Profiles of Human Papillomavirus Detection of the Multinucleated Cells in Cervical Smears
Abstract
:1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. Ethical Approval
2.3. Human Papillomavirus Genotyping on the Whole LBC Samples
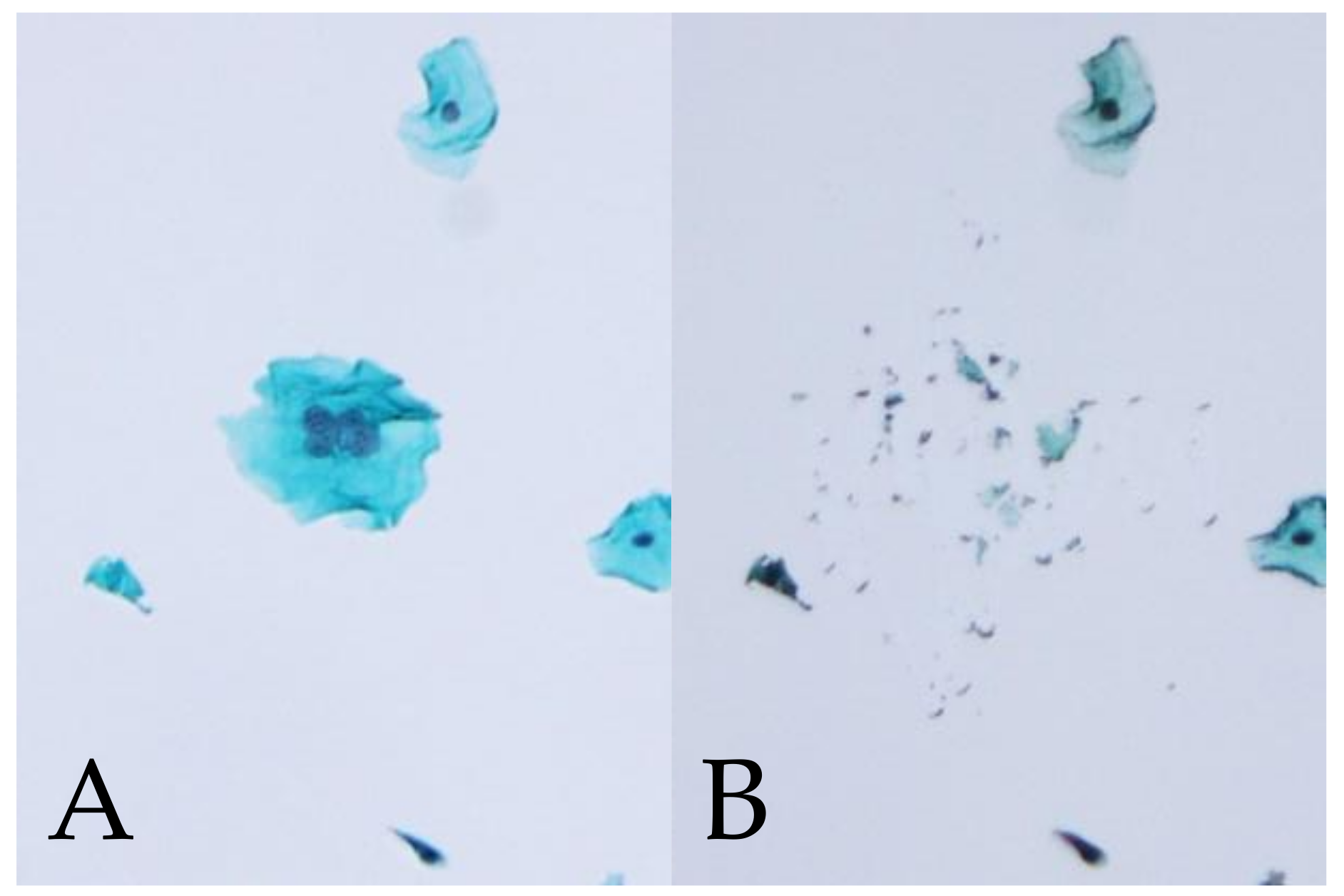
2.4. HPV Genotyping of Manually Microdissected Multinucleated Cells
2.5. Statistical Analysis
3. Results
3.1. Relationships among the Multinucleated Cells, Cytologic Classification, and HPV Genotypes
3.2. HPV Genotype Detection by MD-PCR in Multinucleated Cells for Multiple Infection
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Munoz, N.; Castellsagué, X.; de González, A.B.; Gissmann, L. HPV in the etiology of human cancer. Vaccine 2006, 24, S1–S10. [Google Scholar] [CrossRef]
- Molijn, A.; Kleter, B.; Quint, W.; van Doorn, L.-J. Molecular diagnosis of human papillomavirus (HPV) infections. J. Clin. Virol. 2005, 32, 43–51. [Google Scholar] [CrossRef]
- Castellsague, X.; Diaz, M.; de Sanjose, S.; Munoz, N.; Herrero, R.; Franceschi, S.; Peeling, R.W.; Ashley, R.; Smith, J.S.; Snijders, P.J.; et al. Worldwide human papillomavirus etiology of cervical adenocarcinoma and its cofactors: Implications for screening and prevention. J. Natl. Cancer Inst. 2006, 98, 303–315. [Google Scholar] [CrossRef] [PubMed]
- De Sanjose, S.; Quint, W.G.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- An, H.J.; Cho, N.H.; Lee, S.Y.; Kim, I.H.; Lee, C.; Kim, S.J.; Mun, M.S.; Kim, S.H.; Jeong, J.K. Correlation of cervical carcinoma and precancerous lesions with human papillomavirus (HPV) genotypes detected with the HPV DNA chip microarray method. Cancer 2003, 97, 1672–1680. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-García, A.; Ponce-de-León-Rosales, S.; Cantú-de-León, D.; Fragoso-Ontiveros, V.; Martínez-Ramírez, I.; Orozco-Colín, A.; Mohar, A.; Lizano, M. Impact of human papillomavirus coinfections on the risk of high-grade squamous intraepithelial lesion and cervical cancer. Gynecol. Oncol. 2014, 134, 534–539. [Google Scholar] [CrossRef]
- Bollmann, M.; Bánkfalvi, A.; Trosic, A.; Speich, N.; Schmittt, C.; Bollmann, R. Can we detect cervical human papillomavirus (HPV) infection by cytomorphology alone? Diagnostic value of non-classic cytological signs of HPV effect in minimally abnormal Pap tests. Cytopathology 2005, 16, 13–21. [Google Scholar] [CrossRef]
- Okodo, M.; Okayama, K.; Teruya, K.; Kimura, H.; Noji, N.; Ishii, Y.; Fujii, M.; Oda, M.; Sasagawa, T. Koilocytic changes are not elicited by human papillomavirus genotypes with higher oncogenic potential. J. Med. Virol. 2020, 92, 3766–3773. [Google Scholar] [CrossRef]
- Roteli-Martins, C.M.; Alves, V.A.; Santos, R.T.; Martinez, E.Z.; Syrjänen, K.J.; Derchain, S.F. Value of morphological criteria in diagnosing cervical HPV lesions confirmed by in situ hybridization and hybrid capture assay. Pathol. Res. Pract. 2001, 197, 677–682. [Google Scholar] [CrossRef]
- Duensing, S.; Lee, L.Y.; Duensing, A.; Basile, J.; Piboonniyom, S.; Gonzalez, S.; Crum, C.P.; Munger, K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 2000, 97, 10002–10007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duensing, S.; Munger, K. Mechanisms of genomic instability in human cancer: Insights from studies with human papillomavirus oncoproteins. Int. J. Cancer 2004, 109, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Duensing, S.; Münger, K. Human papillomaviruses and centrosome duplication errors: Modeling the origins of genomic instability. Oncogene 2002, 21, 6241–6248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbert, A.; Bergeron, C.; Wiener, H.; Schenck, U.; Klinkhamer, P.; Bulten, J.; Arbyn, M. European guidelines for quality assurance in cervical cancer screening: Recommendations for cervical cytology terminology. Cytopathology 2007, 18, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Arbyn, M.; Paraskevaidis, E.; Martin-Hirsch, P.; Prendiville, W.; Dillner, J. Clinical utility of HPV-DNA detection: Triage of minor cervical lesions, follow-up of women treated for high-grade CIN: An update of pooled evidence. Gynecol. Oncol. 2005, 99, S7–S11. [Google Scholar] [CrossRef]
- Nayar, R.; Wilbur, D.C. The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Massad, L.S.; Einstein, M.H.; Huh, W.K.; Katki, H.A.; Kinney, W.K.; Schiffman, M.; Solomon, D.; Wentzensen, N.; Lawson, H.W. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. J. Low. Genit. Tract Dis. 2013, 17, S1–S27. [Google Scholar] [CrossRef] [Green Version]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Okodo, M.; Okayama, K.; Teruya, K.; Sasagawa, T. Uniplex E6/E7 PCR method detecting E6 or E7 genes in 39 human papillomavirus types. J. Med. Virol. 2018, 90, 981–988. [Google Scholar] [CrossRef]
- Cancer, I.A.F.R.O. A Review of Human Carcinogens: Personal Habits and Indoor Combustions; World Health Organization: Geneva, Switzerland, 2012.
- Noji, N.; Okayama, K.; Oda, M.; Shimada, A.; Okodo, M. Human papillomavirus infection status of single cells isolated from cervical cytology specimens by simple manual microdissection. J. Med. Virol. 2021, 93, 5084–5094. [Google Scholar] [CrossRef]
- Bernard, E.; Pons-Salort, M.; Favre, M.; Heard, I.; Delarocque-Astagneau, E.; Guillemot, D.; Thiébaut, A.C. Comparing human papillomavirus prevalences in women with normal cytology or invasive cervical cancer to rank genotypes according to their oncogenic potential: A meta-analysis of observational studies. BMC Infect. Dis. 2013, 13, 373. [Google Scholar] [CrossRef]
- Brown, H.M.; Knowlton, A.E.; Snavely, E.; Nguyen, B.D.; Richards, T.S.; Grieshaber, S.S. Multinucleation during C. trachomatis infections is caused by the contribution of two effector pathways. PLoS ONE 2014, 9, e100763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, H.M.; Knowlton, A.E.; Grieshaber, S.S. Chlamydial infection induces host cytokinesis failure at abscission. Cell. Microbiol. 2012, 14, 1554–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, C.W.; Esfahani, F.M. Pregnancy-related changes: A retrospective review of 278 cervical smears. Diagn. Cytopathol. 1997, 17, 99–107. [Google Scholar] [CrossRef]
- Lee, C.; Laimins, L.A. The differentiation-dependent life cycle of human papillomaviruses in keratinocytes. In The Papillomaviruses; Springer: Berlin/Heidelberg, Germany, 2007; pp. 45–67. [Google Scholar]
| MNC | ASC-US | LSIL | ASC-H | HSIL |
|---|---|---|---|---|
| n = 137 | n = 202 | n = 54 | n = 258 | |
| Present | 19.7% | 50.5% | 14.8% | 38.0% |
| (27/137) | (102/202) | (8/54) | (98/258) | |
| Absent | 80.3% | 49.5% | 85.2% | 62.0% |
| (110/137) | (100/202) | (46/54) | (160/258) |
| MNC | High-Risk HPV Genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 18 | 31 | 33 | 35 | 39 | 45 | 51 | 52 | 56 | 58 | 59 | |
| n = 143 | n = 35 | n = 53 | n = 23 | n = 5 | n = 71 | n = 6 | n = 72 | n = 151 | n = 81 | n = 143 | n = 9 | |
| Present | 49.0% | 34.3% | 43.4% | 21.7% | 60.0% | 45.1% | 50.0% | 38.9% | 29.1% | 55.6% | 28.0% | 44.4% |
| (70/143) | (12/35) | (23/53) | (5/23) | (3/5) | (32/71) | (3/6) | (28/72) | (44/151) | (45/81) | (40/143) | (4/9) | |
| Absent | 51.0% | 65.7% | 56.6% | 78.3% | 40.0% | 54.9% | 50.0% | 61.1% | 70.9% | 44.4% | 72.0% | 55.6% |
| (73/143) | (23/35) | (30/53) | (18/23) | (2/5) | (39/71) | (3/6) | (44/72) | (107/151) | (36/81) | (103/143) | (5/9) | |
| MNC | Other HPV Genotypes | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 11 | 26 | 30 | 34 | 40 | 42 | 44 | 53 | 54 | 55 | 61 | 62 | ||
| n = 22 | n = 4 | n = 0 | n = 14 | n = 21 | n = 14 | n = 58 | n = 10 | n = 59 | n = 21 | n = 25 | n = 30 | n = 44 | ||
| Present | 50.0% | 50.0% | – | 28.6% | 66.7% | 21.4% | 24.1% | 20.0% | 45.8% | 19.0% | 28.0% | 26.7% | 34.1% | |
| (11/22) | (2/4) | (4/14) | (14/21) | (3/14) | (14/58) | (2/10) | (27/59) | (4/21) | (7/25) | (8/30) | (15/44) | |||
| Absent | 50.0% | 50.0% | – | 71.4% | 33.3% | 78.6% | 75.9% | 80.0% | 54.2% | 81.0% | 72.0% | 73.3% | 65.9% | |
| (11/22) | (2/4) | (10/14) | (7/21) | (11/14) | (44/58) | (8/10) | (32/59) | (17/21) | (18/25) | (22/30) | (29/44) | |||
| MNC | Other HPV Genotypes | |||||||||||||
| 66 | 67 | 68 | 69 | 70 | 71 | 73 | 74 | 81 | 82 | 84 | 85 | 89 | 90 | |
| n = 22 | n = 15 | n = 33 | n = 2 | n = 10 | n = 24 | n = 9 | n = 51 | n = 24 | n = 39 | n = 17 | n = 0 | n = 17 | n = 37 | |
| Present | 40.9% | 66.7% | 18.2% | 0.0% | 50.0% | 50.0% | 55.6% | 39.2% | 29.2% | 30.8% | 23.5% | – | 47.1% | 43.2% |
| (9/22) | (10/15) | (6/33) | (0/2) | (5/10) | (12/24) | (5/9) | (20/51) | (7/24) | (12/39) | (4/17) | (8/17) | (16/37) | ||
| Absent | 59.1% | 33.3% | 81.8% | 100.0% | 50.0% | 50.0% | 44.4% | 60.8% | 70.8% | 69.2% | 76.5% | – | 52.9% | 56.8% |
| (13/22) | (5/15) | (27/33) | (2/2) | (5/10) | (12/24) | (4/9) | (31/51) | (17/24) | (27/39) | (13/17) | (9/17) | (21/37) | ||
| Case No. | Cytologic Classification | HPV Genotype | |
|---|---|---|---|
| WL-PCR | MD-PCR (MD-PCR Positive/MD Samples) | ||
| 1 | HSIL | 16, 82 | – (0/1) |
| 2 | HSIL | 16, 58, 30 | – (0/3) |
| 3 | LSIL | 16, 34 | 16 (6/8), 34 (1/8) |
| 4 | LSIL | 34, 6, 42, 61, 62, 67, 74 | 34 (3/3) |
| 5 | LSIL | 56, 6, 90 | 56 (3/6) |
| 6 | ASC-US | 56, 39, 73 | 56 (3/6) |
| 7 | HSIL | 56, 51, 30, 34, 53, 67 | 56 (2/6) |
| 8 | LSIL | 56, 6 | 56 (3/8) |
| 9 | LSIL | 56, 55, 74 | 56 (4/9) |
| 10 | LSIL | 56, 90 | – (0/3) |
| 11 | LSIL | 56, 55, 74 | – (0/1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okayama, K.; Sasagawa, T.; Teruya, K.; Oda, M.; Fujii, M.; Kimura, H.; Okodo, M. Profiles of Human Papillomavirus Detection of the Multinucleated Cells in Cervical Smears. Microorganisms 2021, 9, 1575. https://doi.org/10.3390/microorganisms9081575
Okayama K, Sasagawa T, Teruya K, Oda M, Fujii M, Kimura H, Okodo M. Profiles of Human Papillomavirus Detection of the Multinucleated Cells in Cervical Smears. Microorganisms. 2021; 9(8):1575. https://doi.org/10.3390/microorganisms9081575
Chicago/Turabian StyleOkayama, Kaori, Toshiyuki Sasagawa, Koji Teruya, Mizue Oda, Masahiko Fujii, Hirokazu Kimura, and Mitsuaki Okodo. 2021. "Profiles of Human Papillomavirus Detection of the Multinucleated Cells in Cervical Smears" Microorganisms 9, no. 8: 1575. https://doi.org/10.3390/microorganisms9081575
APA StyleOkayama, K., Sasagawa, T., Teruya, K., Oda, M., Fujii, M., Kimura, H., & Okodo, M. (2021). Profiles of Human Papillomavirus Detection of the Multinucleated Cells in Cervical Smears. Microorganisms, 9(8), 1575. https://doi.org/10.3390/microorganisms9081575







