H7N7 (HPAI) | Chicken | Lethbridge, Victoria (3) | 21 February 2014 |
| H5N2 (LPAI) | Turkey | Lethbridge, Victoria (1) Bairnsdale, Victoria (1) | 26 June 2013 |
| H7N6 (LPAI) | Emu | Kerang, Victoria (1) | 2007 |
| Year | Avian Influenza Subtype | Description of Outbreak |
|---|---|---|
| 1976 | H7N7 (LPAI, HPAI) | In a combined broiler and egg farm in the outer suburbs of Melbourne, HPAI A/chicken/Victoria/76 was isolated. LPAI H7 isolated from a separate duck farm (A/duck/Victoria/76). |
| 1985 | H7N7 (HPAI) | Occurred in a chicken farm near Bendigo, Victoria H7N7 (A/chicken/Victoria/85) was isolated. Tests showed this virus to be highly virulent and not substantially different from A/chicken/Victoria/76 [20]. |
| 1992 | H7N3 (HPAI) | Chicken farm near Bendigo, Victoria (different location to the 1985 outbreak) H7N3 (A/chicken/Victoria/92/1) isolated and determined to be HPAI virus. |
| 1994 | H7N3 (HPAI) | Egg farm in Brisbane, Queensland H7N3 (A/chicken/Qld/94) was isolated but different to the H7N3 virus isolated from Victoria in 1992. |
| 1997 | H7N4 (HPAI) | H7N4 was isolated from two chicken broiler-breeder farms and one emu farm near Tamworth, NSW. Viruses isolated from the three properties all had an identical HA cleavage site amino acid sequence. This sequence was also identical to that of the virus identified from the 1994 Queensland outbreak. Control measures resulted in the destruction of 310,565 chickens, 1,232,074 chicken eggs, 261 emu chicks and 147 emu eggs [21]. |
| 2012 | H7N7 (HPAI) | Free range egg laying farm in Maitland, Lower Hunter Valley, NSW. 45,000 birds destroyed as a control measure. |
| 2013 | H7N2 (HPAI) | Two properties infected with HPAI H7N2 with 100% correlation of the same virus isolated from each property. The source of infection at the first property was confirmed to be from wild birds. Control measures on both properties resulted in the destruction of 471,380 birds [17,22]. |
| 2020 | H7N7 (HPAI) H5N2 (LPAI) H7N6 (LPAI) | An outbreak of three different viral strains of AIV across six different farms in Victoria. Three egg farms (HPAI H7N7), two turkey farms (LPAI H5N2) and one emu farm (LPAI H7N6). OIE reports indicate more than 433,000 birds were destroyed as a control measure [17]. |
| Segment | Organ | Tissue | Qualitative Level of Sialic-Acid Receptor Expression |
|---|---|---|---|
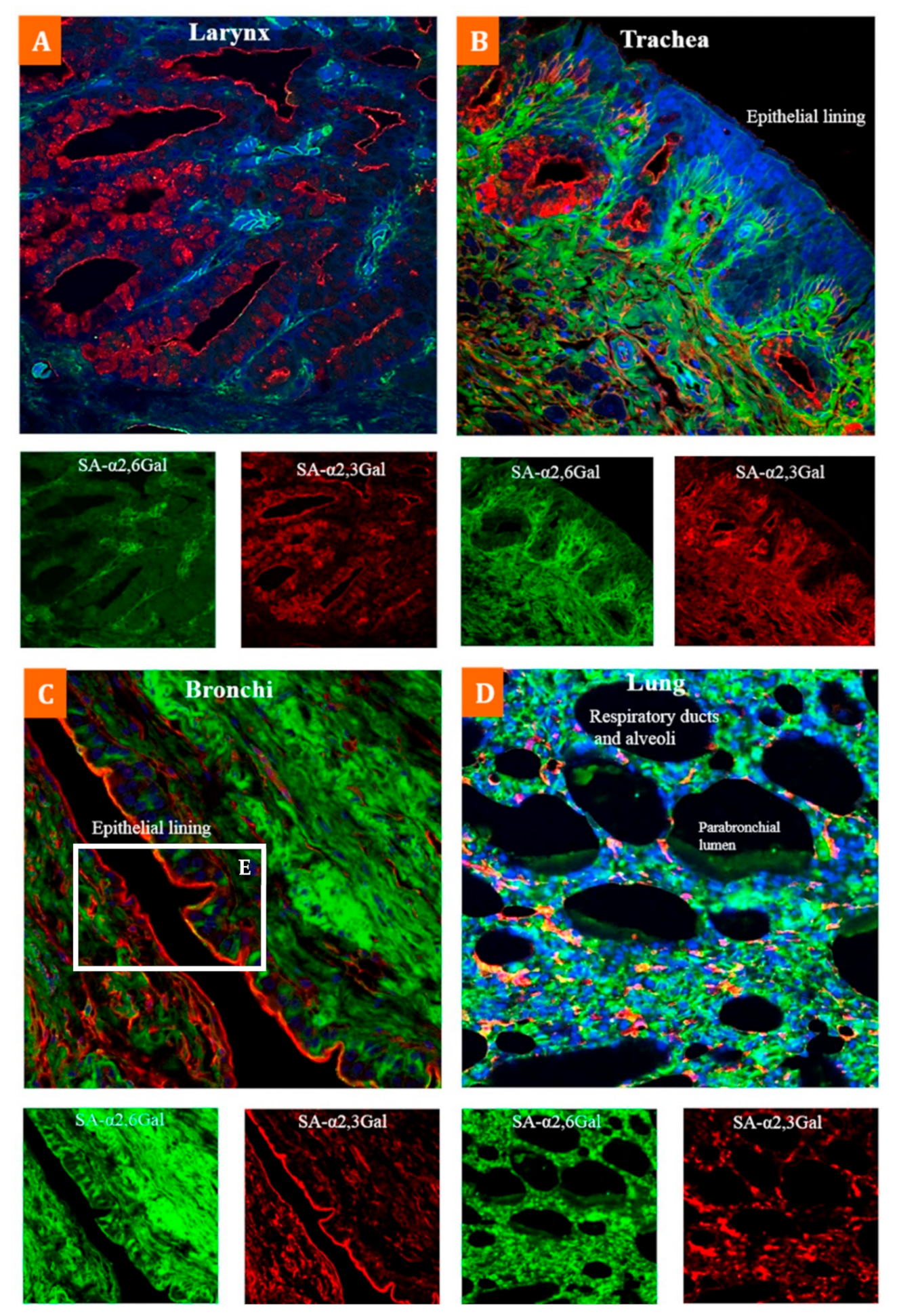
| Respiratory Tract | Larynx-mucosa | α-2,3 + α-2,6 | |
| Trachea-mucosa | α-2,3 + α-2,6 | ||
| Bronchi-mucosa | α-2,3 + α-2,6 | ||
| Alveoli-mucosa | α-2,3 + α-2,6 | ||
| Ciliated and non-ciliated epithelial cells, goblet cells | α-2,3 + α-2,6 | ||
| Submucosa | Higher α-2,6 | ||
| Digestive Tract | Proventriculus-Duodenum | Epithelial cells | α-2,6 dominant throughout |
| Lower portion of small intestine | Some parts of epithelial cells and mainly goblet cells | α-2,3 confined | |
| Large intestine | Epithelial lining of villi and goblet cells | Predominantly α-2,3 | |
| Luminal region of villi | α-2,6 weakly expressed, concentrated here | ||
| Duodenum-Colon | Increased expression of α-2,3 | ||
| Caecal tonsil Lymphoid organ | Abundant expression of α-2,3 + α-2,6 | ||
| Other Major Organs | Kidney | Vascular endothelial wall | α-2,3 + α-2,6 |
| Tubules | α-2,6 | ||
| Liver | α-2,3 predominantly, especially endothelium of the portal vein | ||
| Spleen | Capsule | α-2,3 + α-2,6 (uniform) | |
| Pulp region | Weak α-2,6 | ||
| Skin | Endothelial lining of veins | α-2,3 > α-2,6 | |
| Brain | Consistent but sparse distribution of both receptors | ||
| Skeletal Muscle | Around nuclei | α-2,3 + α-2,6 weakly present | |
| Basement membrane of muscle fibres | α-2,3 weakly present | ||
| Heart | α-2,3 + α-2,6 | ||
| Blood capillaries | α-2,3 higher expression |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bisset, A.T.; Hoyne, G.F. An Outbreak of Highly Pathogenic Avian Influenza (H7N7) in Australia and the Potential for Novel Influenza A Viruses to Emerge. Microorganisms 2021, 9, 1639. https://doi.org/10.3390/microorganisms9081639
Bisset AT, Hoyne GF. An Outbreak of Highly Pathogenic Avian Influenza (H7N7) in Australia and the Potential for Novel Influenza A Viruses to Emerge. Microorganisms. 2021; 9(8):1639. https://doi.org/10.3390/microorganisms9081639
Chicago/Turabian StyleBisset, Andrew T., and Gerard F. Hoyne. 2021. "An Outbreak of Highly Pathogenic Avian Influenza (H7N7) in Australia and the Potential for Novel Influenza A Viruses to Emerge" Microorganisms 9, no. 8: 1639. https://doi.org/10.3390/microorganisms9081639
APA StyleBisset, A. T., & Hoyne, G. F. (2021). An Outbreak of Highly Pathogenic Avian Influenza (H7N7) in Australia and the Potential for Novel Influenza A Viruses to Emerge. Microorganisms, 9(8), 1639. https://doi.org/10.3390/microorganisms9081639






