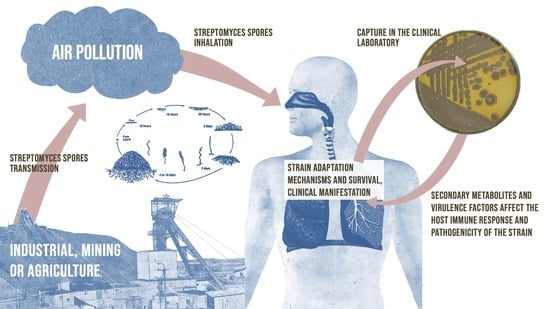
The Genome Analysis of the Human Lung-Associated Streptomyces sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues
Abstract
:1. Introduction
2. Materials and Methods
2.1. Streptomyces sp. TR13141: Isolation Method, Cultivation, and Genome Sequencing
2.2. Morphological Characteristics and Microscopy
2.3. Growth Curve and Carbon Utilization Assay
2.4. Siderophore Production and Identification
2.5. Dataset
2.6. Orthologous Genes and Species Tree Inference
2.7. Biosynthetic Gene Clusters
2.8. Mammalian Cell Entry (mce) Genes and Type VII Secretion System (ESX)
2.9. Genomic Comparison of Streptomyces sp. TR1341, Streptomyces sp. Endophyte_N2, and the Human Associated Streptomyces spp.
3. Results and Discussion
3.1. Streptomyces sp. TR1341: Morphological Characterization and Genome Features
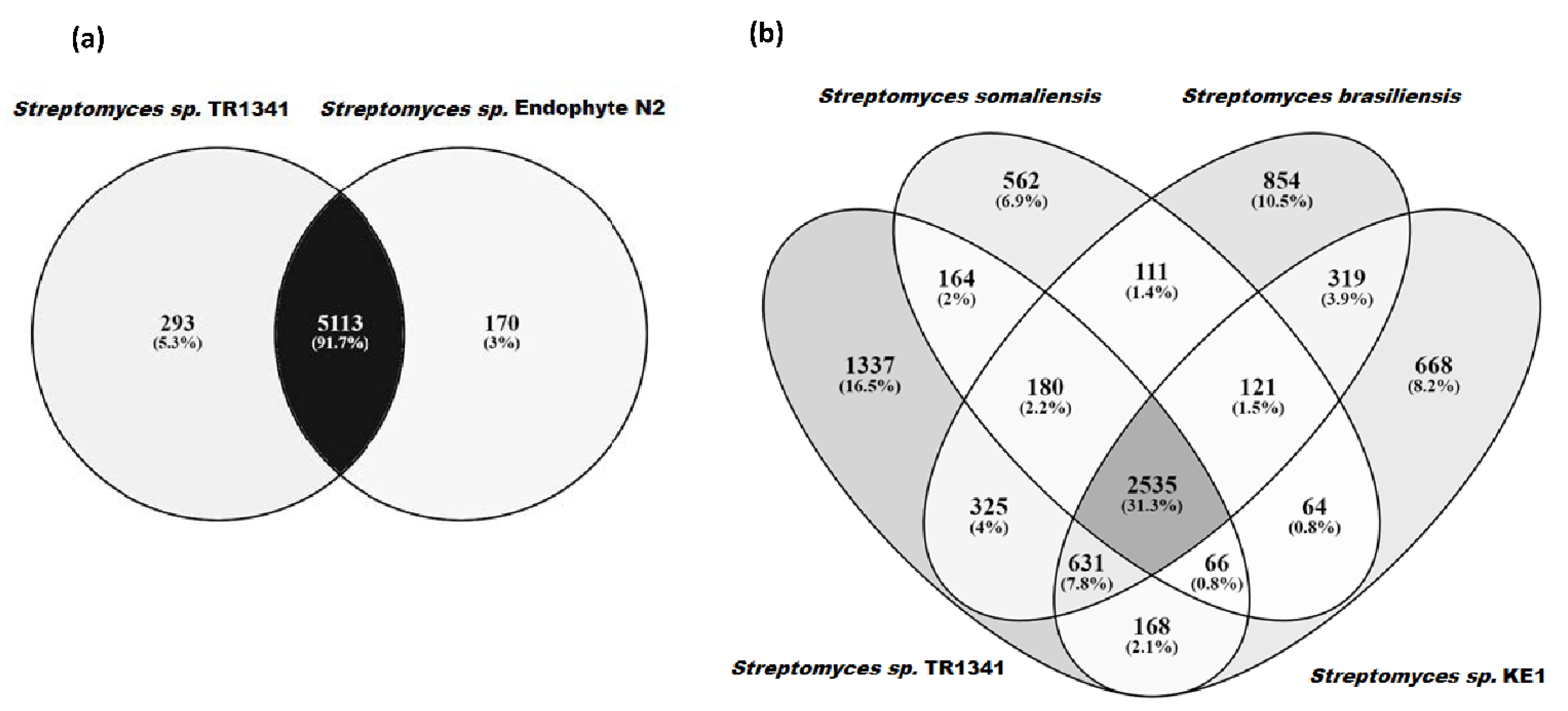
3.2. Orthologous Genes and Tree Inference
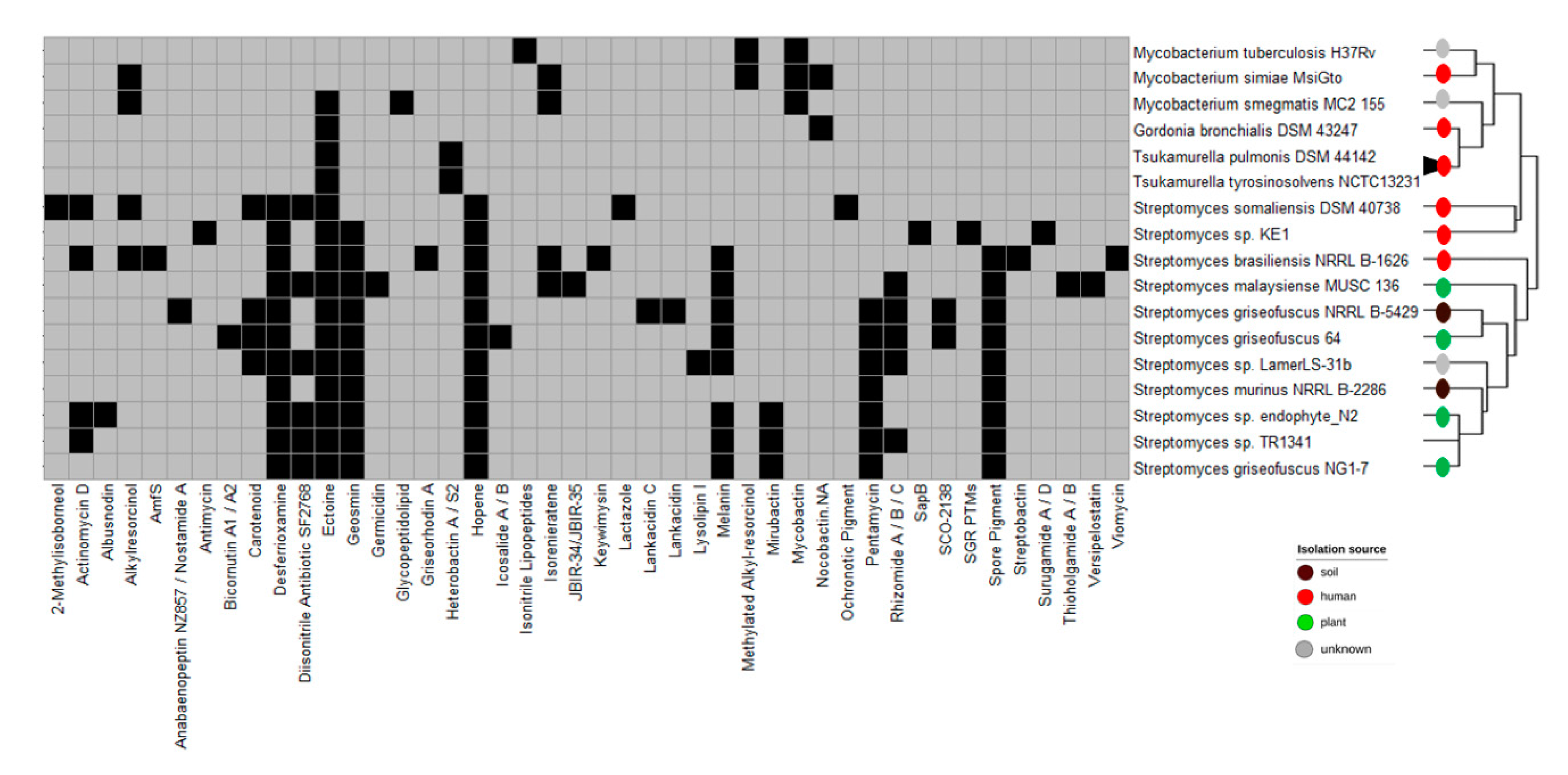
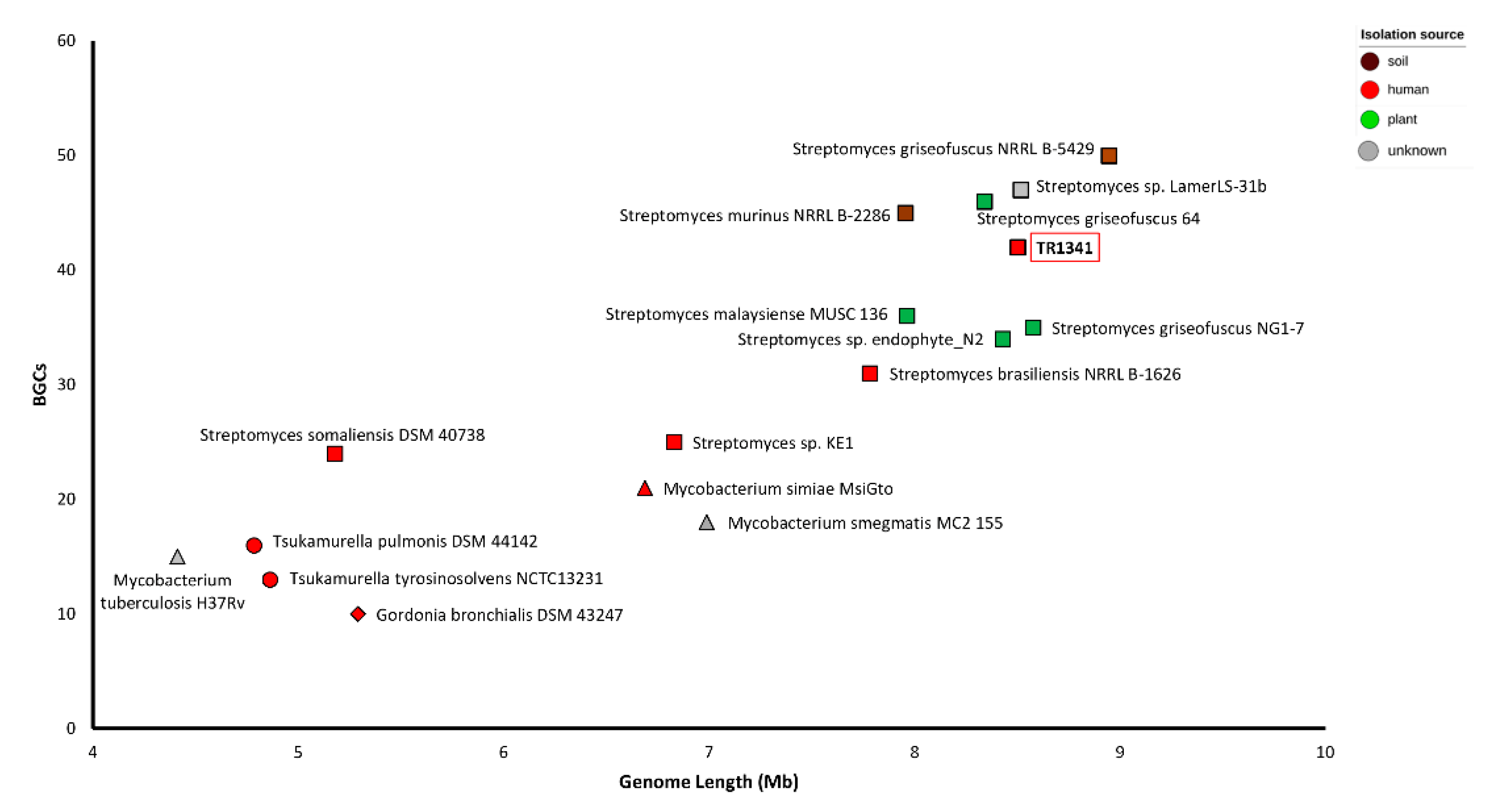
3.3. Biosynthetic Gene Clusters
3.4. Mammalian Cell Entry (mce) Genes and Type VII Secretion System (ESX)
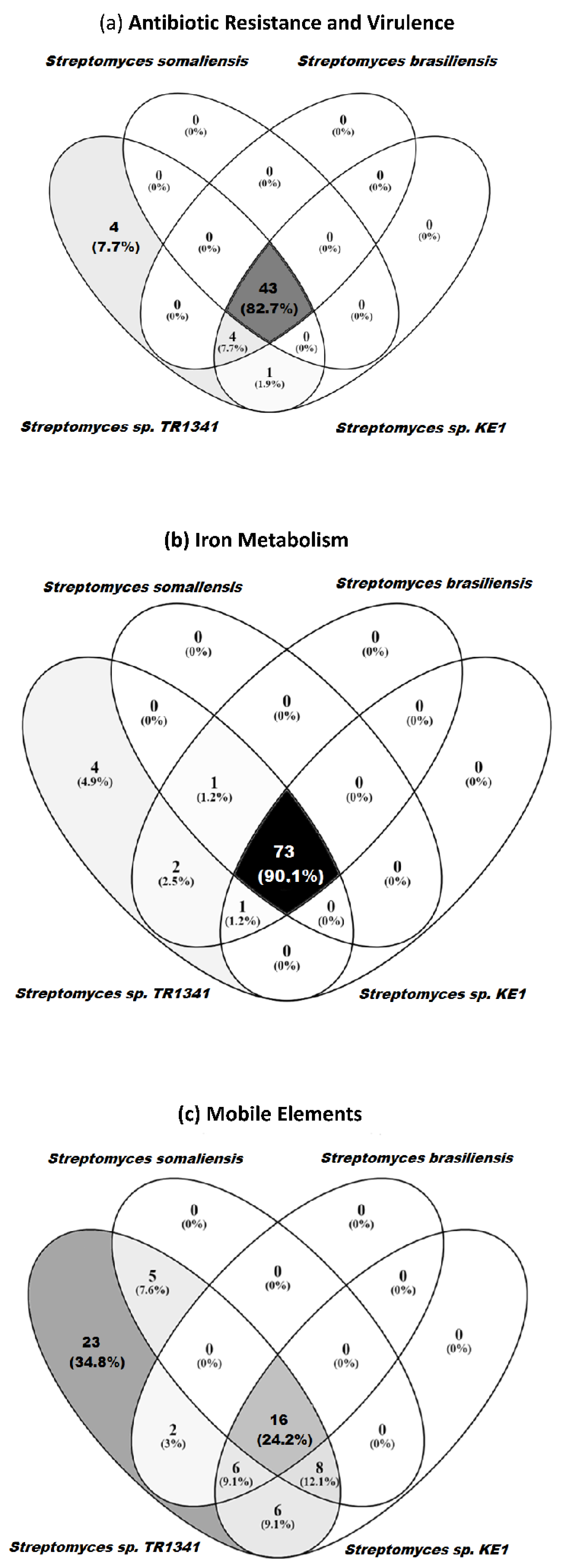
3.5. Genomic Comparison of Streptomyces sp. TR1341, Streptomyces sp. Endophyte_N2, and the Human Associated Streptomyces spp.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; OXford University Press, Inc.: Oxford, UK, 2007; ISBN-13 978-0-19-515066-7. [Google Scholar]
- Müller, R.; Wink, J. Future potential for anti-infectives from bacteria—How to exploit biodiversity and genomic potential. Int. J. Med. Microbiol. 2014, 304, 3–13. [Google Scholar] [CrossRef]
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Chen, M.; Chai, W.; Song, T.; Ma, M.; Lian, X.Y.; Zhang, Z. Anti-glioma Natural Products Downregulating Tumor Glycolytic Enzymes from Marine Actinomycete Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef] [Green Version]
- Kämpfer, P. The Family Streptomycetaceae, Part I: Taxonomy. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 538–604. [Google Scholar]
- Amin, A.; Ahmed, I.; Khalid, N.; Osman, G.; Khan, I.U.; Xiao, M.; Li, W.J. Streptomyces caldifontis sp. nov., isolated from a hot water spring of Tatta Pani, Kotli, Pakistan. Antonie Leeuwenhoek 2017, 110, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Řeháková, K.; Chroňáková, A.; Krištůfek, V.; Kuchtová, B.; Čapková, K.; Scharfen, J.; Čapek, P.; Doležal, J. Bacterial community of cushion plant Thylacospermum ceaspitosum on elevational gradient in the Himalayan cold desert. Front. Microbiol. 2015, 6, 304. [Google Scholar] [CrossRef] [PubMed]
- Chroňáková, A.; Krištůfek, V.; Tichý, M.; Elhottová, D. Biodiversity of Streptomycetes isolated from a succession sequence at a post-mining site and their evidence in Miocene lacustrine sediment. Microbiol. Res. 2010, 165, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Göttler, W.; Herzner, G.; Strohm, E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 2005, 15, 475–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef] [Green Version]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.M.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.R.; Cho, G.; Jeon, C.W.; Weller, D.M.; Thomashow, L.S.; Paulitz, T.C.; Kwak, Y.S. A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun. 2019, 10, 4802. [Google Scholar] [CrossRef] [Green Version]
- Grubbs, K.J.; Surup, F.; Biedermann, P.H.W.; McDonald, B.R.; Klassen, J.L.; Carlson, C.M.; Clardy, J.; Currie, C.R. Cycloheximide-Producing Streptomyces Associated with Xyleborinus saxesenii and Xyleborus affinis Fungus-Farming Ambrosia Beetles. Front. Microbiol. 2020, 11, 2207. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Ramírez, J.M.; Van Der Voort, M.; Raaijmakers, J.M.; Diéguez-Uribeondo, J. Unravelling the microbiome of eggs of the endangered sea turtle Eretmochelys imbricata identifies bacteria with activity against the emerging pathogen Fusarium falciforme. PLoS ONE 2014, 9, e95206. [Google Scholar] [CrossRef]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Sawada, H.; Tanaka, F.; Matsuda, I. Phylogenetic analysis of Streptomyces spp. causing potato scab based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 1996, 46, 476–479. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, J.; Díaz-Cruz, G.; Cheng, Z.; Bignell, D.R.D. Virulence mechanisms of plant-pathogenic Streptomyces species: An updated review. Microbiology 2019, 165, 1025–1040. [Google Scholar] [CrossRef]
- Khalil, M.; Lerat, S.; Beaudoin, N.; Beaulieu, C. The Plant Pathogenic Bacterium Streptomyces scabies Degrades the Aromatic Components of Potato Periderm via the β-Ketoadipate Pathway. Front. Microbiol. 2019, 10, 2795. [Google Scholar] [CrossRef]
- Quintana, E.T.; Wierzbicka, K.; Mackiewicz, P.; Osman, A.; Fahal, A.H.; Hamid, M.E.; Zakrzewska-Czerwinska, J.; Maldonado, L.A.; Goodfellow, M. Streptomyces sudanensis sp. nov., a new pathogen isolated from patients with actinomycetoma. Antonie Leeuwenhoek 2008, 93, 305–313. [Google Scholar] [CrossRef]
- Kirby, R.; Sangal, V.; Tucker, N.P.; Zakrzewska-Czerwińska, J.; Wierzbicka, K.; Herron, P.R.; Chu, C.J.; Chandra, G.; Fahal, A.H.; Goodfellow, M.; et al. Draft genome sequence of the human pathogen Streptomyces somaliensis, a significant cause of actinomycetoma. J. Bacteriol. 2012, 194, 3544–3545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sing, D.; Sing, C.F. Impact of direct soil exposures from airborne dust and geophagy on human health. Int. J. Environ. Res. Public Health 2010, 7, 1205–1223. [Google Scholar] [CrossRef] [Green Version]
- Bolourian, A.; Mojtahedi, Z. Streptomyces, shared microbiome member of soil and gut, as “old friends” against colon cancer. FEMS Microbiol. Ecol. 2018, 94, fiy120. [Google Scholar] [CrossRef]
- Bolourian, A.; Mojtahedi, Z. Immunosuppressants produced by Streptomyces: Evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ. Microbiol. Rep. 2018, 10, 123–126. [Google Scholar] [CrossRef]
- Gallo, R.L.; Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012, 12, 503–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef] [Green Version]
- Herbrík, A.; Corretto, E.; Chroňáková, A.; Langhansová, H.; Petrásková, P.; Hrdý, J.; Čihák, M.; Krištůfek, V.; Bobek, J.; Petříček, M.; et al. A Human Lung-Associated Streptomyces sp. TR1341 Produces Various Secondary Metabolites Responsible for Virulence, Cytotoxicity and Modulation of Immune Response. Front. Microbiol. 2020, 10, 3028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engevik, M.A.; Versalovic, J. Biochemical Features of Beneficial Microbes: Foundations for Therapeutic Microbiology. In Bugs as Drugs; American Society of Microbiology: Washington, DC, USA, 2017; pp. 3–47. [Google Scholar]
- Siddiqui, S.; Anderson, V.L.; Hilligoss, D.M.; Abinun, M.; Kuijpers, T.W.; Masur, H.; Witebsky, F.G.; Shea, Y.R.; Gallin, J.I.; Malech, H.L.; et al. Fulminant mulch pneumonitis: An emergency presentation of chronic granulomatous disease. Clin. Infect. Dis. 2007, 45, 673–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lertcanawanichakul, M.; Chawawisit, K. Identification of Streptomyces spp. isolated from air samples and its cytotoxicity of anti-MRSA bioactive compounds. Biocatal. Agric. Biotechnol. 2019, 20, 101236. [Google Scholar] [CrossRef]
- Kettleson, E.; Kumar, S.; Reponen, T.; Vesper, S.; Méheust, D.; Grinshpun, S.A.; Adhikari, A. Stenotrophomonas, Mycobacterium, and Streptomyces in home dust and air: Associations with moldiness and other home/family characteristics. Indoor Air 2013, 23, 387–396. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofronová, O.; Petrícková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef] [Green Version]
- Huttunen, K.; Hyvärinen, A.; Nevalainen, A.; Komulainen, H.; Hirvonen, M.R. Production of proinflammatory mediators by indoor air bacteria and fungal spores in mouse and human cell lines. Environ. Health Perspect. 2003, 111, 85–92. [Google Scholar] [CrossRef] [Green Version]
- Jussila, J.; Komulainen, H.; Huttunen, K.; Roponen, M.; Hälinen, A.; Hyvärinen, A.; Kosma, V.-M.; Pelkonen, J.; Hirvonen, M.-R. Inflammatory Responses in Mice after Intratracheal Instillation of Spores of Streptomyces californicus Isolated from Indoor Air of a Moldy Building. Toxicol. Appl. Pharmacol. 2001, 171, 61–69. [Google Scholar] [CrossRef]
- Penttinen, P.; Huttunen, K.; Pelkonen, J.; Hirvonen, M.R. The proportions of Streptomyces californicus and Stachybotrys chartarum in simultaneous exposure affect inflammatory responses in mouse RAW264.7 macrophages. Inhal. Toxicol. 2005, 17, 79–85. [Google Scholar] [CrossRef]
- Penttinen, P.; Pelkonen, J.; Huttunen, K.; Hirvonen, M.-R.R. Co-cultivation of Streptomyces californicus and Stachybotrys chartarum stimulates the production of cytostatic compound(s) with immunotoxic properties. Toxicol. Appl. Pharmacol. 2006, 217, 342–351. [Google Scholar] [CrossRef]
- Yacoub, A.T.; Velez, A.P.; Khwaja, S.I.; Sandin, R.L.; Greene, J. Streptomyces pneumonia in an immunocompromised patient: A case report and a review of literature. Infect. Dis. Clin. Pract. 2014, 22, e113–e115. [Google Scholar] [CrossRef]
- Cambier, C.J.; Falkow, S.; Ramakrishnan, L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 2014, 159, 1497–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitter, W.; Houben, E.N.G.; Bottai, D.; Brodin, P.; Brown, E.J.; Cox, J.S.; Derbyshire, K.; Fortune, S.M.; Gao, L.-Y.; Liu, J.; et al. Systematic Genetic Nomenclature for Type VII Secretion Systems. PLoS Pathog. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unnikrishnan, M.; Constantinidou, C.; Palmer, T.; Pallen, M.J. The Enigmatic Esx Proteins: Looking Beyond Mycobacteria. Trends Microbiol. 2017, 25, 192–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gröschel, M.I.; Sayes, F.; Simeone, R.; Majlessi, L.; Brosch, R. ESX secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016, 14, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; García, J.S.; Morbidoni, H.R.; de la Paz Santangelo, M.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [Green Version]
- Fyans, J.K.; Bignell, D.; Loria, R.; Toth, I.; Palmer, T. The ESX/type VII secretion system modulates development, but not virulence, of the plant pathogen Streptomyces scabies. Mol. Plant Pathol. 2013, 14, 119–130. [Google Scholar] [CrossRef]
- Roman, S.A.S.; Facey, P.D.; Fernandez-Martinez, L.; Rodriguez, C.; Vallin, C.; Del Sol, R.; Dyson, P. A heterodimer of EsxA and EsxB is involved in sporulation and is secreted by a type VII secretion system in Streptomyces coelicolor. Microbiology 2010, 156, 1719–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haile, Y.; Caugant, D.A.; Bjune, G.; Wiker, H.G. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol. Med. Microbiol. 2002, 33, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, L.C.; Seipke, R.F.; Prieto, P.; Willemse, J.; Van Wezel, G.P.; Hutchings, M.I.; Hoskisson, P.A. Mammalian cell entry genes in Streptomyces may provide clues to the evolution of bacterial virulence. Sci. Rep. 2013, 3, 1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casali, N.; Riley, L.W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 2007, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Shimono, N.; Morici, L.; Casali, N.; Cantrell, S.; Sidders, B.; Ehrt, S.; Riley, L.W. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. USA 2003, 100, 15918–15923. [Google Scholar] [CrossRef] [Green Version]
- Zhang, V.; Nemeth, E.; Kim, A. Iron in lung pathology. Pharmaceuticals 2019, 12, 30. [Google Scholar] [CrossRef] [Green Version]
- Terra, L.; Dyson, P.; Ratcliffe, N.; Castro, H.C.; Vicente, A.C.P. Biotechnological Potential of Streptomyces Siderophores as New Antibiotics. Curr. Med. Chem. 2021, 28, 1407–1421. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [Green Version]
- Kronstad, J.W.; Caza, M. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 4, 80. [Google Scholar]
- Parrow, N.L.; Fleming, R.E.; Minnick, M.F. Sequestration and scavenging of iron in infection. Infect. Immun. 2013, 81, 3503–3514. [Google Scholar] [CrossRef] [Green Version]
- Braun, V.; Pramanik, A.; Gwinner, T.; Köberle, M.; Bohn, E. Sideromycins: Tools and antibiotics. Biometals 2009, 22, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore production by actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibel-ski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Kieser, T. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000; ISBN 9780708406236. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Milagres, A.M.F.; Machuca, A.; Napoleão, D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 1999, 37, 1–6. [Google Scholar] [CrossRef]
- Sidebottom, A.M.; Karty, J.A.; Carlson, E.E. Accurate Mass MS/MS/MS Analysis of Siderophores Ferrioxamine B and E1 by Collision-Induced Dissociation Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 1899–1902. [Google Scholar] [CrossRef] [Green Version]
- Senges, C.H.R.; Al-Dilaimi, A.; Marchbank, D.H.; Wibberg, D.; Winkler, A.; Haltli, B.; Nowrousian, M.; Kalinowski, J.; Kerr, R.G.; Bandow, J.E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. USA 2018, 115, 2490–2495. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [Green Version]
- Emms, D.M.; Kelly, S. STAG: Species Tree Inference from All Genes. bioRxiv 2018, 267914. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef] [PubMed]
- Mungan, M.D.; Alanjary, M.; Blin, K.; Weber, T.; Medema, M.H.; Ziemert, N. ARTS 2.0: Feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Res. 2020, 48, W546–W552. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [Green Version]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. Available online: http://bioinfogp.cnnb.csic.es/tools/venny/index.html (accessed on 9 September 2020).
- Chiesi USA, Inc. Bronchitol ® Inhaled Dry Powder Mannitol (DPM) for Adult Patients with Cystic Fibrosis; Chiesi USA, Inc.: Cary, NC, USA, 2019. [Google Scholar]
- Mitri, C.; Xu, Z.; Bardin, P.; Corvol, H.; Touqui, L.; Tabary, O. Novel Anti-Inflammatory Approaches for Cystic Fibrosis Lung Disease: Identification of Molecular Targets and Design of Innovative Therapies. Front. Pharmacol. 2020, 11, 1096. [Google Scholar] [CrossRef] [PubMed]
- Nevitt, S.J.; Thornton, J.; Murray, C.S.; Dwyer, T. Inhaled mannitol for cystic fibrosis. Cochrane Database Syst. Rev. 2020, 2020, CD008649. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.J.; Wang, J.H.; Bu, X.L.; Yu, H.L.; Li, P.; Ou, H.Y.; He, Y.; Di Xu, F.; Hu, X.Y.; Zhu, X.M.; et al. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci. Rep. 2016, 6, 18977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ian, E.; Malko, D.B.; Sekurova, O.N.; Bredholt, H.; Rückert, C.; Borisova, M.E.; Albersmeier, A.; Kalinowski, J.; Gelfand, M.S.; Zotchev, S.B. Genomics of Sponge-Associated Streptomyces spp. Closely Related to Streptomyces albus J1074: Insights into Marine Adaptation and Secondary Metabolite Biosynthesis Potential. PLoS ONE 2014, 9, e96719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomihama, T.; Nishi, Y.; Sakai, M.; Ikenaga, M.; Okubo, T.; Ikeda, S. Draft genome sequences of Streptomyces scabiei S58, Streptomyces turgidiscabies T45, and Streptomyces acidiscabies a10, the pathogens of potato common scab, isolated in Japan. Genome Announc. 2016, 4, e00062-16. [Google Scholar] [CrossRef] [Green Version]
- Worsley, S.F.; Newitt, J.; Rassbach, J.; Batey, S.F.D.; Holmes, N.A.; Murrell, J.C.; Wilkinson, B.; Hutchings, M.I. Streptomyces endophytes promote host health and enhance growth across plant species. Appl. Environ. Microbiol. 2020, 86, e01053-20. [Google Scholar] [CrossRef]
- Vicente, C.M.; Santos-Aberturas, J.; Payero, T.D.; Barreales, E.G.; de Pedro, A.; Aparicio, J.F. PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 2014, 98, 9311–9324. [Google Scholar] [CrossRef] [PubMed]
- Payero, T.D.; Vicente, C.M.; Rumbero, Á.; Barreales, E.G.; Santos-Aberturas, J.; de Pedro, A.; Aparicio, J.F. Functional analysis of filipin tailoring genes from Streptomyces filipinensis reveals alternative routes in filipin III biosynthesis and yields bioactive derivatives. Microb. Cell Factories 2015, 14, 114. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, G.B.; Brock, T.D.; Ammann, A.; Gottlieb, D.; Carter, H.E. Filipin, an Antifungal Antibiotic: Isolation and Properties. J. Am. Chem. Soc. 1955, 77, 4799–4801. [Google Scholar] [CrossRef]
- Keller, U.; Lang, M.; Crnovcic, I.; Pfennig, F.; Schauwecker, F. The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: A genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 2010, 192, 2583–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crnovčić, I.; Rückert, C.; Semsary, S.; Lang, M.; Kalinowski, J.; Keller, U. Genetic interrelations in the actinomycin biosynthetic gene clusters of Streptomyces antibioticus IMRU 3720 and Streptomyces chrysomallus ATCC11523, producers of actinomycin X and actinomycin C. Adv. Appl. Bioinform. Chem. 2017, 10, 29–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, A.S.; Pearson, A.; Sáenz, J.P.; Marx, C.J. Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis. Org. Geochem. 2010, 41, 1075–1081. [Google Scholar] [CrossRef]
- Doughty, D.M.; Dieterle, M.; Sessions, A.L.; Fischer, W.W.; Newman, D.K. Probing the Subcellular Localization of Hopanoid Lipids in Bacteria Using NanoSIMS. PLoS ONE 2014, 9, e84455. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; WASHIO, T.; SATO, M.; SUZUKI, Y. Cytotoxic Effects of Several Hopanoids on Mouse Leukemia L1210 and P388 Cells. Biol. Pharm. Bull. 1995, 18, 421–423. [Google Scholar] [CrossRef] [Green Version]
- Moreau, R.A.; Hicks, K.B. Bacteriohopanetetrol and Related Compounds Useful for Modulation of Lipoxygenase Activity and Anti-Inflammatory Applications. U.S. Patent No US6177415B1, 23 January 2001. [Google Scholar]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L.; et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar] [CrossRef] [Green Version]
- Giessen, T.W.; Franke, K.B.; Knappe, T.A.; Kraas, F.I.; Bosello, M.; Xie, X.; Linne, U.; Marahiel, M.A. Isolation, structure elucidation, and biosynthesis of an unusual hydroxamic acid ester-containing siderophore from Actinosynnema mirum. J. Nat. Prod. 2012, 75, 905–914. [Google Scholar] [CrossRef]
- Barona-Gómez, F.; Wong, U.; Giannakopulos, A.E.; Derrick, P.J.; Challis, G.L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, M.; Zhang, Q.; Zhang, X.; Yang, P.; Liu, Z.; Deng, Y.; Zhu, Y.; Huang, X.; Han, L.; et al. Diisonitrile Natural Product SF2768 Functions as a Chalkophore That Mediates Copper Acquisition in Streptomyces thioluteus. ACS Chem. Biol. 2017, 12, 3067–3075. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the actinomycin D biosynthetic pathway from marine-derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef]
- Hassan, A.M.E.; Fahal, A.H.; Ahmed, A.O.; Ismail, A.; Veress, B. The immunopathology of actinomycetoma lesions caused by Streptomyces somaliensis. Trans. R. Soc. Trop. Med. Hyg. 2001, 95, 89–92. [Google Scholar] [CrossRef]
- Shah, S.; Briken, V. Modular Organization of the ESX-5 Secretion System in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2016, 6, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumas, E.; Boritsch, E.C.; Vandenbogaert, M.; De La Vega, R.C.R.; Thiberge, J.M.; Caro, V.; Gaillard, J.L.; Heym, B.; Girard-Misguich, F.; Brosch, R.; et al. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 2016, 8, 387–402. [Google Scholar] [CrossRef]
- Ju, K.S.; Gao, J.; Doroghazi, J.R.; Wang, K.K.A.; Thibodeaux, C.J.; Li, S.; Metzger, E.; Fudala, J.; Su, J.; Zhang, J.K.; et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. USA 2015, 112, 12175–12180. [Google Scholar] [CrossRef] [Green Version]
- Koonin, E.V.; Galperin, M.Y. Sequence-Evolution-Function: Computational Approaches in Comparative Genomics. In Sequence-Evolution-Function: Computational Approaches in Comparative Genomics; Springer: Boston, MA, USA, 2003; ISBN 1-40207-274-0. [Google Scholar]
- Karoonuthaisiri, N.; Weaver, D.; Huang, J.; Cohen, S.N.; Kao, C.M. Regional organization of gene expression in Streptomyces coelicolor. Gene 2005, 353, 53–66. [Google Scholar] [CrossRef]
- Gaillard, J.-L.; Berche, P.; Frehei, C.; Gouin, E.; Cossartt, P. Entry of L. monocytogenes into Cells Is Mediated by Internalin, a Repeat Protein Reminiscent of Surface Antigens from Gram-Positive Cocci. Cell 1991, 65, 1127–1141. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uozomi, T.; Beppu, T. Cloning and characterization of the streptothricin-resistance gene which encodes streptothricin acetyltransferase from Streptomyces lavendulae. J. Antibiot. 1986, 39, 688–693. [Google Scholar] [CrossRef] [Green Version]
- Rajasekaran, M.B.; Nilapwar, S.; Andrews, S.C.; Watson, K.A. EfeO-cupredoxins: Major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals 2010, 23, 1. [Google Scholar] [CrossRef]
- Chew, S.Y.; Chee, W.J.Y.; Than, L.T.L. The glyoxylate cycle and alternative carbon metabolism as metabolic adaptation strategies of Candida glabrata: Perspectives from Candida albicans and Saccharomyces cerevisiae. J. Biomed. Sci. 2019, 26, 52. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puckett, S.; Trujillo, C.; Wang, Z.; Eoh, H.; Ioerger, T.R.; Krieger, I.; Sacchettini, J.; Schnappinger, D.; Rhee, K.Y.; Ehrt, S. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, E2225–E2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koedooder, C.; Guéneuguès, A.; Van Geersdaële, R.; Vergé, V.; Bouget, F.-Y.; Labreuche, Y.; Obernosterer, I.; Blain, S. The Role of the Glyoxylate Shunt in the Acclimation to Iron Limitation in Marine Heterotrophic Bacteria. Front. Mar. Sci. 2018, 5, 435. [Google Scholar] [CrossRef] [Green Version]
- Flores-Díaz, M.; Monturiol-Gross, L.; Naylor, C.; Alape-Girón, A.; Flieger, A. Bacterial Sphingomyelinases and Phospholipases as Virulence Factors. Microbiol. Mol. Biol. Rev. 2016, 80, 597–628. [Google Scholar] [CrossRef] [Green Version]
- van der Meer-Janssen, Y.P.M.; van Galen, J.; Batenburg, J.J.; Helms, J.B. Lipids in host-pathogen interactions: Pathogens exploit the complexity of the host cell lipidome. Prog. Lipid Res. 2010, 49, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Bacterial phospholipases and their role in virulence. Trends Microbiol. 1997, 5, 156–161. [Google Scholar] [CrossRef]



| Streptomyces sp. TR1341 | Streptomyces sp. Endophyte_N2 | S. somaliensis DSM 40738 | |
|---|---|---|---|
| Genome size (Mb) | 8,507,620 | 8,428,700 | 5,176,903 |
| Contigs | 170 | 1 | 243 |
| GC content (%) | 71.80 | 71.83 | 74.20 |
| CDS (Prokka) | 7442 | 7253 | 4613 |
| rRNA (5S/16S/23S) | 7/8/3 | 7/7/7 | 6/6/6 |
| tRNA | 90 | 90 | 70 |
| tmRNA | 1 | 1 | 1 |
| CRISPR | 3 | 3 | 1 |
| BGC Type | BGC | Most Similar Known BGC | Genes Showing Similarity (%) |
|---|---|---|---|
| bacteriocin | Region 29.1 | Informatipeptin | 28 |
| Region 6.2 | - | - | |
| betalactone, PKS-like | Region 5.2 | bafilomycin B1 | 11 |
| ectoine | Region 67.1 | Ectoine | 100 |
| hglE-KS | Region 39.1 | Primycin | 5 |
| lanthipeptide | Region 23.1 | - | - |
| melanin | Region 84.1 | Melanin | 60 |
| NRPS | Region 116.1 | rhizomide A/rhizomide B/rhizomide C | 100 |
| Region 139.1 | rhizomide A/rhizomide B/rhizomide C | 100 | |
| Region 15.1 | Mirubactin | 78 | |
| Region 1.2 | diisonitrile antibiotic SF2768 | 66 | |
| Region 76.1 | Stenothricin | 18 | |
| Region 55.1 | acyldepsipeptide 1 | 15 | |
| Region 2.1 | Bleomycin | 9 | |
| Region 25.1 | caniferolide A/caniferolide B/caniferolide C/caniferolide D | 4 | |
| Region 30.2 | - | - | |
| Region 109.1 | - | - | |
| NRPS, betalactone | Region 17.1 | Kirromycin | 16 |
| Region 34.1 | formicamycins A–M | 6 | |
| NRPS, ectoine | Region 3.1 | Showdomycin | 23 |
| NRPS, other | Region 11.1 | actinomycin D | 89 |
| NRPS, siderophore | Region 30.1 | salinosporamide A | 16 |
| NRPS, T1PKS | Region 1.3 | Pentamycin, Filipin | 100 |
| NRPS, T1PKS, transAT-PKS-like | Region 2.2 | cinnabaramide A | 18 |
| NRPS-like | Region 162.1 | rhizomide A/rhizomide B/rhizomide C | 100 |
| Region 18.1 | Paulomycin | 13 | |
| NRPS-like, T1PKS | Region 1.1 | Borrelidin | 9 |
| other, lanthipeptide | Region 10.2 | A-503083 A/A-503083 B/A-503083 E/A-503083 F | 7 |
| PKS-like, T1PKS, other | Region 6.3 | Meilingmycin | 5 |
| siderophore | Region 64.1 | Desferrioxamine | 66 |
| T1PKS | Region 77.1 | Catenulisporolides | 3 |
| T1PKS, NRPS | Region 112.1 | - | - |
| T1PKS, NRPS, terpene | Region 10.1 | Ebelactone | 5 |
| T1PKS, siderophore, NRPS | Region 20.1 | Kinamycin | 22 |
| T2PKS, T1PKS | Region 13.1 | spore pigment | 83 |
| T3PKS | Region 27.1 | Herboxidiene | 7 |
| T3PKS, NRPS | Region 92.1 | A-47934 | 26 |
| terpene | Region 6.1 | Geosmin | 100 |
| Region 26.1 | julichrome Q3-3/julichrome Q3-5 | 25 | |
| Region 24.1 | - | - | |
| Region 43.1 | - | - | |
| terpene, thiopeptide, LAP | Region 5.1 | Hopene | 92 |
| Origin | Organism | Strain | No. MleD | No. MleE | No. Mce Operons |
|---|---|---|---|---|---|
| Soil/plant | Streptomyces sp. | TR1341 | - | - | - |
| Streptomyces sp. | endophyte_N2 | - | - | - | |
| Streptomyces griseofuscus | NG1-7 | - | - | - | |
| Streptomyces griseofuscus | NRRL B-5429 | - | - | - | |
| Streptomyces griseofuscus | g64 | - | - | - | |
| Streptomyces malaysiense | MUSC 136 | - | - | - | |
| Streptomyces murinus | NRRL B-2286 | - | - | - | |
| Streptomyces sp. | LamerLS-31b | - | - | - | |
| Human | Streptomyces sp. | KE1 | 6 | 2 | 1 |
| Streptomyces brasiliensis | NRRL B-1626 | 6 | 2 | 1 | |
| Streptomyces somaliensis | DSM 40738 | - | - | - | |
| Human | Mycobacterium tuberculosis | H37Rv | 24 | 8 | 4 |
| Mycobacterium smegmatis | MC2-155 | 38 | 12 | 6 | |
| Mycobacterium simiae | MsiGto | 56 | 16 | 9 | |
| Human | Gordonia bronchialis | DSM 43247 | 29 | 9 | 5 |
| Human | Tsukamurella pulmonis | DSM 44142 | 25 | 8 | 4 |
| Tsukamurella tyrosinosolvens | NCTC13231 | 19 | 6 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lara, A.C.; Corretto, E.; Kotrbová, L.; Lorenc, F.; Petříčková, K.; Grabic, R.; Chroňáková, A. The Genome Analysis of the Human Lung-Associated Streptomyces sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues. Microorganisms 2021, 9, 1547. https://doi.org/10.3390/microorganisms9081547
Lara AC, Corretto E, Kotrbová L, Lorenc F, Petříčková K, Grabic R, Chroňáková A. The Genome Analysis of the Human Lung-Associated Streptomyces sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues. Microorganisms. 2021; 9(8):1547. https://doi.org/10.3390/microorganisms9081547
Chicago/Turabian StyleLara, Ana Catalina, Erika Corretto, Lucie Kotrbová, František Lorenc, Kateřina Petříčková, Roman Grabic, and Alica Chroňáková. 2021. "The Genome Analysis of the Human Lung-Associated Streptomyces sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues" Microorganisms 9, no. 8: 1547. https://doi.org/10.3390/microorganisms9081547
APA StyleLara, A. C., Corretto, E., Kotrbová, L., Lorenc, F., Petříčková, K., Grabic, R., & Chroňáková, A. (2021). The Genome Analysis of the Human Lung-Associated Streptomyces sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues. Microorganisms, 9(8), 1547. https://doi.org/10.3390/microorganisms9081547