Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure–Function Relationship Studies and Engineering
Abstract
1. Introduction
2. Inventory of Characterized GSs and BRSs
2.1. Native GSs and BRSs Produced by LAB
2.2. Recombinant GSs and BRSs Produced by E. coli
| Enzyme | Organism | Genbank | Specificity | Size (aa) | Reference |
|---|---|---|---|---|---|
| GTF-0 | Lb. reuteri | AAY86923.1 | R | 1781 | [79] |
| GTF-A* | Lb. reuteri | AAU08015.1 | R | 1781 | [80] |
| ASR* | Ln. mesenteroides | CAB65910.2 | A | 2057 | [81] |
| ASR | Ln. citreum | AIM52834.1 | A | 2057 | (Wangpaiboon et al. NP) |
| GTF-SI* | S. mutans | BAA26114.1 | M | 1455 | [82] |
| GFT-ML1 | Lb. reuteri | AAU08004.1 | M | 1772 | [83] |
| GFT-L | S. salivarius | AAC41412.1 | M | 1449 | [84] |
| GTF-J | S. salivarius | AAA26896.1 | M | 1517 | [84] |
| GTF-I | S. sobrinus | BAA02976.1 | M | 1590 | [85] |
| GTF-I | S. downei | AAC63063.1 | M | 1597 | [86] |
| GTF-B | S. mutans | AAA88588.1 | M | 1475 | [87] |
| GTF-I | S. criceti | BAF62338.1 | M | 1461 | [88] |
| GTF-F | S. orisuis | BAF62337.1 | M | 1466 | [88] |
| GFT-D | S. mutans | AAN58619.1 | GS (n.d) | 1462 | [89] |
| GTF-C | S. mutans | AAN58706.1 | GS (n.d) | 1455 | [89] |
| GTF-B | S. mutans | AAN58705.1 | GS (n.d) | 1476 | [89] |
| LcDS | Ln. citreum | BAF96719.1 | D | 1477 | [90] |
| GTF-U | S. sobrinus | BAC07265.1 | D | 1554 | [91] |
| GTF-S | S. downei | AAA26898.1 | D | 1365 | [92] |
| GTF-R | S. oralis | BAA95201.1 | D | 1575 | [93] |
| GTF-M | S. salivarius | AAC41413.1 | D | 1577 | [84] |
| GTF-Kg3 | Lb. fermentum | AAU08008.1 | D | 1595 | [94] |
| GTF-Kg15 | Lb. sakei | AAU08011.1 | D | 1561 | [94] |
| GTF-K | S. salivarius | CAA77898.1 | D | 1599 | [84] |
| GTF-I | S. sobrinus | BAA14241.1 | D | 1575 | [95] |
| GTF-G | S. gordonii | AAC43483.1 | D | 1577 | [96] |
| GFT-D | S. mutans | AAA26895.1 | D | 1430 | [97] |
| GTF-33 | Lb. parabuchneri | AAU08006.1 | D | 1463 | [94] |
| GTF-1971 | Lb. animalis | CCK33644.1 | D | 1585 | (Ruhmkorf et al., NP) |
| GTF-180* | Lb. reuteri | AAU08001.1 | D | 1772 | [94] |
| GTF-1624 | Lb. curvatus | CCK33643.1 | D | 1697 | (Ruhmkorf et al., NP) |
| GTF-106A | Lb. reuteri | ABP88726.1 | D | 1782 | (Kaditzky et al., NP) |
| DSR-X | Ln. mesenteroides | AAQ98615.2 | D | 1485 | [98] |
| DSR-WC | W. cibaria | ACK38203.1 | D | 1472 | [99] |
| DSR-T | Ln. mesenteroides | BAA90527.1 | D | 1015 | [100] |
| DSR-S | Ln. mesenteroides | AAD10952.1 | D | 1527 | [101] |
| DSR-P | Ln. mesenteroides | AAS79426.1 | D | 1454 | [102] |
| DSR-N | Ln. mesenteroides | AFP53921.1 | D | 1527 | (Siddiqui et al. NP) |
| DSR-K39 | W. cibaria | ADB43097.3 | D | 1445 | [60] |
| DSR-F | Ln. citreum | ACY92456.1 | D | 1527 | [103] |
| DSR-D | Ln. mesenteroides | AAG61158.1 | D | 1527 | [104] |
| DSR-C39-2 | W. confusa | CCF30682.1 | D | 1412 | [59] |
| DSR-C | Ln. citreum | CAB76565.1 | D | 1477 | [81] |
| DSR-BCB4 | Ln. citreum | ABF85832.1 | D | 1465 | [105] |
| DSR-B742 | Ln. citreum | AAG38021.1 | D | 1508 | [106] |
| DSR-B | Ln. citreum | AAB95453.1 | D | 1508 | [107] |
| DSR-A | Ln. citreum | AAB40875.1 | D | 1290 | [108] |
| DEX-YG | Ln. mesenteroides | ABC75033.1 | D | 1527 | [109] |
| DEX-T | Ln. citreum | ACA83218.1 | D | 1495 | [69] |
| Wc392-DSR | W. confusa | AHU88292.1 | D | 1423 | (Krajala et al., NP) |
| DSR | Ln. lactis | ACT20911.1 | D | 1500 | (Kim et al., NP) |
| DSR-DP | Ln. citreum | CDX66641.1 | D | 1278 | [71] |
| DSR-M* | Ln. citreum | CDX66895.1 | D | 2065 | [71] |
| wcCab3-DSR | W. confusa | AKE50934.1 | D | 1401 | (Shukla et al., NP) |
| DSR-R | Ln. mesenteroides | AAN38835.1 | D | 1330 | (Kim et al. NP) |
| GTF-P | S. sanguinis | BAF43788.1 | D | 1575 | [95] |
| GTF-Tl | S. sobrinus | AAX76986.1 | D | 1506 | [110] |
| GTF-106B (DSR106.1) | Lb. reuteri | ABP88725 | D | 1883 | [111] |
| DSR | Lb. animalis | CCK33644.1 | D | 1585 | [111] |
| DSR-E | Ln. citreum | CAD22883.1 | D+α-1,2BRS | 2835 | [77] |
| GtfZ | Lb. kunkeei | KRK22577.1 | D+α-1,3BRS | 2621 | [78] |
| Gsy | Ln. mesenteroides | ANJ45894.1 | GS (n.d) | 1466 | [76] |
| BRS-A | Ln. citreum | CDX66896.1 | α-1,2BRS | 1877 | [71] |
| BRS-B | Ln. citreum | CDX65123.1 | α-1,3BRS | 1888 | [21] |
| BRS-C | Ln. fallax | WP_010006776.1 | α-1,3BRS | 1774 | [21] |
| BRS-D | Lb. kunkeei | WP_051592287.1 | α-1,2BRS | 1463 | [21] |
| GBD-CD2* | Ln. citreum | CAD22883.1 | α-1,2BRS | 1694 | [112] |
3. Structure–Function Relationships
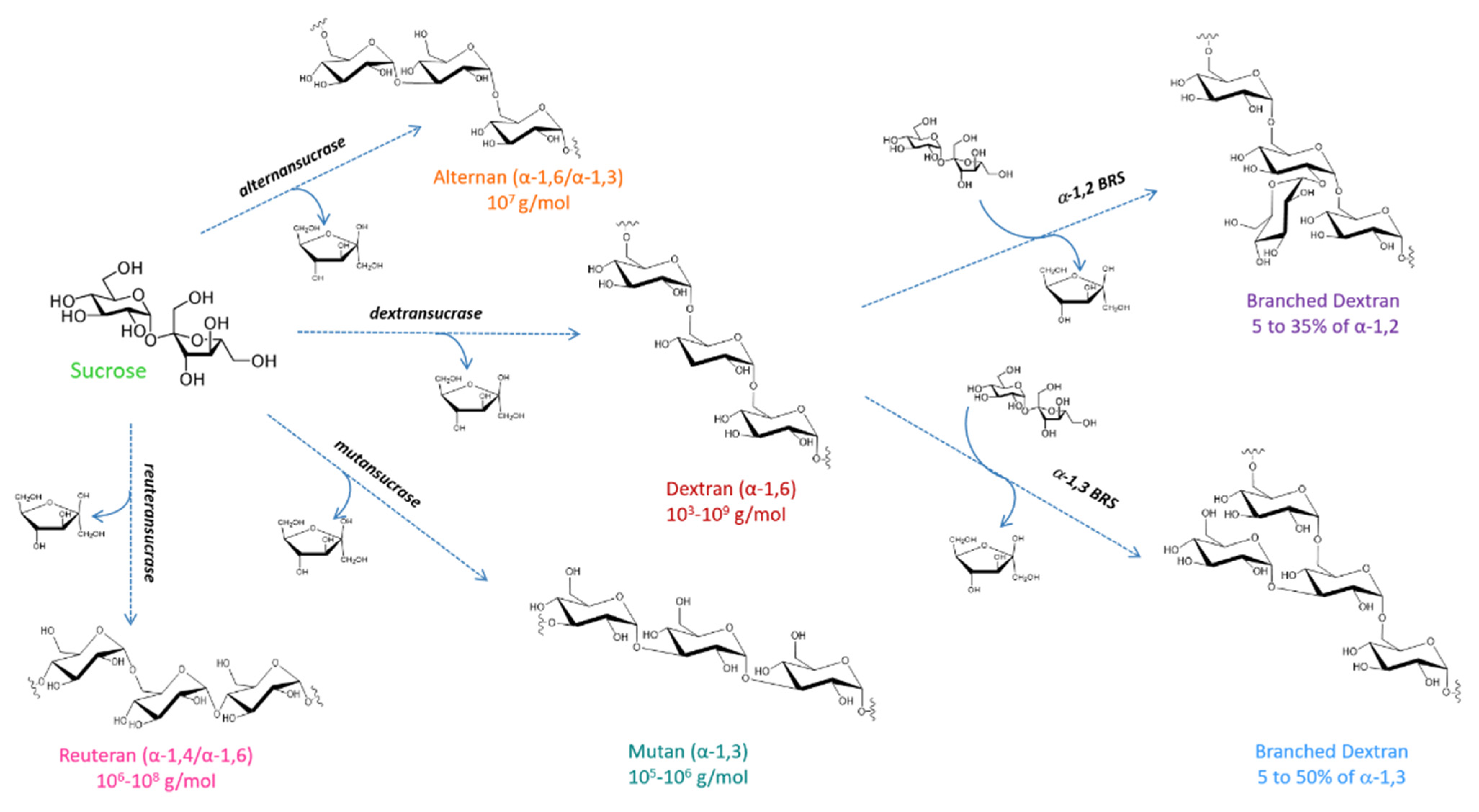
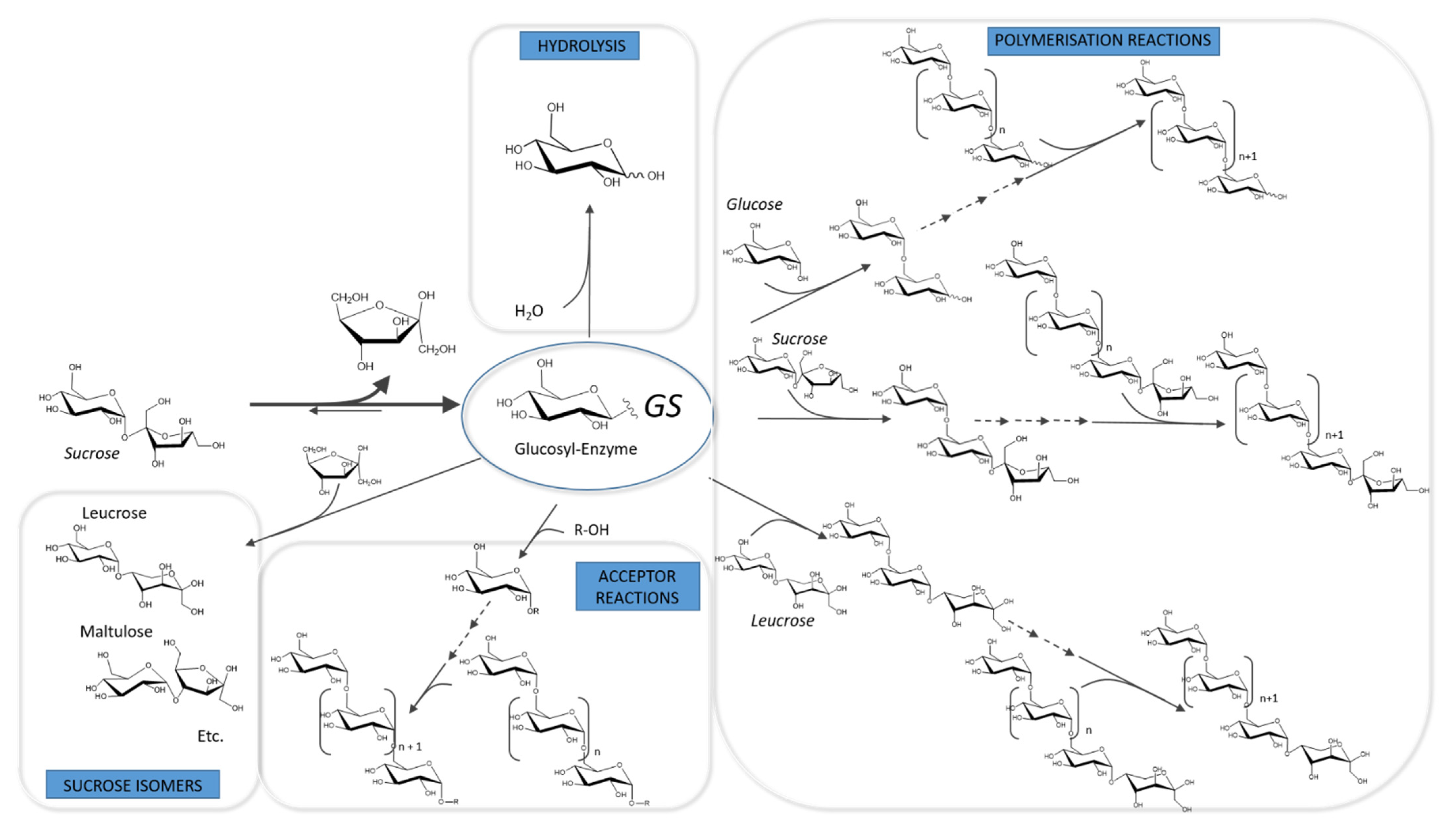
3.1. Catalytic Mechanism and Products
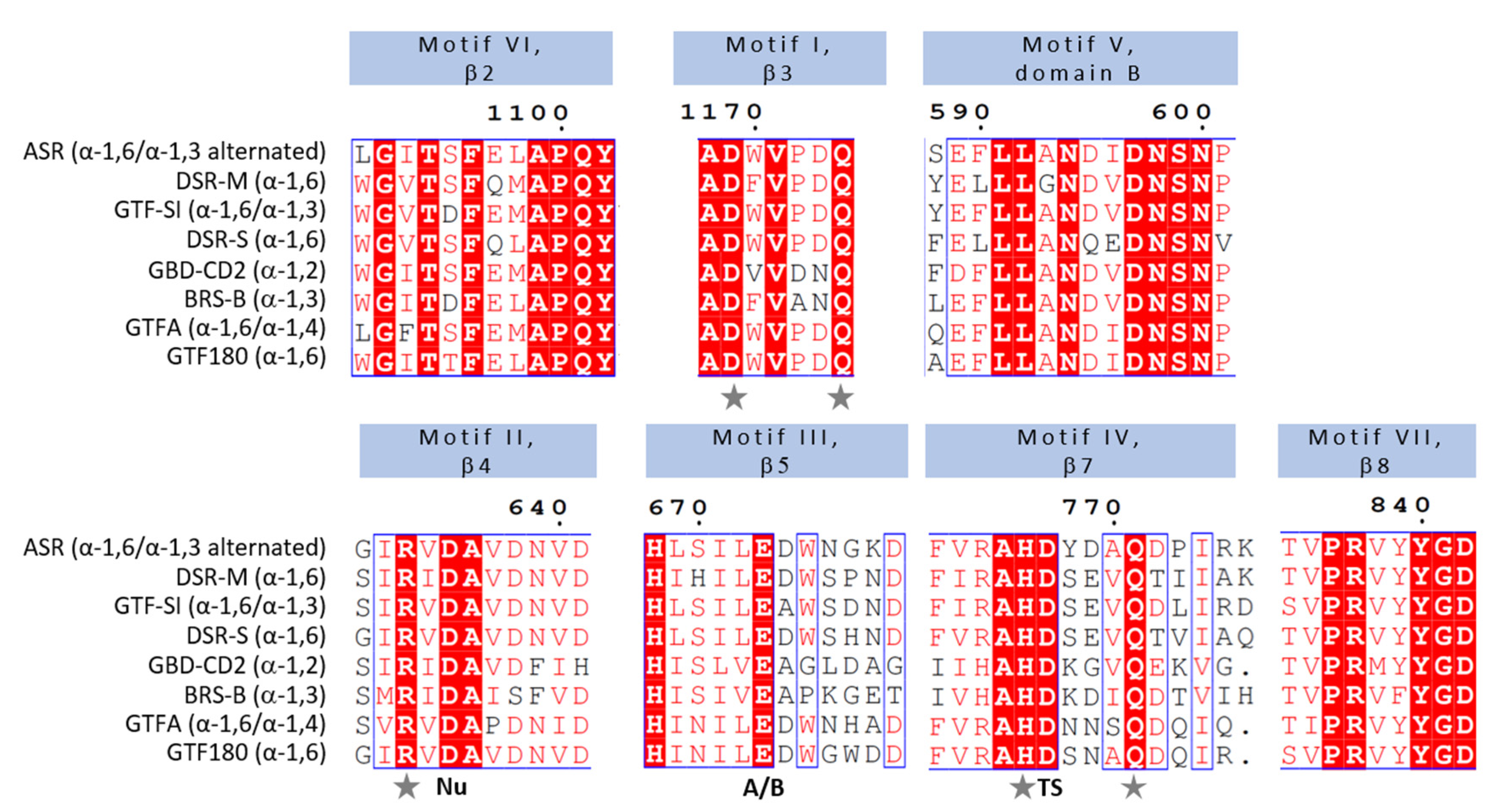
3.2. Mechanistic Insights from Primary Structures
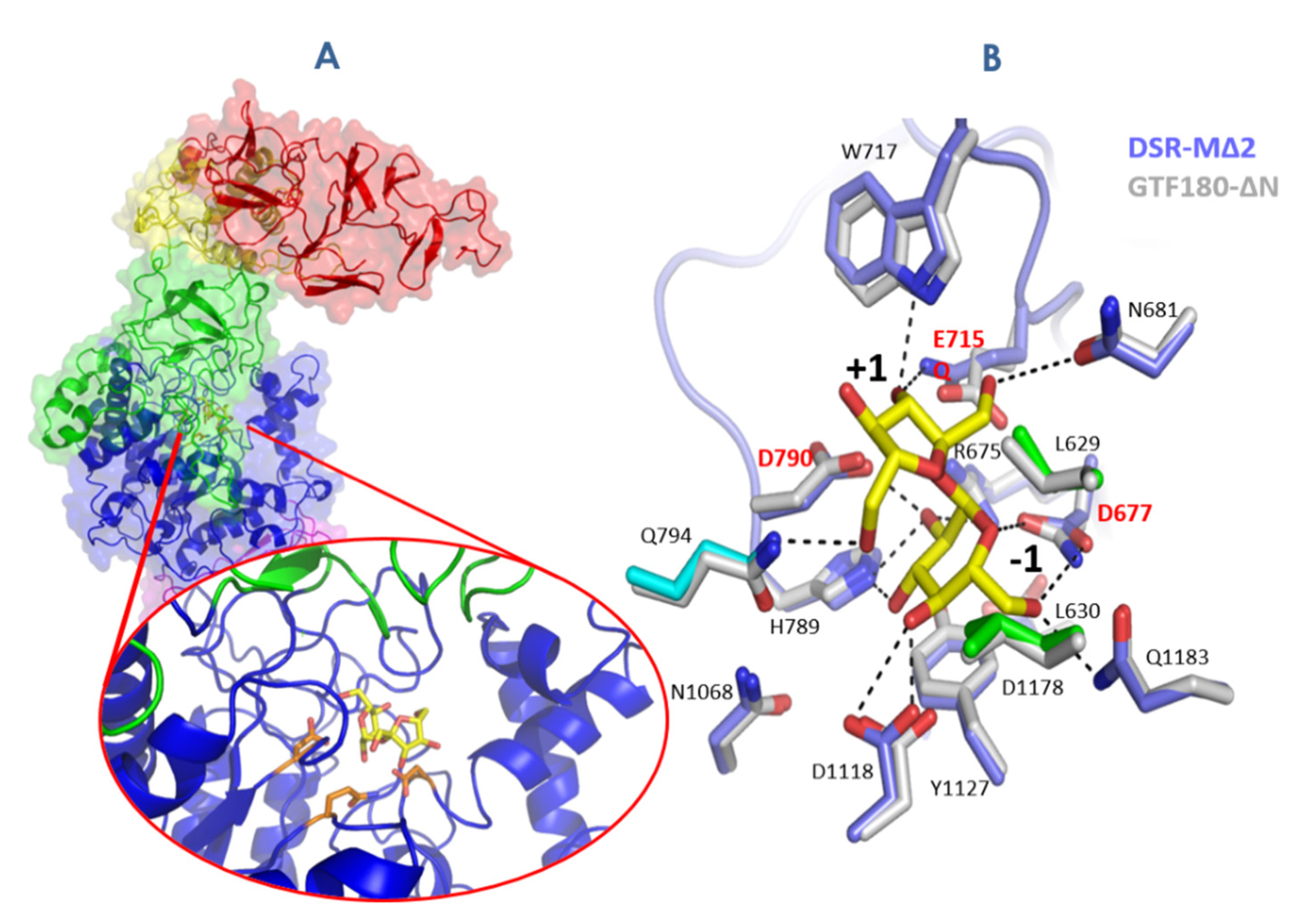
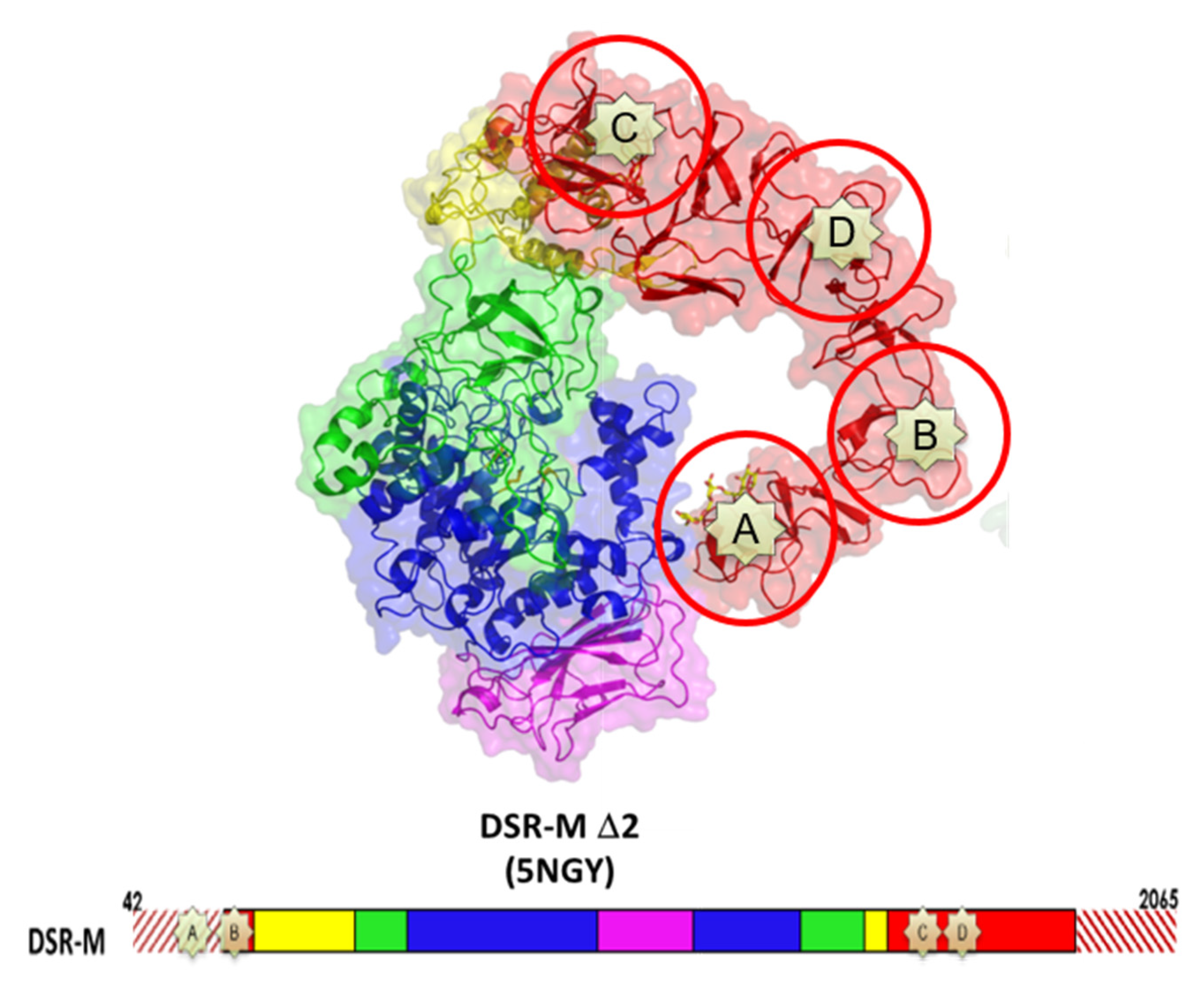
3.3. Going Further with the Three-Dimensional Structures
4. Enzyme Engineering for Man-Made α-Glucans, Oligosaccharides and Glucoconjugates
4.1. Engineering the Linkage Specificity to Diversify Glucan and Oligosaccharide Structures
4.2. GS Engineering for Size-Controlled Polysaccharides and/or Enhance Production of Oligosaccharides
4.3. Engineering GSs and BRSs for Non-Natural Acceptor Glucosylation
5. Outlook
Funding
Conflicts of Interest
References
- Prestegard, J.H.; Liu, J.; Widmalm, G. Oligosaccharides and Polysaccharides. In Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- Freitas, F.; Alves, V.D.; Reis, M.A. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 388–398. [Google Scholar] [CrossRef]
- Moscovici, M. Present and future medical applications of microbial exopolysaccharides. Front. Microbiol. 2015, 6, 1012. [Google Scholar] [CrossRef] [PubMed]
- Roca, C.; Alves, V.D.; Freitas, F.; Reis, M.A.M. Exopolysaccharides enriched in rare sugars: Bacterial sources, production, and applications. Front. Microbiol. 2015, 6, 288. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.J.; Ortiz-Soto, M.E.; Roth, C.; Barnes, W.J.; Seibel, J.; Urbanowicz, B.R.; Pfrengle, F. Enzymatic Synthesis of Artificial Polysaccharides. ACS Sustain. Chem. Eng. 2020, 8, 11853–11871. [Google Scholar] [CrossRef]
- Zhao, L.; Ma, Z.; Yin, J.; Shi, G.; Ding, Z. Biological strategies for oligo/polysaccharide synthesis: Biocatalyst and microbial cell factory. Carbohydr. Polym. 2021, 258, 117695. [Google Scholar] [CrossRef]
- Chen, Z.; Ni, D.; Zhang, W.; Stressler, T.; Mu, W. Lactic acid bacteria-derived α-glucans: From enzymatic synthesis to miscellaneous applications. Biotechnol. Adv. 2021, 47, 107708. [Google Scholar] [CrossRef] [PubMed]
- Shetty, P.R.; Batchu, U.R.; Buddana, S.K.; Sambasiva Rao, K.; Penna, S. A comprehensive review on α-D-Glucans: Structural and functional diversity, derivatization and bioapplications. Carbohydr. Res. 2021, 503, 108297. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, G.; Arumugam, S.; Doble, M. Industrial production and applications of α/β linear and branched glucans. Indian Chem. Eng. 2020, 1–15. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Loesche, W.J. Dental Caries: A Treatable Infection; Automated Diagnostic Documentation, Inc.: Grand Haven, MI, USA, 1993. [Google Scholar]
- Heinze, T.; Liebert, T.; Heublein, B.; Hornig, S. Functional Polymers Based on Dextran. In Polysaccharides II; Klemm, D., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2006; pp. 199–291. [Google Scholar]
- Leathers, T.D. Polysaccharides and Polyamides in the Food Industry. Properties, Production, and Patents; Wiley-Blackwell: Hoboken, NJ, USA, 2005; Volume 1, pp. 233–255. [Google Scholar]
- Vettori, M.H.; Blanco, K.; Cortezi, M.; De Lima, C.; Contiero, J. Dextran: Effect of process parameters on production, purification and molecular weight and recent applications. Diálogos Ciênc 2012, 31, 171–186. [Google Scholar] [CrossRef]
- Hu, Q.; Lu, Y.; Luo, Y. Recent advances in dextran-based drug delivery systems: From fabrication strategies to applications. Carbohydr. Polym. 2021, 264, 1–15. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Padungros, P.; Nakapong, S.; Charoenwongpaiboon, T.; Rejzek, M.; Field, R.A.; Pichyangkura, R. An α-1,6-and α-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and film-forming properties. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Dennes, T.J.; Perticone, A.M.; Paullin, J.L. Cationic Poly Alpha-1,3-Glucan Ethers. Patent US2014070906W, 25 June 2015. [Google Scholar]
- Paullin, J.L.; Perticone, A.M.; Kasat, R.B.; Dennes, T.J. Preparation of Poly Alpha-1,3-Glucan Ethers. Patent US201314107067A, 26 June 2014. [Google Scholar]
- Brison, Y.; Malbert, Y.; Czaplicki, G.; Mourey, L.; Remaud-Siméon, M.; Tranier, S. Structural Insights into the Carbohydrate Binding Ability of an α-(1→2) Branching Sucrase from Glycoside Hydrolase Family 70. J. Biol. Chem. 2016, 291, 7527–7540. [Google Scholar] [CrossRef] [PubMed]
- Moulis, C.; André, I.; Remaud-Siméon, M. GH13 amylosucrases and GH70 branching sucrases, atypical enzymes in their respective families. Cell Mol. Life Sci. CMLS 2016, 73, 2661–2679. [Google Scholar] [CrossRef]
- Vuillemin, M.; Claverie, M.; Brison, Y.; Séverac, E.; Bondy, P.; Morel, S.; Monsan, P.; Moulis, C.; Remaud-Siméon, M. Characterization of the First α-(1→3) Branching Sucrases of the GH70 Family. J. Biol. Chem. 2016, 291, 7687–7702. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids. Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Claverie, M.; Cioci, G.; Vuillemin, M.; Monties, N.; Roblin, P.; Lippens, G.; Remaud-Siméon, M.; Moulis, C. Investigations on the Determinants Responsible for Low Molar Mass Dextran Formation by DSR-M Dextransucrase. ACS Catal. 2017, 7, 7106–7119. [Google Scholar] [CrossRef]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; Leeuwen, S.S.; van Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 2, 250–272. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Meng, X.; Dijkhuizen, L.; Liu, W. Structures, physico-chemical properties, production and (potential) applications of sucrose-derived α-d-glucans synthesized by glucansucrases. Carbohydr. Polym. 2020, 249, 1–12. [Google Scholar] [CrossRef]
- Monsan, P.; Remaud-Siméon, M.; André, I. Transglucosidases as efficient tools for oligosaccharide and glucoconjugate synthesis. Curr. Opin. Microbiol. 2010, 13, 293–300. [Google Scholar] [CrossRef]
- Koepsell, H.J.; Tsuchiya, H.M.; Hellman, N.N.; Kazenko, A.; Hoffman, C.A.; Sharpe, E.S.; Jackson, R.W. Enzymatic Synthesis of Dextran Acceptor Specificity and Chain Initiation. J. Biol. Chem. 1953, 200, 793–801. [Google Scholar] [CrossRef]
- Gangoiti, J.; Corwin, S.F.; Lamothe, L.M.; Vafiadi, C.; Hamaker, B.R.; Dijkhuizen, L. Synthesis of novel α-glucans with potential health benefits through controlled glucose release in the human gastrointestinal tract. Crit. Rev. Food. Sci. Nutr. 2020, 60, 123–146. [Google Scholar] [CrossRef]
- Djouzi, Z.; Andrieux, C.; Pelenc, V.; Somarriba, S.; Popot, F.; Paul, F.; Monsan, P.; Szylit, O. Degradation and fermentation of α-gluco-oligosaccharides by bacterial strains from human colon: In vitro and in vivo studies in gnotobiotic rats. J. Appl. Bacteriol. 1995, 79, 117–127. [Google Scholar] [CrossRef]
- Holt, S.M.; Skory, C.; Cote, G. Growth Assessment of Bifidobacterium on Glucansucrase-Derived Oligosaccharides. In Proceedings of the 1st International Electronic Conference on Microbiology, Online, 2–30 November 2020. [Google Scholar] [CrossRef]
- Holt, S.M.; Teresi, J.M.; Côté, G.L. Influence of alternansucrase-derived oligosaccharides and other carbohydrates on α-galactosidase and α-glucosidase activity in Bifidobacterium adolescentis. Lett. Appl. Microbiol. 2008, 46, 73–79. [Google Scholar] [CrossRef]
- Holt, S.M.; Miller-Fosmore, C.M.; Côté, G.L. Growth of various intestinal bacteria on alternansucrase-derived oligosaccharides. Lett. Appl. Microbiol. 2005, 40, 385–390. [Google Scholar] [CrossRef]
- Sanz, M.L.; Côté, G.L.; Gibson, G.R.; Rastall, R.A. Prebiotic properties of alternansucrase maltose-acceptor oligosaccharides. J. Agric. Food Chem. 2005, 53, 5911–5916. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jang, J.-K.; Park, Y.-S. Production Optimization, Structural Analysis, and Prebiotic- and Anti-Inflammatory Effects of Gluco-Oligosaccharides Produced by Leuconostoc lactis SBC001. Microorganisms 2021, 9, 3229. [Google Scholar] [CrossRef] [PubMed]
- Dols, M.; Remaud Siméon, M.; Willemot, R.-M.; Vignon, M.R.; Monsan, P.F. Structural characterization of the maltose acceptor-products synthesized by Leuconostoc mesenteroides NRRL B-1299 dextransucra Structural characterization of the maltose acceptor-products synthesized by Leuconostoc mesenteroides NRRL B-1299 dextransucrase se. Carbohydr. Res. 1997, 305, 549–559. [Google Scholar] [CrossRef]
- Hasselwander, O.; DiCosimo, R.; You, Z.; Cheng, Q.; Rothman, S.C.; Suwannakham, S.; Baer, Z.C.; Roesch, B.M.; Ruebling-Jass, K.D.; Lai, J.P.; et al. Development of dietary soluble fibres by enzymatic synthesis and assessment of their digestibility in in vitro, animal and randomised clinical trial models. Int. J. Food Sci. Nutr. 2017, 68, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.L.; Gibson, G.R.; Rastall, R.A. Influence of disaccharide structure on prebiotic selectivity in vitro. J. Agric. Food Chem. 2005, 53, 5192–5199. [Google Scholar] [CrossRef] [PubMed]
- Sarbini, S.R.; Kolida, S.; Naeye, T.; Einerhand, A.W.; Gibson, G.R.; Rastall, R.A. The prebiotic effect of α-1,2 branched, low molecular weight dextran in the batch and continuous faecal fermentation system. J. Funct. Foods 2013, 5, 1938–1946. [Google Scholar] [CrossRef]
- Sarbini, S.R.; Kolida, S.; Naeye, T.; Einerhand, A.; Brison, Y.; Remaud-Siméon, M.; Monsan, P.; Gibson, G.R.; Rastall, R.A. In vitro fermentation of linear and α-1,2-branched dextrans by the human fecal microbiota. Appl. Environ. Microbiol. 2011, 77, 5307–5315. [Google Scholar] [CrossRef] [PubMed]
- Malbert, Y.; Moulis, C.; Brison, Y.; Morel, S.; André, I.; Remaud-Siméon, M. Engineering a branching sucrase for flavonoid glucoside diversification. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Morel, S.; Andre, I.; Brison, Y.; Cambon, E.; Malbert, Y.; Pompon, D.; Remaud-Siméon, M.; Urban, P. Novel Flavonoids O-a-glucosylated on the B Cycle, Method for the Production Thereof and Uses. Patent US2017107242A1, 20 April 2017. [Google Scholar]
- Meulenbeld, G.H.; Hartmans, S. Transglycosylation by Streptococcus mutans GS-5 glucosyltransferase-D: Acceptor specificity and engineering of reaction conditions. Biotechnol. Bioeng. 2000, 70, 363–369. [Google Scholar] [CrossRef]
- Meulenbeld, G.H.; Zuilhof, H.; van Veldhuizen, A.; van den Heuvel, R.H.H.; Hartmans, S. Enhanced (+)-Catechin Transglucosylating Activity of Streptococcus mutans GS-5 Glucosyltransferase-D due to Fructose Removal. Appl. Environ. Microbiol. 1999, 65, 4141–4147. [Google Scholar] [CrossRef]
- André, I.; Grelier, S.; Guieysse, D.; Lafraya, A.; Monsan, P.; Moulis, C.; Peruch, F.; Remaud-Siméon, M.; Vuillemin, M. Enzymatic Production of Glycosylated Synthons. Patent US201615559193A, 13 September 2018. [Google Scholar]
- Grimaud, F.; Faucard, P.; Tarquis, L.; Pizzut-Serin, S.; Roblin, P.; Morel, S.; Gall, S.L.; Falourd, X.; Rolland-Sabaté, A.; Lourdin, D.; et al. Enzymatic synthesis of polysaccharide-based copolymers. Green Chem. 2018, 20, 4012–4022. [Google Scholar] [CrossRef]
- Gangoiti, J.; van Leeuwen, S.S.; Gerwig, G.J.; Duboux, S.; Vafiadi, C.; Pijning, T.; Dijkhuizen, L. 4,3-α-Glucanotransferase, a novel reaction specificity in glycoside hydrolase family 70 and clan GH-H. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Leemhuis, H.; Dijkman, W.P.; Dobruchowska, J.M.; Pijning, T.; Grijpstra, P.; Kralj, S.; Kamerling, J.P.; Dijkhuizen, L. 4,6-α-Glucanotransferase activity occurs more widespread in Lactobacillus strains and constitutes a separate GH70 subfamily. Appl. Microbiol. Biotechnol. 2013, 97, 181–193. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Bai, Y.; Pijning, T.; Van Leeuwen, S.S.; Dijkhuizen, L. Structure-function relationships of family GH70 glucansucrase and 4,6-α-glucanotransferase enzymes, and their evolutionary relationships with family GH13 enzymes. Cell Mol. Life Sci. CMLS 2016, 73, 2681–2706. [Google Scholar] [CrossRef] [PubMed]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnol. Adv. 2018, 36, 196–207. [Google Scholar] [CrossRef]
- Pasteur, L. On the viscous fermentation and the butyrous fermentation. Bull Soc. Chim. 1861, 11, 30–31. [Google Scholar]
- Scheibler, C. Investigation on the nature of the gelatinous excretion (so-called frog’s spawn) which is observed in production of beet-sugar juices. Z. Ver. Dtsch. Zucker Ind. 1874, 24, 309–319. [Google Scholar]
- Hestrin, S.; Avineri-Shapiro, S. Mechanism of Polysaccharide Production from Sucrose. Nature 1943, 152, 49–50. [Google Scholar] [CrossRef]
- Van Tieghem, P. On sugar-mill gum. Ann. Sci. Nature Bot. Biol. Veg. 1878, 7, 180–203. [Google Scholar]
- Jeanes, A.; Haynes, W.C.; Wilham, C.A.; Rankin, J.C.; Melvin, E.H.; Austin, M.J.; Cluskey, J.E.; Fisher, B.E.; Tsuchiya, H.M.; Rist, C.E. Characterization and classification of dextrans from ninety-six strains of bacteria. J. Am. Chem. Soc. 1954, 76, 5041–5052. [Google Scholar] [CrossRef]
- Monsan, P.; Bozonnet, S.; Albenne, C.; Joucla, G.; Willemot, R.-M.; Remaud-Siméon, M. Homopolysaccharides from lactic acid bacteria. Int. Dairy J. 2001, 11, 675–685. [Google Scholar] [CrossRef]
- Côté, G.L.; Skory, C.D.; Unser, S.M.; Rich, J.O. The production of glucans via glucansucrases from Lactobacillus satsumensis isolated from a fermented beverage starter culture. Appl. Microbiol. Biotechnol. 2013, 97, 7265–7273. [Google Scholar] [CrossRef]
- Bechtner, J.; Wefers, D.; Schmid, J.; Vogel, R.F.; Jakob, F. Identification and comparison of two closely related dextransucrases released by water kefir borne Lactobacillus hordei TMW 1.1822 and Lactobacillus nagelii TMW 1.1827. Microbiology 2019, 165, 956–966. [Google Scholar] [CrossRef]
- Vasileva, T.; Bivolarski, V.; Michailova, G.; Salim, A.; Rabadjiev, Y.; Ivanova, I.; Iliev, I. Glucansucrases produced by fructophilic lactic acid bacteria Lactobacillus kunkeei H3 and H25 isolated from honeybees. J. Basic. Microbiol. 2017, 57, 68–77. [Google Scholar] [CrossRef]
- Amari, M.; Arango, L.F.G.; Gabriel, V.; Robert, H.; Morel, S.; Moulis, C.; Gabriel, B.; Remaud-Siméon, M.; Fontagné-Faucher, C. Characterization of a novel dextransucrase from Weissella confusa isolated from sourdough. Appl. Microbiol. Biotechnol. 2013, 97, 5413–5422. [Google Scholar] [CrossRef]
- Bounaix, M.-S.; Robert, H.; Gabriel, V.; Morel, S.; Remaud-Siméon, M.; Gabriel, B.; Fontagné-Faucher, C. Characterization of dextran-producing Weissella strains isolated from sourdoughs and evidence of constitutive dextransucrase expression. FEMS Microbiol. Lett. 2010, 311, 18–26. [Google Scholar] [CrossRef]
- Dimopoulou, M.; Vuillemin, M.; Campbell-Sills, H.; Lucas, P.M.; Ballestra, P.; Miot-Sertier, C.; Favier, M.; Coulon, J.; Moine, V.; Doco, T.; et al. Exopolysaccharide (EPS) Synthesis by Oenococcus oeni: From Genes to Phenotypes. PLoS ONE 2014, 9, e98898. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Goyal, A. Novel dextran from Pediococcus pentosaceus CRAG3 isolated from fermented cucumber with anti-cancer properties. Int. J. Biol. Macromol. 2013, 62, 352–357. [Google Scholar] [CrossRef]
- Shukla, S.; Verma, A.K.; Kajala, I.; Nyyssolä, A.; Baruah, R.; Katina, K.; Juvonen, R.; Tenkanen, M.; Goyal, A. Structure modeling and functional analysis of recombinant dextransucrase from Weissella confusa Cab3 expressed in Lactococcus lactis. Prep. Biochem. Biotechnol. 2016, 46, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Côté, G.L.; Robyt, J.F. The formation of α-D-(1→3) branch linkages by an exocellular glucansucrase from Leuconostoc mesenteroides NRRL B-742. Carbohydr. Res. 1983, 119, 141–156. [Google Scholar] [CrossRef]
- Côté, G.L.; Robyt, J.F. Isolation and partial characterization of an extracellular glucansucrase from Leuconostoc mesenteroides NRRL B-1355 that synthesizes an alternating (1→6),(1→3)-α-D-glucan. Carbohydr. Res. 1982, 101, 57–74. [Google Scholar] [CrossRef]
- Sato, S.; Koga, T.; Inoue, M. Isolation and some properties of extracellular D-glucosyltransferases and D-fructosyltransferases from Streptococcus mutans serotypes c, e, and f. Carbohydr. Res. 1984, 134, 293–304. [Google Scholar] [CrossRef]
- Aoki, H.; Shiroza, T.; Hayakawa, M.; Sato, S.; Kuramitsu, H.K. Cloning of a Streptococcus mutans glucosyltransferase gene coding for insoluble glucan synthesis. Infect. Immun. 1986, 53, 587–594. [Google Scholar] [CrossRef]
- Biswas, S.; Biswas, I. Complete genome sequence of Streptococcus mutans GS-5, a serotype c strain. J. Bacteriol. 2012, 194, 4787–4788. [Google Scholar] [CrossRef]
- Kim, J.F.; Jeong, H.; Lee, J.-S.; Choi, S.-H.; Ha, M.; Hur, C.-G.; Kim, J.-S.; Lee, S.; Park, H.-S.; Park, Y.-H.; et al. Complete Genome Sequence of Leuconostoc citreum KM20. J. Bacteriol. 2008, 190, 3093–3094. [Google Scholar] [CrossRef] [PubMed]
- Laguerre, S.; Amari, M.; Vuillemin, M.; Robert, H.; Loux, V.; Klopp, C.; Morel, S.; Gabriel, B.; Remaud-Siméon, M.; Gabriel, V.; et al. Genome sequences of three Leuconostoc citreum strains, LBAE C10, LBAE C11, and LBAE E16, isolated from wheat sourdoughs. J. Bacteriol. 2012, 194, 1610–1611. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Passerini, D.; Vuillemin, M.; Ufarté, L.; Morel, S.; Loux, V.; Fontagné-Faucher, C.; Monsan, P.; Remaud-Siméon, M.; Moulis, C. Inventory of the GH70 enzymes encoded by Leuconostoc citreum NRRL B-1299—Identification of three novel α-transglucosylases. FEBS J. 2015, 282, 2115–2130. [Google Scholar] [CrossRef]
- Passerini, D.; Vuillemin, M.; Laguerre, S.; Amari, M.; Loux, V.; Gabriel, V.; Robert, H.; Morel, S.; Monsan, P.; Gabriel, B.; et al. Complete Genome Sequence of Leuconostoc citreum Strain NRRL B-742. Genome. Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Münkel, F.; Bechtner, J.; Eckel, V.; Fischer, A.; Herbi, F.; Jakob, F.; Wefers, D. Detailed Structural Characterization of Glucans Produced by Glucansucrases from Leuconostoc citreum TMW 2.1194. J. Agric. Food Chem. 2019, 67, 6856–6866. [Google Scholar] [CrossRef]
- Münkel, F.; Wefers, D. Fine structures of different dextrans assessed by isolation and characterization of endo-dextranase liberated isomalto-oligosaccharides. Carbohydr. Polym. 2019, 215, 296–306. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Waiyaseesang, N.; Panpetch, P.; Charoenwongpaiboon, T.; Nepogodiev, S.A.; Ekgasit, S.; Field, R.A.; Pichayangkura, R. Characterisation of insoluble α-1,3-/α-1,6 mixed linkage glucan produced in addition to soluble α-1,6-linked dextran by glucansucrase (DEX-N) from Leuconostoc citreum ABK-1. Int. J. Biol. Macromol. 2020, 152, 473–482. [Google Scholar] [CrossRef]
- Yan, M.; Wang, B.-H.; Xu, X.; Chang, P.; Hang, F.; Wu, Z.; You, C.; Liu, Z. Molecular and functional study of a branching sucrase-like glucansucrase reveals an evolutionary intermediate between two subfamilies of the GH70 enzymes. Appl. Environ. Microbiol. 2018, 84, 1–13. [Google Scholar] [CrossRef]
- Fabre, E.; Bozonnet, S.; Arcache, A.; Willemot, R.-M.; Vignon, M.; Monsan, P.; Remaud-Siméon, M. Role of the two catalytic domains of DSR-E dextransucrase and their involvement in the formation of highly α-1,2 branched dextran. J. Bacteriol. 2005, 187, 296–303. [Google Scholar] [CrossRef]
- Meng, X.; Gangoiti, J.; Wang, X.; Grijpstra, P.; van Leeuwen, S.S.; Pijning, T.; Dijkhuizen, L. Biochemical characterization of a GH70 protein from Lactobacillus kunkeei DSM 12361 with two catalytic domains involving branching sucrase activity. Appl. Microbiol. Biotechnol. 2018, 102, 7935–7950. [Google Scholar] [CrossRef] [PubMed]
- Kralj, S.; Stripling, E.; Sanders, P.; van Geel-Schutten, G.H.; Dijkhuizen, L. Highly Hydrolytic Reuteransucrase from Probiotic Lactobacillus reuteri Strain ATCC 55730. Appl. Environ. Microbiol. 2005, 71, 3942–3950. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, G.H.; Rahaoui, H.; Leer, R.J.; Faber, E.J.; van der Maarel, M.; Dijkhuizen, L. Molecular characterization of a novel glucosyltransferase from Lactobacillus reuteri strain 121 synthesizing a unique, highly branched glucan with alpha-(1 -> 4) and alpha-(1 -> 6) glucosidic bonds. Appl. Environ. Microbiol. 2002, 68, 4283–4291. [Google Scholar] [CrossRef] [PubMed]
- Argüello-Morales, M.A.; Remaud-Simeon, M.; Pizzut, S.; Sarçabal, P.; Willemot, R.; Monsan, P. Sequence analysis of the gene encoding alternansucrase, a sucrose glucosyltransferase from Leuconostoc mesenteroides NRRL B-1355. FEMS Microbiol. Lett. 2000, 182, 81–85. [Google Scholar] [CrossRef]
- Fujiwara, T.; Terao, Y.; Hoshino, T.; Kawabata, S.; Ooshima, T.; Sobue, S.; Kimura, S.; Hamada, S. Molecular analyses of glucosyltransferase genes among strains of Streptococcus mutans. FEMS Microbiol. Lett. 1998, 161, 331–336. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, G.H.; Dondorff, M.M.G.; Kirsanovs, S.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Glucan synthesis in the genus Lactobacillus: Isolation and characterization of glucansucrase genes, enzymes and glucan products from six different strains. Microbiology 2004, 150, 3681–3690. [Google Scholar] [CrossRef]
- Simpson, C.L.; Cheetham, N.W.H.; Jacques, N.A. Four glucosyltransferases, GtfJ.; GtfK.; GtfL and GtfM, from Streptococcus salivarius ATCC 25975. Microbiology 1995, 141, 1451–1460. [Google Scholar] [CrossRef]
- Sato, S.; Inoue, M.; Hanada, N.; Aizawa, Y.; Isobe, Y.; Katayama, T. DNA sequence of the glucosyltransferase gene of serotype d Streptococcus sobrinus. Mitochondrial. DNA 1993, 4, 19–27. [Google Scholar] [CrossRef]
- Ferretti, J.J.; Gilpin, M.L.; Russell, R.R. Nucleotide sequence of a glucosyltransferase gene from Streptococcus sobrinus MFe28. J. Bacteriol. 1987, 169, 4271–4278. [Google Scholar] [CrossRef]
- Shiroza, T.; Ueda, S.; Kuramitsu, H.K. Sequence analysis of the gtfB gene from Streptococcus mutans. J. Bacteriol. 1987, 169, 4263–4270. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki-Kuwahara, N.; Takada, K.; Hirasawa, M. Sequence and phylogenetic analyses of novel glucosyltransferase genes of mutans streptococci isolated from pig oral cavity. J. Microbiol. 2008, 46, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Ajdić, D.; McShan, W.M.; McLaughlin, R.E.; Savić, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef]
- Yi, A.-R.; Lee, S.-R.; Jang, M.-U.; Park, J.-M.; Eom, H.-J.; Han, N.-S.; Kim, T.-J. Cloning of Dextransucrase Gene from Leuconostoc citreum HJ-P4 and Its High-Level Expression in, E. coli by Low Temperature Induction. J. Microbiol. Biotechnol. 2009, 19, 829–835. [Google Scholar] [CrossRef] [PubMed]
- Hanada, N.; Fukushima, K.; Nomura, Y.; Senpuku, H.; Hayakawa, M.; Mukasa, H.; Shiroza, T.; Abiko, Y. Cloning and nucleotide sequence analysis of the Streptococcus sobrinus gtfU gene that produces a highly branched water-soluble glucan. Biochim. Biophys. Acta 2002, 1570, 75–79. [Google Scholar] [CrossRef]
- Gilmore, K.S.; Russell, R.R.; Ferretti, J.J. Analysis of the Streptococcus downei gtfS gene, which specifies a glucosyltransferase that synthesizes soluble glucans. Infect. Immun. 1990, 58, 2452–2458. [Google Scholar] [CrossRef]
- Fujiwara, T.; Hoshino, T.; Ooshima, T.; Sobue, S.; Hamada, S. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 2000, 68, 2475–2483. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, G.H.; van der Maarel, M.; Dijkhuizen, L. Biochemical and molecular characterization of Lactobacillus reuteri 121 reuteransucrase. Microbiol. Sgm 2004, 150, 2099–2112. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, T.; Fujiwara, T.; Kawabata, S. Evolution of Cariogenic Character in Streptococcus mutans: Horizontal Transmission of Glycosyl Hydrolase Family 70 Genes. Sci. Rep. 2012, 2, 1–7. [Google Scholar] [CrossRef]
- Vickerman, M.M.; Sulavik, M.C.; Nowak, J.D.; Gardner, N.M.; Jones, G.W.; Clewell, D.B. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene, gtfG. DNA Seq. J. DNA Seq. Map 1997, 7, 83–95. [Google Scholar] [CrossRef]
- Shimamura, A.; Nakano, Y.J.; Mukasa, H.; Kuramitsu, H.K. Identification of amino acid residues in Streptococcus mutans glucosyltransferases influencing the structure of the glucan product. J. Bacteriol. 1994, 176, 4845–4850. [Google Scholar] [CrossRef] [PubMed]
- Yalin, Y.; Jin, L.; Jianhua, W.; Da, T.; Zigang, T. Expression and characterization of dextransucrase gene dsrX from Leuconostoc mesenteroides in Escherichia coli. J. Biotechnol. 2008, 133, 505–512. [Google Scholar] [CrossRef]
- Kang, H.-K.; Oh, J.-S.; Kim, D. Molecular characterization and expression analysis of the glucansucrase DSRWC from Weissella cibaria synthesizing a alpha(1-->6) glucan. FEMS Microbiol. Lett. 2009, 292, 33–41. [Google Scholar] [CrossRef]
- Funane, K.; Mizuno, K.; Takahara, H.; Kobayashi, M. Gene encoding a dextransucrase-like protein in Leuconostoc mesenteroides NRRL B-512F. Biosci. Biotechnol. Biochem. 2000, 64, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Monchois, V.; Remaud-Simeon, M.; Russell, R.R.B.; Monsan, P.; Willemot, R.-M. Characterization of Leuconostoc mesenteroides NRRL B-512F dextransucrase (DSRS) and identification of amino-acid residues playing a key role in enzyme activity. Appl. Microbiol. Biotechnol. 1997, 48, 465–472. [Google Scholar] [CrossRef]
- Olvera, C.; Fernández-Vázquez, J.L.; Ledezma-Candanoza, L.; López-Munguía, A. Role of the C-terminal region of dextransucrase from Leuconostoc mesenteroides IBT-PQ in cell anchoring. Microbiol. Read. Engl. 2007, 153, 3994–4002. [Google Scholar] [CrossRef]
- Fraga Vidal, R.; Moulis, C.; Escalier, P.; Remaud-Siméon, M.; Monsan, P. Isolation of a Gene from Leuconostoc citreum B/110-1-2 Encoding a Novel Dextransucrase Enzyme. Curr. Microbiol. 2011, 62, 1260–1266. [Google Scholar] [CrossRef] [PubMed]
- Neubauer, H.; Bauché, A.; Mollet, B. Molecular characterization and expression analysis of the dextransucrase DsrD of Leuconostoc mesenteroides Lcc4 in homologous and heterologous Lactococcus lactis cultures. Microbiology 2003, 149, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-K.; Kim, Y.-M.; Kim, D.-M. Functional, genetic, and bioinformatic characterization of dextransucrase (DSRBCB4) gene in Leuconostoc mesenteroides B-1299CB4. J. Microbiol. Biotechnol. 2008, 18, 1050–1058. [Google Scholar] [PubMed]
- Kim, H.; Kim, D.; Ryu, H.J.; Robyt, J.F. Cloning and sequencing of the alpha-1 -> 6 dextransucrase gene from Leuconostoc mensenteroides B-742C. B. J. Microbiol. Biotechnol. 2000, 10, 559–563. [Google Scholar]
- Monchois, V.; Remaud-Siméon, M.; Monsan, P.; Willemot, R.-M. Cloning and sequencing of a gene coding for an extracellular dextransucrase (DSRB) from Leuconostoc mesenteroides NRRL B-1299 synthesizing only a α(1–6) glucan. FEMS Microbiol. Lett. 1998, 159, 307–315. [Google Scholar] [CrossRef]
- Monchois, V.; Willemot, R.-M.; Remaud-Simeon, M.; Croux, C.; Monsan, P. Cloning and sequencing of a gene coding for a novel dextransucrase from Leuconostoc mesenteroides NRRL B-1299 synthesizing only α(1–6) and α(1–3) linkages. Gene 1996, 182, 23–32. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Y.; Zhu, C.; Zhu, B.; Wang, Y. Cloning, sequencing and expression of a dextransucrase gene (dexYG) from Leuconostoc mesenteroides. Biotechnol. Lett. 2008, 30, 1441–1446. [Google Scholar] [CrossRef] [PubMed]
- Hanada, N.; Isobe, Y.; Aizawa, Y.; Katayama, T.; Sato, S.; Inoue, M. Nucleotide sequence analysis of the gtfT gene from Streptococcus sobrinus OMZ176. Infect. Immun. 1993, 61, 2096–2103. [Google Scholar] [CrossRef] [PubMed]
- Rühmkorf, C.; Bork, C.; Mischnick, P.; Rübsam, H.; Becker, T.; Vogel, R.F. Identification of Lactobacillus curvatus TMW 1.624 dextransucrase and comparative characterization with Lactobacillus reuteri TMW 1.106 and Lactobacillus animalis TMW 1.971 dextransucrases. Food Microbiol. 2013, 34, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Brison, Y.; Fabre, E.; Moulis, C.; Portais, J.-C.; Monsan, P.; Remaud-Siméon, M. Synthesis of dextrans with controlled amounts of α-1,2 linkages using the transglucosidase GBD–CD2. Appl. Microbiol. Biotechnol. 2009, 86, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Mooser, G.; Hefta, S.A.; Paxton, R.J.; Shively, J.E.; Lee, T.D. Isolation and sequence of an active-site peptide containing a catalytic aspartic acid from two Streptococcus sobrinus alpha-glucosyltransferases. J. Biol. Chem. 1991, 266, 8916–8922. [Google Scholar] [CrossRef]
- Mooser, G.; Iwaoka, K.R. Sucrose 6-alpha-D-glucosyltransferase from Streptococcus sobrinus: Characterization of a glucosyl-enzyme complex. Biochemistry 1989, 28, 443–449. [Google Scholar] [CrossRef]
- Koshland, D.E. Stereochemistry and the Mechanism of Enzymatic Reactions. Biol. Rev. 1953, 28, 416–436. [Google Scholar] [CrossRef]
- Davies, G.J.; Wilson, K.S.; Henrissat, B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997, 321, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Luzio, G.A.; Mayer, R.M. The hydrolysis of sucrose by dextransucrase. Carbohydr. Res. 1983, 111, 311–318. [Google Scholar] [CrossRef]
- Daudé, D.; Remaud-Siméon, M.; André, I. Sucrose analogs: An attractive (bio)source for glycodiversification. Nat. Prod. Rep. 2012, 29, 945–960. [Google Scholar] [CrossRef]
- Moulis, C.; Joucla, G.; Harrison, D.; Fabre, E.; Potocki-Veronese, G.; Monsan, P.; Remaud-Siméon, M. Understanding the polymerization mechanism of glycoside-hydrolase family 70 glucansucrases. J. Biol. Chem. 2006, 281, 31254–31267. [Google Scholar] [CrossRef]
- Cheetham, N.W.H.; Slodki, M.E.; Walker, G.J. Structure of the linear, low molecular weight dextran synthesized by a D-glucosyltransferase (GTF-S3) of Streptococcus sobrinus. Carbohydr. Polym. 1991, 16, 341–353. [Google Scholar] [CrossRef]
- Dobruchowska, J.M.; Meng, X.; Leemhuis, H.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Gluco-oligomers initially formed by the reuteransucrase enzyme of Lactobacillus reuteri 121 incubated with sucrose and malto-oligosaccharides. Glycobiology 2013, 23, 1084–1096. [Google Scholar] [CrossRef]
- Côté, G.L.; Skory, C.D. Isomelezitose formation by glucansucrases. Carbohydr. Res. 2017, 439, 57–60. [Google Scholar] [CrossRef][Green Version]
- Joucla, G.; Pizzut-Serin, S.; Monsan, P.; Remaud-Siméon, M. Construction of a fully active truncated alternansucrase partially deleted of its carboxy-terminal domain. FEBS Lett. 2006, 580, 763–768. [Google Scholar] [CrossRef]
- Molina, M.; Moulis, C.; Monties, N.; Pizzut-Serin, S.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud Siméon, M. Deciphering an undecided enzyme: Investigations of the structural determinants involved in the linkage specificity of alternansucrase. ACS Catal. 2019, 9, 2222–2237. [Google Scholar] [CrossRef]
- Komatsu, H.; Abe, Y.; Eguchi, K.; Matsuno, H.; Matsuoka, Y.; Sadakane, T.; Inoue, T.; Fukui, K.; Kodama, T. Kinetics of dextran-independent α-(1→3)-glucan synthesis by Streptococcus sobrinus glucosyltransferase I. FEBS J. 2011, 278, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Bivolarski, V.; Vasileva, T.; Gabriel, V.; Iliev, I. Synthesis of glucooligosaccharides with prebiotic potential by glucansucrase URE 13–300 acceptor reactions with maltose, raffinose and lactose. Eng. Life Sci. 2018, 18, 904–913. [Google Scholar] [CrossRef]
- Díez-Municio, M.; Montilla, A.; Jimeno, M.L.; Corzo, N.; Olano, A.; Moreno, F.J. Synthesis and Characterization of a Potential Prebiotic Trisaccharide from Cheese Whey Permeate and Sucrose by Leuconostoc mesenteroides Dextransucrase. J. Agric. Food Chem. 2012, 60, 1945–1953. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.T.; Dijkhuizen, L.; van Leeuwen, S.S. Structural characterization of glucosylated lactose derivatives synthesized by the Lactobacillus reuteri GtfA and Gtf180 glucansucrase enzymes. Carbohydr. Res. 2017, 449, 59–64. [Google Scholar] [CrossRef]
- Robyt, J.F.; Eklund, S.H. Relative, quantitative effects of acceptors in the reaction of Leuconostoc mesenteroides B-512F dextransucrase. Carbohydr. Res. 1983, 121, 279–286. [Google Scholar] [CrossRef]
- Shi, Q.; Juvonen, M.; Hou, Y.; Kajala, I.; Nyyssölä, A.; Maina, N.H.; Maaheimo, H.; Virkki, L.; Tenkanen, M. Lactose- and cellobiose-derived branched trisaccharides and a sucrose-containing trisaccharide produced by acceptor reactions of Weissella confusa dextransucrase. Food Chem. 2016, 190, 226–236. [Google Scholar] [CrossRef]
- Bertrand, A.; Morel, S.; Lefoulon, F.; Rolland, Y.; Monsan, P.; Remaud-Siméon, M. Leuconostoc mesenteroides glucansucrase synthesis of flavonoid glucosides by acceptor reactions in aqueous-organic solvents. Carbohydr. Res. 2006, 341, 855–863. [Google Scholar] [CrossRef]
- Seo, E.-S.; Lee, J.-H.; Park, J.-Y.; Kim, D.; Han, H.-J.; Robyt, J.F. Enzymatic synthesis and anti-coagulant effect of salicin analogs by using the Leuconostoc mesenteroides glucansucrase acceptor reaction. J. Biotechnol. 2005, 117, 31–38. [Google Scholar] [CrossRef]
- Te Poele, E.M.; Grijpstra, P.; van Leeuwen, S.S.; Dijkhuizen, L. Glucosylation of Catechol with the GTFA Glucansucrase Enzyme from Lactobacillus reuteri and Sucrose as Donor Substrate. Bioconjug. Chem. 2016, 27, 937–946. [Google Scholar] [CrossRef]
- Gerwig, G.J.; te Poele, E.M.; Dijkhuizen, L.; Kamerling, J.P. Structural analysis of rebaudioside A derivatives obtained by Lactobacillus reuteri 180 glucansucrase-catalyzed trans-α-glucosylation. Carbohydr. Res. 2017, 440–441. [Google Scholar] [CrossRef] [PubMed]
- Te Poele, E.M.; Devlamynck, T.; Jäger, M.; Gerwig, G.J.; de Walle, D.V.; Dewettinck, K.; Hirsch, A.K.H.; Kamerling, J.P.; Soetaert, W.; Dijkhuizen, L. Glucansucrase (mutant) enzymes from Lactobacillus reuteri 180 efficiently transglucosylate Stevia component rebaudioside A, resulting in a superior taste. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Richard, G.; Morel, S.; Willemot, R.-M.; Monsan, P.; Remaud-Siméon, M. Glucosylation of α-butyl- and α-octyl-D-glucopyranosides by dextransucrase and alternansucrase from Leuconostoc mesenteroides. Carbohydr. Res. 2003, 338, 855–864. [Google Scholar] [CrossRef]
- Benkoulouche, M.; Ben Imeddourene, A.; Barel, L.-A.; Le Heiget, G.; Pizzut, S.; Kulyk, H.; Bellvert, F.; Bozonnet, S.; Mulard, L.A.; Remaud-Siméon, M.; et al. Redirecting substrate regioselectivity using engineered ΔN 123 -GBD-CD2 branching sucrases for the production of pentasaccharide repeating units of Shigella flexneri 3a, 4a and 4b haptens. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Benkoulouche, M.; Barel, L.-A.; Le Heiget, G.; Ben Imeddourene, A.; Le Guen, Y.; Monties, N.; Guerreiro, C.; Remaud-Siméon, M.; Moulis, C.; et al. Convergent Chemoenzymatic Strategy to Deliver a Diversity of Shigella flexneri Serotype-Specific O-Antigen Segments from a Unique Lightly Protected Tetrasaccharide Core. J. Org. Chem. 2021, 86, 2058–2075. [Google Scholar] [CrossRef]
- Salamone, S.; Guerreiro, C.; Cambon, E.; André, I.; Remaud-Siméon, M.; Mulard, L.A. Programmed chemo-enzymatic synthesis of the oligosaccharide component of a carbohydrate-based antibacterial vaccine candidate. Chem. Commun. 2015, 51, 2581–2584. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.J.; Jones, K.R.; Kuramitsu, H.K.; Macrina, F.L. Molecular cloning and characterization of the glucosyltransferase C gene (gtfC) from Streptococcus mutans LM7. Infect. Immun. 1987, 55, 2176–2182. [Google Scholar] [CrossRef]
- MacGregor, E.A.; Jespersen, H.M.; Svensson, B. A circularly permuted α-amylase-type α/β-barrel structure in glucan-synthesizing glucosyltransferases. FEBS Lett. 1996, 378, 263–266. [Google Scholar] [CrossRef]
- Janeček, S. How many conserved sequences regions are there in the α-amylase family? Biologia 2002, 57, 29–41. [Google Scholar]
- Janeček, S. α-amylase family: Molecular biology and evolution. Prog. Biophys. Mol. Biol. 1997, 67, 67–97. [Google Scholar] [CrossRef]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, H.M.; Ann MacGregor, E.; Henrissat, B.; Sierks, M.R.; Svensson, B. Starch- and glycogen-debranching and branching enzymes: Prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J. Protein Chem. 1993, 12, 791–805. [Google Scholar] [CrossRef]
- Swistowska, A.M.; Gronert, S.; Wittrock, S.; Collisi, W.; Hecht, H.-J.; Hofer, B. Identification of structural determinants for substrate binding and turnover by glucosyltransferase R supports the permutation hypothesis. FEBS Lett. 2007, 581, 4036–4042. [Google Scholar] [CrossRef] [PubMed]
- Tsumori, H.; Minami, T.; Kuramitsu, H.K. Identification of essential amino acids in the Streptococcus mutans glucosyltransferases. J. Bacteriol. 1997, 179, 3391–3396. [Google Scholar] [CrossRef]
- Monchois, V.; Vignon, M.; Escalier, P.-C.; Svensson, B.; Russell, R.R.B. Involvement of Gln937 of Streptococcus downei GTF-I glucansucrase in transition-state stabilization. Eur. J. Biochem. 2000, 267, 4127–4136. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, H.-B.; Li, M.-Q.; Hu, X.-Q.; Li, Y. Functional analysis of truncated and site-directed mutagenesis dextransucrases to produce different type dextrans. Enzyme Microb. Technol. 2017, 102, 26–34. [Google Scholar] [CrossRef]
- Wittrock, S.; Swistowska, A.M.; Collisi, W.; Hofmann, B.; Hecht, H.-J.; Hofer, B. Re- or displacement of invariant residues in the C-terminal half of the catalytic domain strongly affects catalysis by glucosyltransferase R. FEBS Lett. 2008, 582, 491–496. [Google Scholar] [CrossRef][Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef]
- Meng, X.; Pijning, T.; Dobruchowska, J.M.; Gerwig, G.J.; Dijkhuizen, L. Characterization of the functional roles of amino acid residues in acceptor binding subsite +1 in the active site of the glucansucrase GTF180 enzyme of Lactobacillus reuteri 180. J. Biol. Chem. 2015, 290, 30131–30141. [Google Scholar] [CrossRef]
- Brison, Y.; Pijning, T.; Malbert, Y.; Fabre, É.; Mourey, L.; Morel, S.; Potocki-Véronèse, G.; Monsan, P.; Tranier, S.; Remaud-Siméon, M.; et al. Functional and structural characterization of α-(1->2) branching sucrase derived from DSR-E glucansucrase. J. Biol. Chem. 2012, 287, 7915–7924. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Guionnet, M.; Schörghuber, J.; Lichtenecker, R.; Moulis, C.; Remaud-Siméon, M.; Lippens, G. Futile Encounter Engineering of the DSR-M Dextransucrase Modifies the Resulting Polymer Length. Biochemistry 2019, 58, 2853–2859. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Pijning, T.; Tietema, M.; Dobruchowska, J.M.; Yin, H.; Gerwig, G.J.; Kralj, S.; Dijkhuizen, L. Characterization of the glucansucrase GTF180 W1065 mutant enzymes producing polysaccharides and oligosaccharides with altered linkage composition. Food Chem. 2017, 217, 81–90. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Sitthiyotha, T.; Chunsrivirot, S.; Charoenwongpaiboon, T.; Pichyangkura, R. Unravelling Regioselectivity of Leuconostoc citreum ABK-1 Alternansucrase by Acceptor Site Engineering. Int. J. Mol. Sci. 2021, 22, 3229. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Ito, S.; Shimamura, T.; Weyand, S.; Kawarasaki, Y.; Misaka, T.; Abe, K.; Kobayashi, T.; Cameron, A.D.; Iwata, S. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J. Mol. Biol. 2011, 408, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Vujičić-Žagar, A.; Pijning, T.; Kralj, S.; López, C.A.; Eeuwema, W.; Dijkhuizen, L.; Dijkstra, B.W. Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 21406–21411. [Google Scholar] [CrossRef]
- Pijning, T.; Vujičić-Žagar, A.; Kralj, S.; Dijkhuizen, L.; Dijkstra, B.W. Flexibility of truncated and full-length glucansucrase GTF180 enzymes from Lactobacillus reuteri 180. FEBS J. 2014, 281, 2159–2171. [Google Scholar] [CrossRef]
- Pijning, T.; Vujičić-Žagar, A.; Kralj, S.; Dijkhuizen, L.; Dijkstra, B.W. Structure of the α-1,6/α-1,4-specific glucansucrase GTFA from Lactobacillus reuteri 121. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. 2012, 68, 1448–1454. [Google Scholar] [CrossRef] [PubMed]
- Osorio, M.I.; Zúñiga, M.A.; Mendoza, F.; Jaña, G.A.; Jiménez, V.A. Modulation of glucan-enzyme interactions by domain V in GTF-SI from Streptococcus mutans. Proteins Struct. Funct. Bioinforma. 2019, 87, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Ben Imeddourene, A.; Esque, J.; André, I. Combining multi-scale modelling methods to decipher molecular motions of a branching sucrase from glycoside-hydrolase family 70. PLoS ONE 2018, 13, e0201323. [Google Scholar] [CrossRef]
- Meng, X.; Dobruchowska, J.M.; Pijning, T.; López, C.A.; Kamerling, J.P.; Dijkhuizen, L. Residue Leu940 has a crucial role in the linkage and reaction specificity of the glucansucrase GTF180 of the probiotic bacterium Lactobacillus reuteri 180. J. Biol. Chem. 2014, 289, 32773–32782. [Google Scholar] [CrossRef]
- Côté, G.L.; Dunlap, C.A.; Vermillion, K.E.; Skory, C.D. Production of isomelezitose from sucrose by engineered glucansucrases. Amylase 2017, 1, 82–93. [Google Scholar] [CrossRef]
- Molina, M.; Moulis, C.; Monties, N.; Guieysse, D.; Morel, S.; Cioci, G.; Remaud-Siméon, M. A specific oligosaccharide-binding site in the alternansucrase catalytic domain mediates alternan elongation. J. Biol. Chem. 2020, 295, 9474–9489. [Google Scholar] [CrossRef]
- Giffard, P.M.; Jacques, N.A. Definition of a fundamental repeating unit in streptococcal glucosyltransferase glucan-binding regions and related sequences. J. Dent. Res. 1994, 73, 1133–1141. [Google Scholar] [CrossRef]
- Monchois, V.; Arguello-Morales, M.; Russell, R.R.B. Isolation of an Active Catalytic Core of Streptococcus downei MFe28 GTF-I Glucosyltransferase. J. Bacteriol. 1999, 181, 2290–2292. [Google Scholar] [CrossRef]
- Claverie, M.; Cioci, G.; Vuillemin, M.; Bondy, P.; Remaud-Simeon, M.; Moulis, C. Processivity of dextransucrases synthesizing very high molar mass dextran is mediated by sugar-binding pockets in domain V. J. Biol. Chem. 2020, 295, 5602–5613. [Google Scholar] [CrossRef] [PubMed]
- Giffard, P.M.; Allen, D.M.; Milward, C.P.; Simpson, C.L.; Jacques, N.A. Sequence of the gtfK gene of Streptococcus salivarius ATCC 25975 and evolution of the gtf genes of oral Streptococci. J. Gen. Microbiol. 1993, 139, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Monchois, V.; Vignon, M.; Russell, R.R.B. Isolation of key amino acid residues at the N-terminal end of the core region Streptococcus downei glucansucrase, GTF-I. Appl. Microbiol. Biotechnol. 1999, 52, 660–665. [Google Scholar] [CrossRef]
- Komatsu, H.; Katayama, M.; Sawada, M.; Hirata, Y.; Mori, M.; Inoue, T.; Fukui, K.; Fukada, H.; Kodama, T. Thermodynamics of the Binding of the C-Terminal Repeat Domain of Streptococcus sobrinus Glucosyltransferase-I to Dextran. Biochemistry 2007, 46, 8436–8444. [Google Scholar] [CrossRef] [PubMed]
- Lis, M.; Shiroza, T.; Kuramitsu, H.K. Role of C-terminal direct repeating units of the Streptococcus mutans glucosyltransferase-S in glucan binding. Appl. Environ. Microbiol. 1995, 61, 2040–2042. [Google Scholar] [CrossRef] [PubMed]
- Funane, K.; Ishii, T.; Terasawa, K.; Yamamoto, T.; Kobayashi, M. Construction of Chimeric Glucansucrases for Analyzing Substrate-binding Regions That Affect the Structure of Glucan Products. Biosci. Biotechnol. Biochem. 2004, 68, 1912–1920. [Google Scholar] [CrossRef]
- Suwannarangsee, S.; Moulis, C.; Potocki-Veronese, G.; Monsan, P.; Remaud-Simeon, M.; Chulalaksananukul, W. Search for a dextransucrase minimal motif involved in dextran binding. FEBS Lett. 2007, 581, 4675–4680. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.S.H.; Joucla, G.; Remaud-Simeon, M.; Russell, R.R.B. Conserved Repeat Motifs and Glucan Binding by Glucansucrases of Oral Streptococci and Leuconostoc mesenteroides. J. Bacteriol. 2004, 186, 8301–8308. [Google Scholar] [CrossRef]
- Wong, C.; Hefta, S.A.; Paxton, R.J.; Shively, J.E.; Mooser, G. Size and subdomain architecture of the glucan-binding domain of sucrose:3-alpha-D-glucosyltransferase from Streptococcus sobrinus. Infect. Immun. 1990, 58, 2165–2170. [Google Scholar] [CrossRef]
- Singh, J.S.; Taylor, K.G.; Doyle, R.J. Essential amino acids involved in glucan-dependent aggregation of Streptococcus sobrinus. Carbohydr. Res. 1993, 244, 137–147. [Google Scholar] [CrossRef]
- Wren, B.W.; Russell, R.R.; Tabaqchali, S. Antigenic cross-reactivity and functional inhibition by antibodies to Clostridium difficile toxin, A.; Streptococcus mutans glucan-binding protein, and a synthetic peptide. Infect. Immun. 1991, 59, 3151–3155. [Google Scholar] [CrossRef]
- Monchois, V.; Willemot, R.M.; Monsan, P. Glucansucrases: Mechanism of action and structure-function relationships. FEMS Microbiol. Rev. 1999, 23, 131–151. [Google Scholar] [CrossRef]
- Wu, S.; Snajdrova, R.; Moore, J.C.; Baldenius, K.; Bornscheuer, U.T. Biocatalysis: Enzymatic Synthesis for Industrial Applications. Angew. Chem. Int. Ed. 2021, 60, 88–119. [Google Scholar] [CrossRef]
- Joucla, G. Caractérisation de l’alternane-Saccharase de Leuconostoc Mesenteroides NRRL B-1355: Approche Rationnelle et al.Éatoire Pour La Conception de Nouvelles Glucane-Saccharases. Ph.D. Thesis, INSA/Toulouse University, Toulouse, France, 2003. [Google Scholar]
- Li, M.; Zhang, H.; Li, Y.; Hu, X.; Yang, J. The thermoduric effects of site-directed mutagenesis of proline and lysine on dextransucrase from Leuconostoc mesenteroides 0326. Int. J. Biol. Macromol. 2018, 107, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.J.; Macrina, F.L. Molecular organization and expression of the gtfA gene of Streptococcus mutans LM7. Infect. Immun. 1986, 54, 77–84. [Google Scholar] [CrossRef]
- Pucci, M.J.; Macrina, F.L. Cloned gtfA gene of Streptococcus mutans LM7 alters glucan synthesis in Streptococcus sanguis. Infect. Immun. 1985, 48, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Robeson, J.P.; Barletta, R.G.; Curtiss, R. Expression of a Streptococcus mutans glucosyltransferase gene in Escherichia coli. J. Bacteriol. 1983, 153, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.R.B.; Gilpin, M.L.; Mukasa, H.; Dougan, G.Y. Characterization of Glucosyltransferase Expressed from a Streptococcus sobrinus Gene Cloned in Escherichia Coli. Microbiology 1987, 133, 935–944. [Google Scholar] [CrossRef]
- Abo, H.; Matsumura, T.; Kodama, T.; Ohta, H.; Fukui, K.; Kato, K.; Kagawa, H. Peptide sequences for sucrose splitting and glucan binding within Streptococcus sobrinus glucosyltransferase (water-insoluble glucan synthetase). J. Bacteriol. 1991, 173, 989–996. [Google Scholar] [CrossRef]
- Moulis, C. Ingénierie rationnelle de la dextrane-saccharase DSR-S: Compréhension du mécanisme de polymérisation pour la synthèse de dextranes de taille contrôlée. Ph.D. Thesis, INSA/Toulouse University, Toulouse, France, 2006. [Google Scholar]
- Monchois, V.; Reverte, A.; Remaud-Simeon, M.; Monsan, P.; Willemot, R.-M. Effect of Leuconostoc mesenteroides NRRL B-512F Dextransucrase Carboxy-Terminal Deletions on Dextran and Oligosaccharide Synthesis. Appl. Environ. Microbiol. 1998, 64, 1644–1649. [Google Scholar] [CrossRef]
- Kralj, S.; van Leeuwen, S.S.; Valk, V.; Eeuwema, W.; Kamerling, J.P.; Dijkhuizen, L. Hybrid reuteransucrase enzymes reveal regions important for glucosidic linkage specificity and the transglucosylation/hydrolysis ratio. FEBS J. 2008, 275, 6002–6010. [Google Scholar] [CrossRef][Green Version]
- Hellmuth, H.; Wittrock, S.; Kralj, S.; Dijkhuizen, L.; Hofer, B.; Seibel, J. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: Toward tailor-made polymer and oligosaccharide products. Biochemistry 2008, 47, 6678–6684. [Google Scholar] [CrossRef]
- Kang, H.K.; Kimura, A.; Kim, D. Bioengineering of Leuconostoc mesenteroides Glucansucrases That Gives Selected Bond Formation for Glucan Synthesis and/or Acceptor-Product Synthesis. J. Agric. Food Chem. 2011, 59, 4148–4155. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Eeuwema, W.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural characterization of bioengineered α-D-glucans produced by mutant glucansucrase GTF180 enzymes of Lactobacillus reuteri strain 180. Biomacromolecules 2009, 10, 580–588. [Google Scholar] [CrossRef]
- van Leeuwen, S.S.; Kralj, S.; Gerwig, G.J.; Dijkhuizen, L.; Kamerling, J.P. Structural Analysis of Bioengineered α-d-Glucan Produced by a Triple Mutant of the Glucansucrase GTF180 Enzyme from Lactobacillus reuteri Strain 180: Generation of (α1→4) Linkages in a Native (1→3)(1→6)-α-d-Glucan. Biomacromolecules 2008, 9, 2251–2258. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.Y.; Gänzle, M.G. Site Directed Mutagenesis of Dextransucrase DsrM from Weissella cibaria: Transformation to a Reuteransucrase. J. Agric. Food Chem. 2016, 64, 6848–6855. [Google Scholar] [CrossRef]
- Kralj, S.; van Geel-Schutten, I.G.H.; Faber, E.J.; van der Maarel, M.J.E.C.; Dijkhuizen, L. Rational Transformation of Lactobacillus reuteri 121 Reuteransucrase into a Dextransucrase. Biochemistry 2005, 44, 9206–9216. [Google Scholar] [CrossRef]
- Côté, G.L.; Skory, C.D. Effects of mutations at threonine-654 on the insoluble glucan synthesized by Leuconostoc mesenteroides NRRL B-1118 glucansucrase. Appl. Microbiol. Biotechnol. 2014, 98, 6651–6658. [Google Scholar] [CrossRef]
- Münkel, F.; Fischer, A.; Wefers, D. Structural characterization of mixed-linkage α-glucans produced by mutants of Lactobacillus reuteri TMW 1.106 dextransucrase. Carbohydr. Polym. 2020, 231, 115697. [Google Scholar] [CrossRef] [PubMed]
- Irague, R.; Tarquis, L.; André, I.; Moulis, C.; Morel, S.; Monsan, P.; Potocki-Véronèse, G.; Remaud-Siméon, M. Combinatorial Engineering of Dextransucrase Specificity. PLoS ONE 2013, 8, e77837. [Google Scholar] [CrossRef]
- Irague, R.; Massou, S.; Moulis, C.; Saurel, O.; Milon, A.; Monsan, P.; Remaud-Siméon, M.; Portais, J.-C.; Potocki-Véronèse, G. NMR-Based Structural Glycomics for High-Throughput Screening of Carbohydrate-Active Enzyme Specificity. Anal. Chem. 2011, 83, 1202–1206. [Google Scholar] [CrossRef]
- Meng, X.; Dobruchowska, J.M.; Pijning, T.; Gerwig, G.J.; Dijkhuizen, L. Synthesis of New Hyperbranched α-Glucans from Sucrose by Lactobacillus reuteri 180 Glucansucrase Mutants. J. Agric. Food Chem. 2016, 64, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, S.; Zhang, H.; Li, Y.; Hu, X. Characterization of the inserted mutagenesis dextransucrases from Leuconostoc mesenteroides 0326 to produce hyperbranched dextran. Int. J. Biol. Macromol. 2018, 112, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Bender, B.J.; Cisneros, A.; Duran, A.M.; Finn, J.A.; Fu, D.; Lokits, A.D.; Mueller, B.K.; Sangha, A.K.; Sauer, M.F.; Sevy, A.M.; et al. Protocols for Molecular Modeling with Rosetta3 and RosettaScripts. Biochemistry 2016, 55, 4748–4763. [Google Scholar] [CrossRef]
- Marques, S.M.; Planas-Iglesias, J.; Damborsky, J. Web-based tools for computational enzyme design. Curr. Opin. Struct. Biol. 2021, 69, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Planas-Iglesias, J.; Marques, S.M.; Pinto, G.P.; Musil, M.; Stourac, J.; Damborsky, J.; Bednar, D. Computational design of enzymes for biotechnological applications. Biotechnol. Adv. 2021, 47, 107696. [Google Scholar] [CrossRef] [PubMed]
- Devlamynck, T.; Te Poele, E.M.; Meng, X.; van Leeuwen, S.S.; Dijkhuizen, L. Glucansucrase Gtf180-ΔN of Lactobacillus reuteri 180: Enzyme and reaction engineering for improved glycosylation of non-carbohydrate molecules. Appl. Microbiol. Biotechnol. 2016, 100, 7529–7539. [Google Scholar] [CrossRef] [PubMed]
- Devlamynck, T.; Te Poele, E.M.; Quataert, K.; Gerwig, G.J.; van de Walle, D.; Dewettinck, K.; Kamerling, J.P.; Soetaert, W.; Dijkhuizen, L. Trans-α-glucosylation of stevioside by the mutant glucansucrase enzyme Gtf180-ΔN-Q1140E improves its taste profile. Food Chem. 2019, 272, 653–662. [Google Scholar] [CrossRef]
- Klingel, T.; Hadamjetz, M.; Fischer, A.; Wefers, D. Glucosylation of flavonoids and flavonoid glycosides by mutant dextransucrase from Lactobacillus reuteri TMW 1.106. Carbohydr. Res. 2019, 483, 107741. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.-H.; Lee, J.-H.; Jhon, D.-Y.; Jun, W.-J.; Kang, S.-S.; Sim, J.; Choi, H.; Moon, J.-H.; Kim, D. Synthesis and characterization of novel quercetin-α-d-glucopyranosides using glucansucrase from Leuconostoc mesenteroides. Enzyme Microb. Technol. 2007, 40, 1124–1129. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, Y.; Jia, Y.; Wang, W.; Li, Y.; Lu, S.; Jin, J.-M.; Tang, S.-Y. Engineering a Carbohydrate-processing Transglycosidase into Glycosyltransferase for Natural Product Glycodiversification. Sci. Rep. 2016, 6, 21051. [Google Scholar] [CrossRef]
- Mazurenko, S.; Prokop, Z.; Damborsky, J. Machine Learning in Enzyme Engineering. ACS Catal. 2020, 10, 1210–1223. [Google Scholar] [CrossRef]
- Hofmann, J.; Pagel, K. Glycan Analysis by Ion Mobility–Mass Spectrometry. Angew. Chem. Int. Ed. 2017, 56, 8342–8349. [Google Scholar] [CrossRef]
- Ollivier, S.; Tarquis, L.; Fanuel, M.; Li, A.; Durand, J.; Laville, E.; Potocki-Veronese, G.; Ropartz, D.; Rogniaux, H. Anomeric Retention of Carbohydrates in Multistage Cyclic Ion Mobility (IMSn): De Novo Structural Elucidation of Enzymatically Produced Mannosides. Anal. Chem. 2021, 93, 6254–6261. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Delbianco, M.; Anggara, K.; Michnowicz, T.; Pardo-Vargas, A.; Bharate, P.; Sen, S.; Pristl, M.; Rauschenbach, S.; Schlickum, U.; et al. Imaging single glycans. Nature 2020, 582, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, M.; Malbert, Y.; Laguerre, S.; Remaud-Siméon, M.; Moulis, C. Optimizing the production of an α-(1→2) branching sucrase in Escherichia coli using statistical design. Appl. Microbiol. Biotechnol. 2014, 98, 5173–5184. [Google Scholar] [CrossRef]
- Malten, M.; Hollmann, R.; Deckwer, W.-D.; Jahn, D. Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol. Bioeng. 2005, 89, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Skory, C.D.; Côté, G.L. Secreted expression of Leuconostoc mesenteroides glucansucrase in Lactococcus lactis for the production of insoluble glucans. Appl. Microbiol. Biotechnol. 2015, 99, 10001–10010. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina, M.; Cioci, G.; Moulis, C.; Séverac, E.; Remaud-Siméon, M. Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure–Function Relationship Studies and Engineering. Microorganisms 2021, 9, 1607. https://doi.org/10.3390/microorganisms9081607
Molina M, Cioci G, Moulis C, Séverac E, Remaud-Siméon M. Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure–Function Relationship Studies and Engineering. Microorganisms. 2021; 9(8):1607. https://doi.org/10.3390/microorganisms9081607
Chicago/Turabian StyleMolina, Manon, Gianluca Cioci, Claire Moulis, Etienne Séverac, and Magali Remaud-Siméon. 2021. "Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure–Function Relationship Studies and Engineering" Microorganisms 9, no. 8: 1607. https://doi.org/10.3390/microorganisms9081607
APA StyleMolina, M., Cioci, G., Moulis, C., Séverac, E., & Remaud-Siméon, M. (2021). Bacterial α-Glucan and Branching Sucrases from GH70 Family: Discovery, Structure–Function Relationship Studies and Engineering. Microorganisms, 9(8), 1607. https://doi.org/10.3390/microorganisms9081607





