Genome-Wide Mapping Reveals Complex Regulatory Activities of BfmR in Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Construction of Vectors
2.3. Construction of P. aeruginosa ΔczcRS and ΔphoBΔbfmS Mutants
2.4. Expression and Purifications of Recombinant Proteins CzcR-His6 and His6-BfmR
2.5. Chromatin Immunoprecipitation (ChIP)
2.6. ChIP-seq Library Construction, Sequencing, and Data Analysis
2.7. Motif Detection
2.8. RNA-seq and Data Analysis
2.9. ChIP-Quantitative Polymerase Chain Reaction (ChIP-qPCR)
2.10. Electrophoretic Mobility Shift Assay (EMSA)
2.11. Dye Primer-Based DNase I Footprinting Assay
2.12. Monitoring Gene Expression by lux-Based Reporters
2.13. Western Blot Analysis
2.14. Measurement of Inorganic Phosphate Level and Bacteria Growth in the Medium
2.15. Measurement of Intracellular Polyphosphate (polyP)
2.16. Biofilm Formation Assays
3. Results
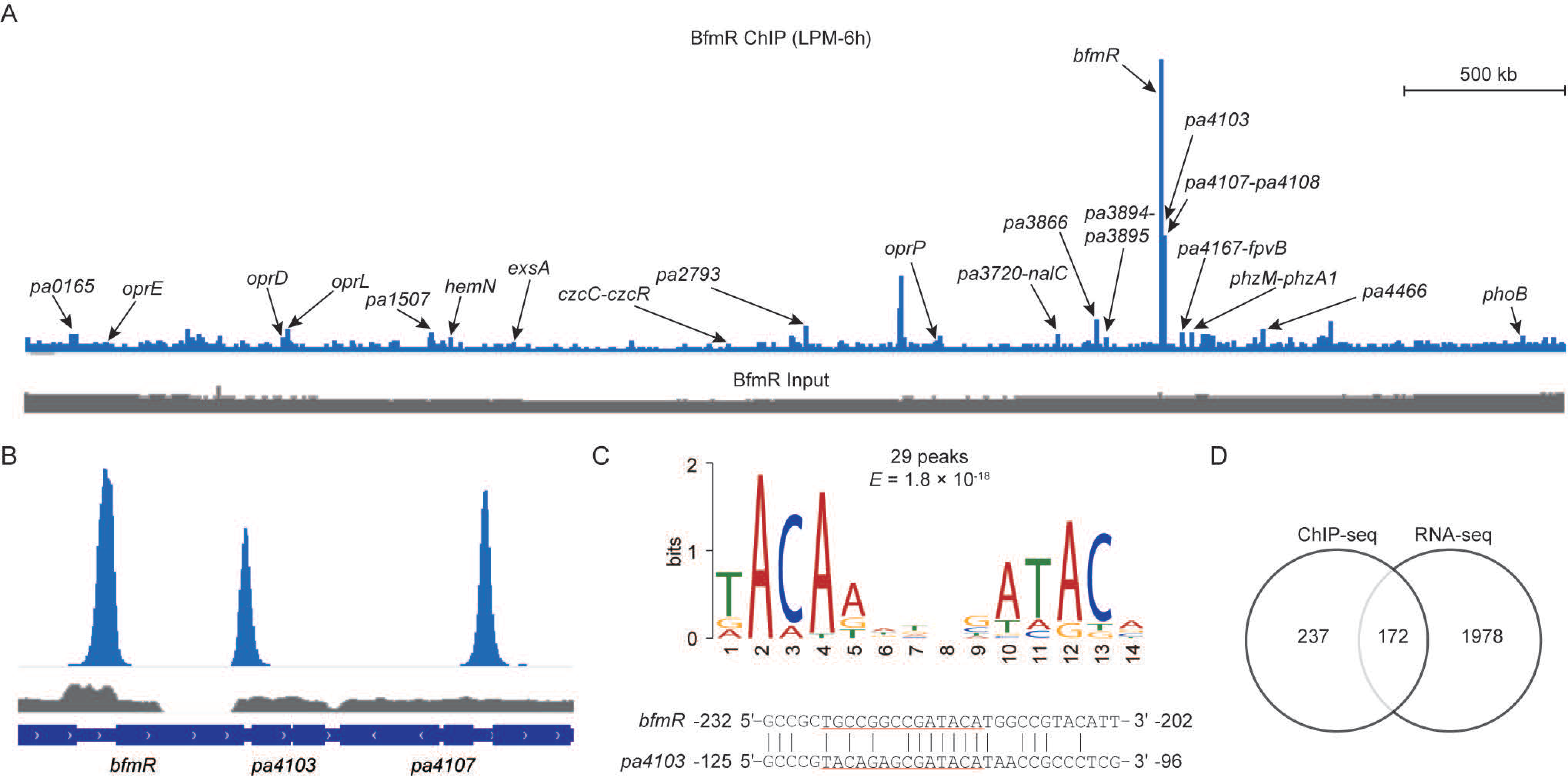
3.1. Genome-Wide Binding Patterns of BfmR in P. aeruginosa MPAO1
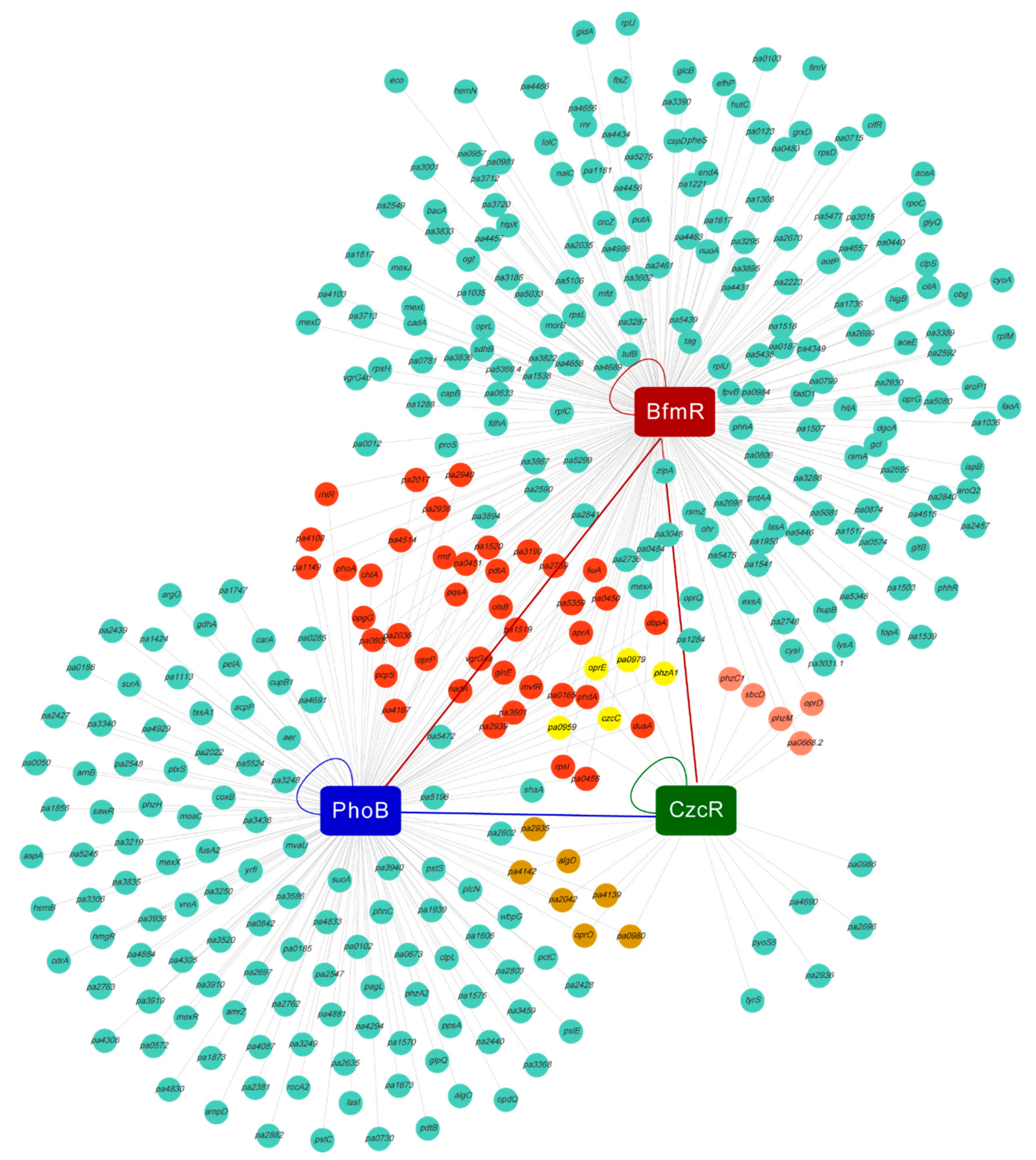
3.2. Identification of BfmR-Regulated Genes
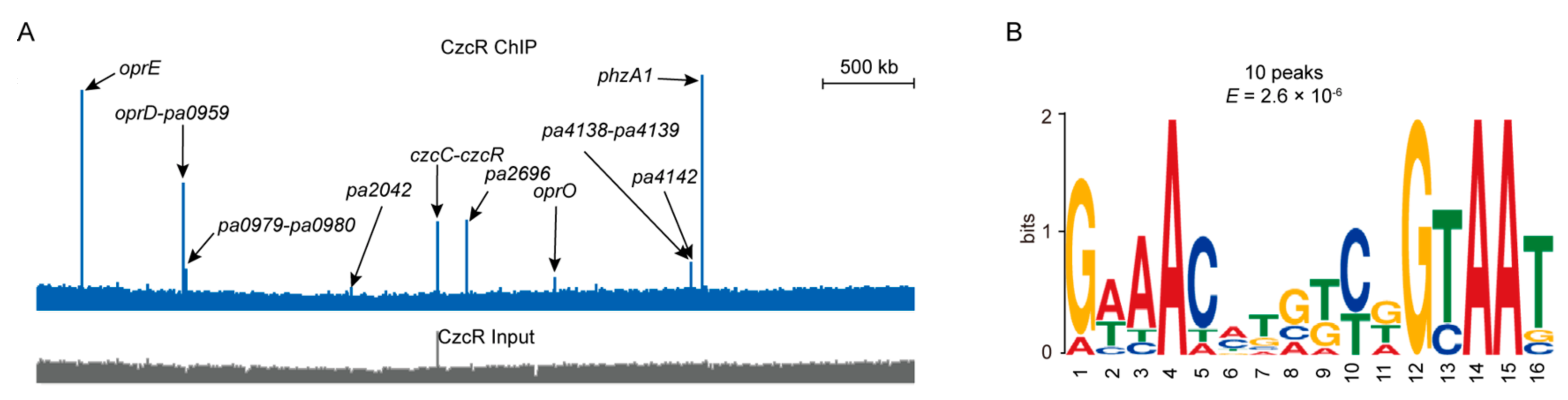
3.3. BfmR Binds to the Promoters of CzcR-Targeted Genes
3.4. BfmR Binds to and Induces exsA Promoter
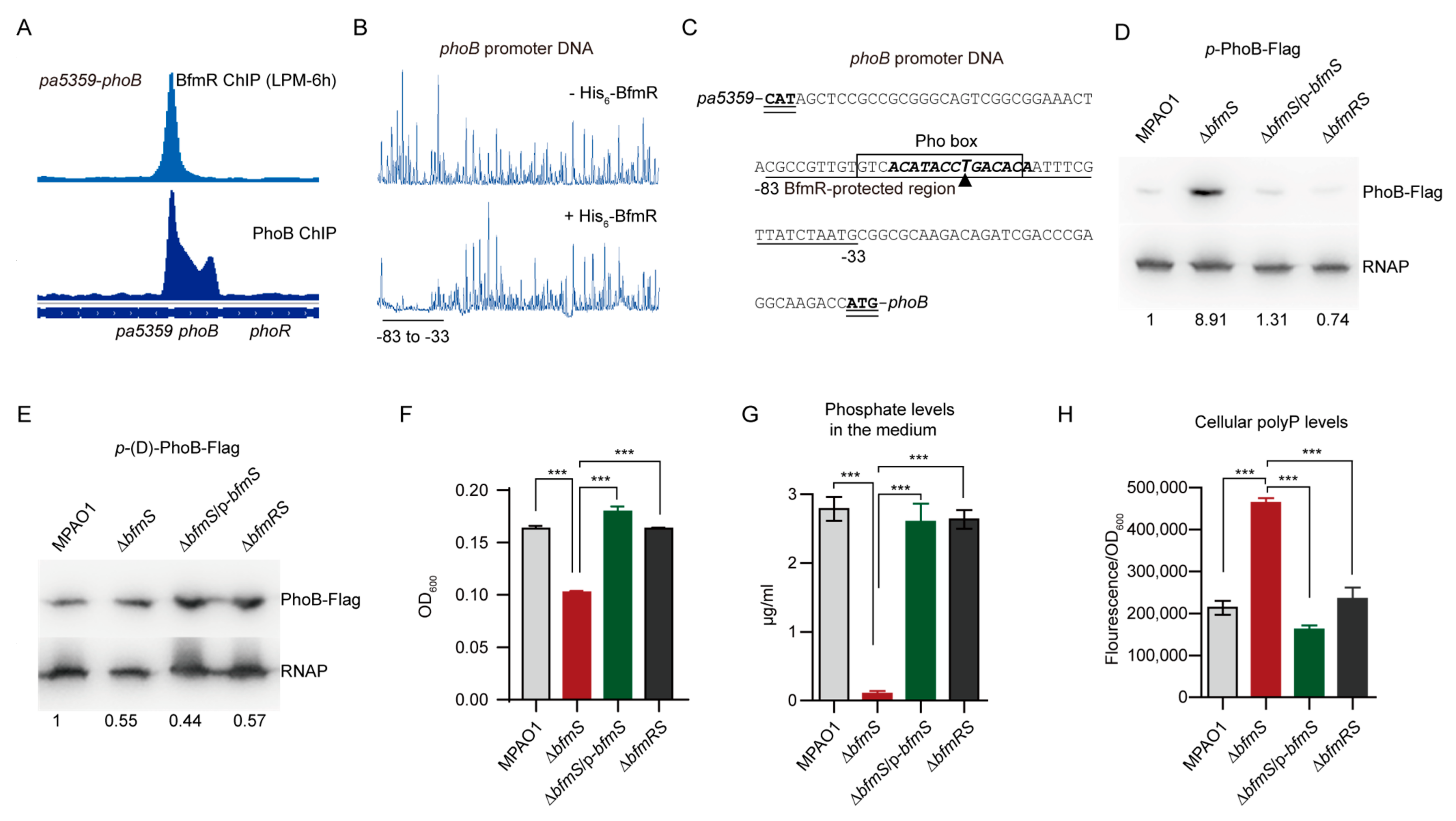
3.5. BfmR Binds to the Promoter of a Number of PhoB-Targeted Genes
3.6. BfmR Directly Activates phoB
3.7. BfmR Boosts Pi Depletion and Inorganic Polyphosphate (polyP) Accumulation
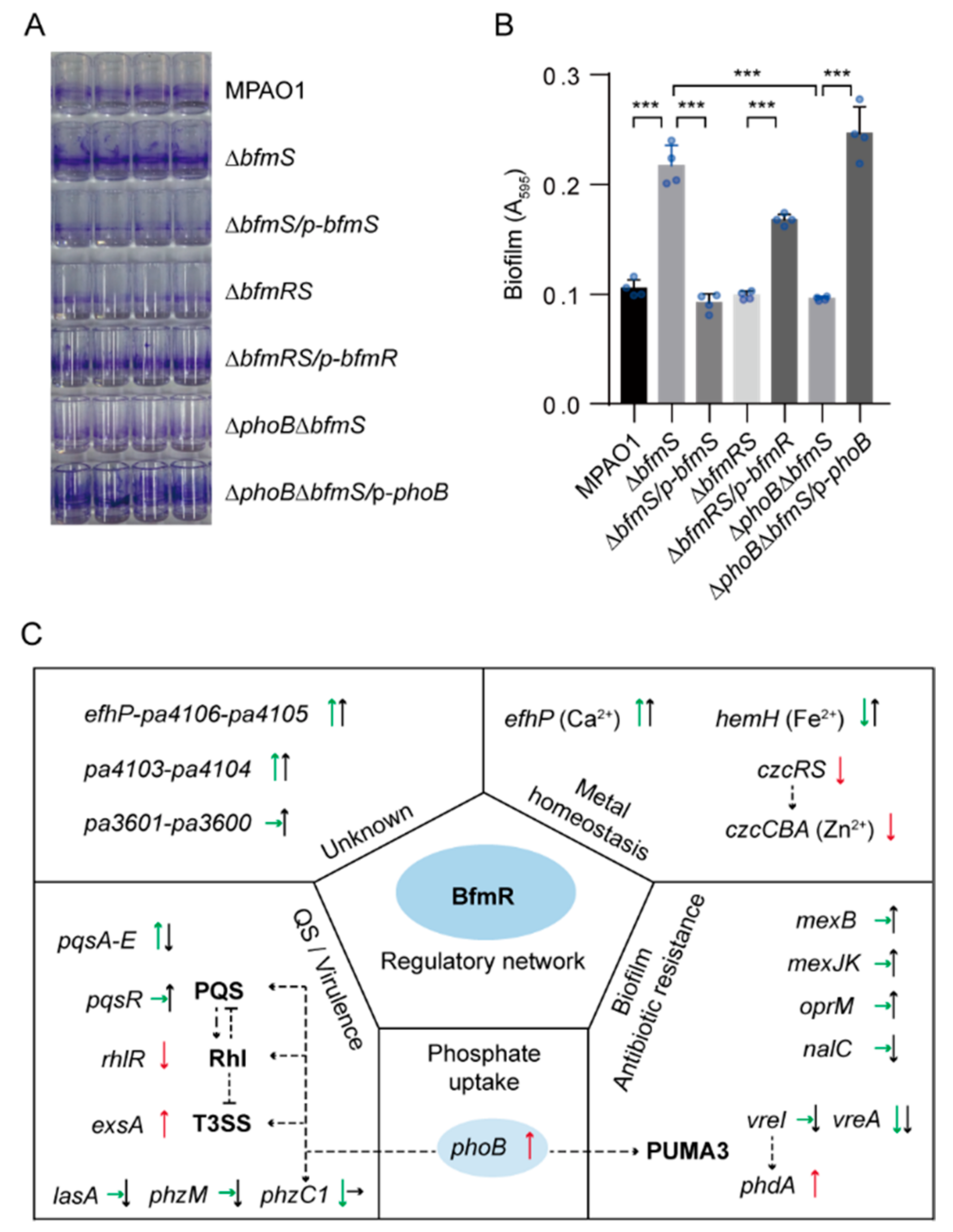
3.8. PhoB Is Required for BfmR-Dependent Biofilm Formation
4. Discussions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 2004, 32, 470–485. [Google Scholar] [CrossRef]
- Willyard, C. The drug-resistant bacteria that pose the greatest health threats. Nature 2017, 543, 15. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Curran, C.S.; Bolig, T.; Torabi-Parizi, P. Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 2018, 197, 708–727. [Google Scholar] [CrossRef]
- Cao, Q.; Yang, N.; Wang, Y.; Xu, C.; Zhang, X.; Fan, K.; Chen, F.; Liang, H.; Zhang, Y.; Deng, X.; et al. Mutation-Induced remodeling of the BfmRS two-component system in Pseudomonas aeruginosa clinical isolates. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, Q.; Cao, Q.; Gan, J.; Sun, N.; Yang, C.-G.; Bae, T.; Wu, M.; Lan, L. Histamine activates hinK to promote the virulence of Pseudomonas aeruginosa. Sci. Bull. 2021, in press. [Google Scholar]
- Bodey, G.P.; Bolivar, R.; Fainstein, V.; Jadeja, L. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, 279–313. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Schneper, L.; Kumari, H.; Mathee, K. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res. 2013, 41, 1–20. [Google Scholar] [CrossRef]
- Cao, Q.; Wang, Y.; Chen, F.; Xia, Y.; Lou, J.; Zhang, X.; Yang, N.; Sun, X.; Zhang, Q.; Zhuo, C.; et al. A novel signal transduction pathway that modulates rhl quorum sensing and bacterial virulence in Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1004340. [Google Scholar] [CrossRef] [PubMed]
- Rodrigue, A.; Quentin, Y.; Lazdunski, A.; Méjean, V.; Foglino, M. Two-Component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000, 8, 498–504. [Google Scholar] [CrossRef]
- Gooderham, W.J.; Hancock, R.E. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2009, 33, 279–294. [Google Scholar] [CrossRef]
- Laub, M.T.; Goulian, M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007, 41, 121–145. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component signal transduction. Annu. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef] [PubMed]
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-Component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009, 5, e1000668. [Google Scholar] [CrossRef]
- Petrova, O.E.; Schurr, J.R.; Schurr, M.J.; Sauer, K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol. Microbiol. 2011, 81, 767–783. [Google Scholar] [CrossRef] [PubMed]
- Marvig, R.L.; Johansen, H.K.; Molin, S.; Jelsbak, L. Genome analysis of a transmissible lineage of Pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 2013, 9, e1003741. [Google Scholar] [CrossRef]
- Damkiaer, S.; Yang, L.; Molin, S.; Jelsbak, L. Evolutionary remodeling of global regulatory networks during long-term bacterial adaptation to human hosts. Proc. Natl. Acad. Sci. USA 2013, 110, 7766–7771. [Google Scholar] [CrossRef]
- Yang, L.; Jelsbak, L.; Marvig, R.L.; Damkiær, S.; Workman, C.T.; Rau, M.H.; Hansen, S.K.; Folkesson, A.; Johansen, H.K.; Ciofu, O.; et al. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. USA 2011, 108, 7481–7486. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, M.A.; Alwood, A.; Thaipisuttikul, I.; Spencer, D.; Haugen, E.; Ernst, S.; Will, O.; Kaul, R.; Raymond, C.; Levy, R.; et al. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 14339–14344. [Google Scholar] [CrossRef] [PubMed]
- Brint, J.M.; Ohman, D.E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 1995, 177, 7155–7163. [Google Scholar] [CrossRef]
- Köhler, T.; Curty, L.K.; Barja, F.; van Delden, C.; Pechère, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef]
- Jansons, I.; Touchie, G.; Sharp, R.; Almquist, K.; Farinha, M.A.; Lam, J.S.; Kropinski, A.M. Deletion and transposon mutagenesis and sequence analysis of the pRO1600 OriR region found in the broad-host-range plasmids of the pQF series. Plasmid 1994, 31, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Becher, A.; Schweizer, H.P. Integration-Proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 2000, 29, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, H.P.; Hoang, T.T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 1995, 158, 15–22. [Google Scholar] [CrossRef]
- Ding, Y.; Liu, X.; Chen, F.; Di, H.; Xu, B.; Zhou, L.; Deng, X.; Wu, M.; Yang, C.-G.; Lan, L. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2014, 111, E4981–E4990. [Google Scholar] [CrossRef]
- Chen, F.; Di, H.; Wang, Y.; Cao, Q.; Xu, B.; Zhang, X.; Yang, N.; Liu, G.; Yang, C.-G.; Xu, Y.; et al. Small-Molecule targeting of a diapophytoene desaturase inhibits S. aureus virulence. Nat. Chem. Biol. 2016, 12, 174–179. [Google Scholar] [CrossRef]
- Jones, C.J.; Newsom, D.; Kelly, B.; Irie, Y.; Jennings, L.K.; Xu, B.; Limoli, D.H.; Harrison, J.J.; Parsek, M.R.; White, P.; et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1003984. [Google Scholar] [CrossRef]
- Blasco, B.; Chen, J.M.; Hartkoorn, R.; Sala, C.; Uplekar, S.; Rougemont, J.; Pojer, F.; Cole, S.T. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog. 2012, 8, e1002621. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-Seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Blythe, S.A.; Reid, C.D.; Kessler, D.S.; Klein, P.S. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn. 2009, 238, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Taneyhill, L.A.; Adams, M.S. Investigating regulatory factors and their DNA binding affinities through real time quantitative PCR (RT-QPCR) and chromatin immunoprecipitation (ChIP) assays. Methods Cell Biol. 2008, 87, 367–389. [Google Scholar] [PubMed]
- Gentsch, G.E.; Patrushev, I.; Smith, J.C. Genome-Wide snapshot of chromatin regulators and states in Xenopus embryos by ChIP-Seq. J. Vis. Exp. 2015, 96. [Google Scholar] [CrossRef]
- Lan, L.; Murray, T.S.; Kazmierczak, B.I.; He, C. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 2010, 75, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.; Karkhoff-Schweizer, R.R.; Kutchma, A.J.; Schweizer, H.P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 1998, 212, 77–86. [Google Scholar] [CrossRef]
- Yang, N.; Ding, S.; Chen, F.; Zhang, X.; Xia, Y.; Di, H.; Cao, Q.; Deng, X.; Wu, M.; Wong, C.C.L.; et al. The Crc protein participates in down-regulation of the Lon gene to promote rhamnolipid production and rhl quorum sensing in Pseudomonas aeruginosa. Mol. Microbiol. 2015, 96, 526–547. [Google Scholar] [CrossRef]
- Yang, N.; Cao, Q.; Hu, S.; Xu, C.; Fan, K.; Chen, F.; Yang, C.-G.; Liang, H.; Wu, M.; Bae, T.; et al. Alteration of protein homeostasis mediates the interaction of Pseudomonas aeruginosa with Staphylococcus aureus. Mol. Microbiol. 2020, 114, 423–442. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cao, Q.; Lan, L. Glucose-Binding of Periplasmic Protein GltB Activates GtrS-GltR Two-Component System in Pseudomonas aeruginosa. Microorganisms 2021, 9, 447. [Google Scholar] [CrossRef]
- Aschar-Sobbi, R.; Abramov, A.Y.; Diao, C.; Kargacin, M.E.; Kargacin, G.J.; French, R.J.; Pavlov, E. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 2008, 18, 859–866. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Varadarajan, A.R.; Allan, R.N.; Valentin, J.D.P.; Ocampo, O.E.C.; Somerville, V.; Pietsch, F.; Buhmann, M.T.; West, J.; Skipp, P.J.; van der Mei, H.C. An integrated model system to gain mechanistic insights into biofilm-associated antimicrobial resistance in Pseudomonas aeruginosa MPAO1. NPJ Biofilms Microbiomes 2020, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- McKenna, J.A.; Wing, H.J. The antiactivator of type III secretion, OspD1, Is transcriptionally regulated by VirB and H-NS from Remote Sequences in Shigella flexneri. J. Bacteriol. 2020, 202. [Google Scholar] [CrossRef]
- Dieppois, G.; Ducret, V.; Caille, O.; Perron, K. The transcriptional regulator CzcR modulates antibiotic resistance and quorum sensing in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e38148. [Google Scholar] [CrossRef]
- Caille, O.; Rossier, C.; Perron, K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J. Bacteriol. 2007, 189, 4561–4568. [Google Scholar] [CrossRef]
- Perron, K.; Caille, O.; Rossier, C.; Van Delden, C.; Dumas, J.-L.; Kohler, T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 8761–8768. [Google Scholar] [CrossRef]
- Garber, M.E.; Rajeev, L.; Kazakov, A.E.; Trinh, J.; Masuno, D.; Thompson, M.G.; Kaplan, N.; Luk, J.; Novichkov, P.S.; Mukhopadhyay, A. Multiple signaling systems target a core set of transition metal homeostasis genes using similar binding motifs. Mol. Microbiol. 2018, 107, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.T.; van der Lelie, D.; Springael, D.; Romling, U.; Ahmed, N.; Mergeay, M. Identification of a gene cluster, czr, involved in cadmium and zinc resistance in Pseudomonas aeruginosa. Gene 1999, 238, 417–425. [Google Scholar] [CrossRef]
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 2931. [Google Scholar] [CrossRef]
- Jensen, V.; Löns, D.; Zaoui, C.; Bredenbruch, F.; Meissner, A.; Dieterich, G.; Münch, R.; Häussler, S. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 2006, 188, 8601–8606. [Google Scholar] [CrossRef] [PubMed]
- Marsden, A.E.; Intile, P.J.; Schulmeyer, K.H.; Simmons-Patterson, E.; Urbanowski, M.L.; Wolfgang, M.C.; Yahr, T.L. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J. Bacteriol. 2016, 198, 1442–1450. [Google Scholar] [CrossRef] [PubMed]
- Yahr, T.L.; Frank, D.W. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 3832–3838. [Google Scholar] [CrossRef]
- Liang, H.; Deng, X.; Li, X.; Ye, Y.; Wu, M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 2014, 42, 10307–10320. [Google Scholar] [CrossRef]
- Bielecki, P.; Jensen, V.; Schulze, W.; Godeke, J.; Strehmel, J.; Eckweiler, D.; Nicolai, T.; Bielecka, A.; Wille, T.; Gerlach, R.G.; et al. Cross talk between the response regulators PhoB and TctD allows for the integration of diverse environmental signals in Pseudomonas aeruginosa. Nucleic Acids Res. 2015, 43, 6413–6425. [Google Scholar] [CrossRef]
- Siehnel, R.J.; Egli, C.; Hancock, R.E. Polyphosphate-selective porin OprO of Pseudomonas aeruginosa: Expression, purification and sequence. Mol. Microbiol. 1992, 6, 2319–2326. [Google Scholar] [CrossRef] [PubMed]
- Quesada, J.M.; Otero-Asman, J.R.; Bastiaansen, K.C.; Civantos, C.; Llamas, M.A. The activity of the Pseudomonas aeruginosa virulence regulator σ(VreI) is modulated by the anti-σ factor VreR and the transcription factor PhoB. Front. Microbiol. 2016, 7, 1159. [Google Scholar] [CrossRef]
- Faure, L.M.; Llamas, M.A.; Bastiaansen, K.C.; de Bentzmann, S.; Bigot, S. Phosphate starvation relayed by PhoB activates the expression of the Pseudomonas aeruginosa sigmavreI ECF factor and its target genes. Microbiology 2013, 159, 1315–1327. [Google Scholar] [CrossRef]
- Meng, X.; Ahator, S.D.; Zhang, L.H. Molecular mechanisms of phosphate stress activation of Pseudomonas aeruginosa quorum sensing systems. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Siehnel, R.J.; Worobec, E.A.; Hancock, R.E. Regulation of components of the Pseudomonas aeruginosa phosphate-starvation-inducible regulon in Escherichia coli. Mol. Microbiol. 1988, 2, 347–352. [Google Scholar] [CrossRef]
- Blus-Kadosh, I.; Zilka, A.; Yerushalmi, G.; Banin, E. The effect of pstS and phoB on quorum sensing and swarming motility in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e74444. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.J.; Wanner, B.L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 2010, 13, 198–203. [Google Scholar] [CrossRef] [PubMed]
- De Almeida, L.G.; Ortiz, J.H.; Schneider, R.P.; Spira, B. phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl. Environ. Microbiol. 2015, 81, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Sarkisova, S.A.; Lotlikar, S.R.; Guragain, M.; Kubat, R.; Cloud, J.; Franklin, M.J. A Pseudomonas aeruginosa EF-Hand protein, EfhP (PA4107), modulates stress responses and virulence at high calcium concentration. PLoS ONE 2014, 9, e98985. [Google Scholar] [CrossRef] [PubMed]
- Broder, U.N.; Jaeger, T.; Jenal, U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016, 2, 16184. [Google Scholar] [CrossRef]
- Haddad, A.; Jensen, V.; Becker, T.; Haussler, S. The Pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2019, 1, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, K.; Cao, Q.; Lan, L. Genome-Wide Mapping Reveals Complex Regulatory Activities of BfmR in Pseudomonas aeruginosa. Microorganisms 2021, 9, 485. https://doi.org/10.3390/microorganisms9030485
Fan K, Cao Q, Lan L. Genome-Wide Mapping Reveals Complex Regulatory Activities of BfmR in Pseudomonas aeruginosa. Microorganisms. 2021; 9(3):485. https://doi.org/10.3390/microorganisms9030485
Chicago/Turabian StyleFan, Ke, Qiao Cao, and Lefu Lan. 2021. "Genome-Wide Mapping Reveals Complex Regulatory Activities of BfmR in Pseudomonas aeruginosa" Microorganisms 9, no. 3: 485. https://doi.org/10.3390/microorganisms9030485
APA StyleFan, K., Cao, Q., & Lan, L. (2021). Genome-Wide Mapping Reveals Complex Regulatory Activities of BfmR in Pseudomonas aeruginosa. Microorganisms, 9(3), 485. https://doi.org/10.3390/microorganisms9030485






