Phagotrophic Protists and Their Associates: Evidence for Preferential Grazing in an Abiotically Driven Soil Ecosystem
Abstract
1. Introduction
2. Methods
2.1. Shotgun Metagenome Sequencing and rDNA Identification
2.2. Constructing Association Networks
2.3. Comparing the Influence of Abiotic and Biotic Factors on Protist Distribution
2.4. Inferring Biological Significance from Network Associations
3. Results
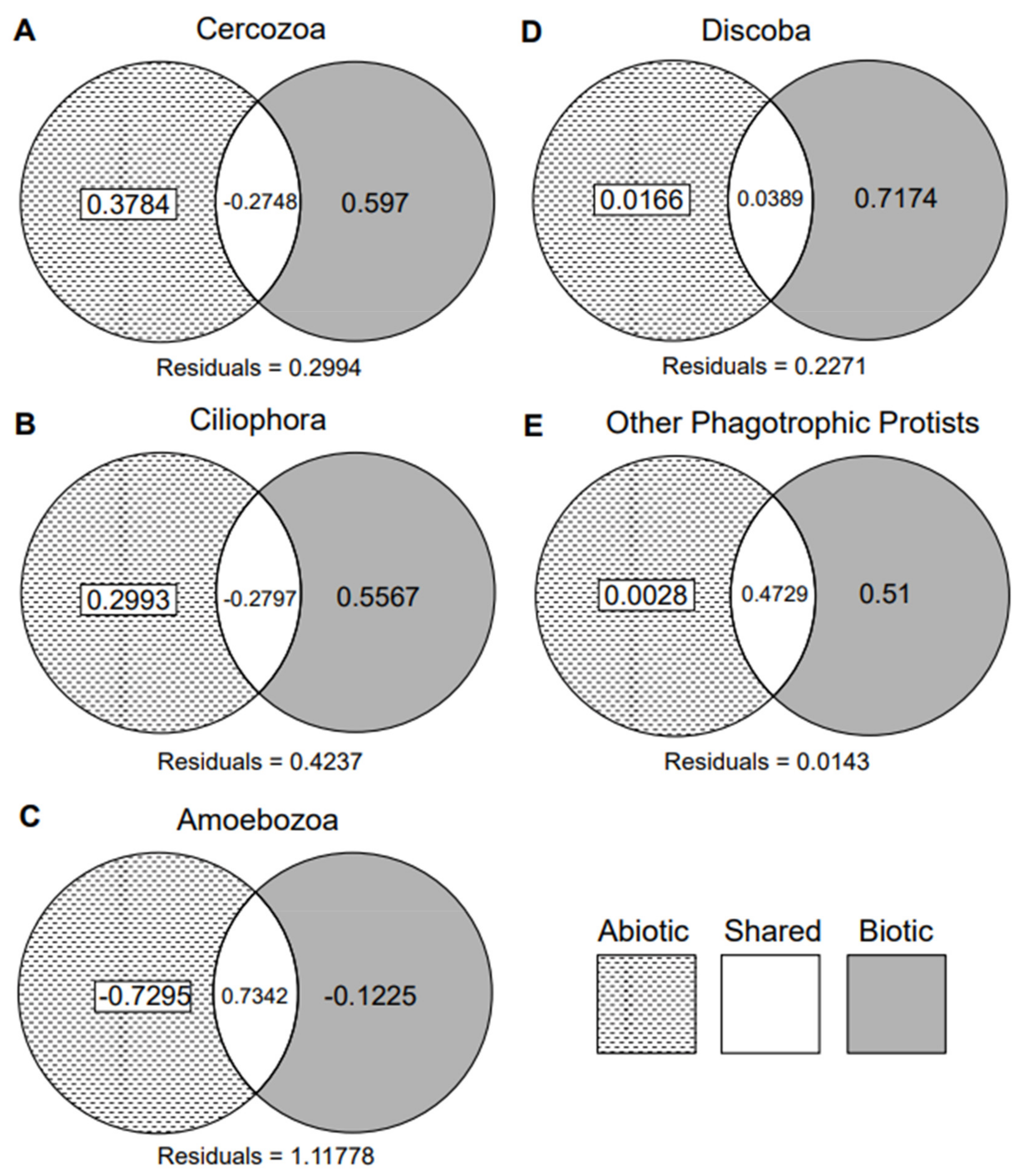
3.1. Biotic vs. Abiotic Drivers
3.2. Trends in Network Analyses
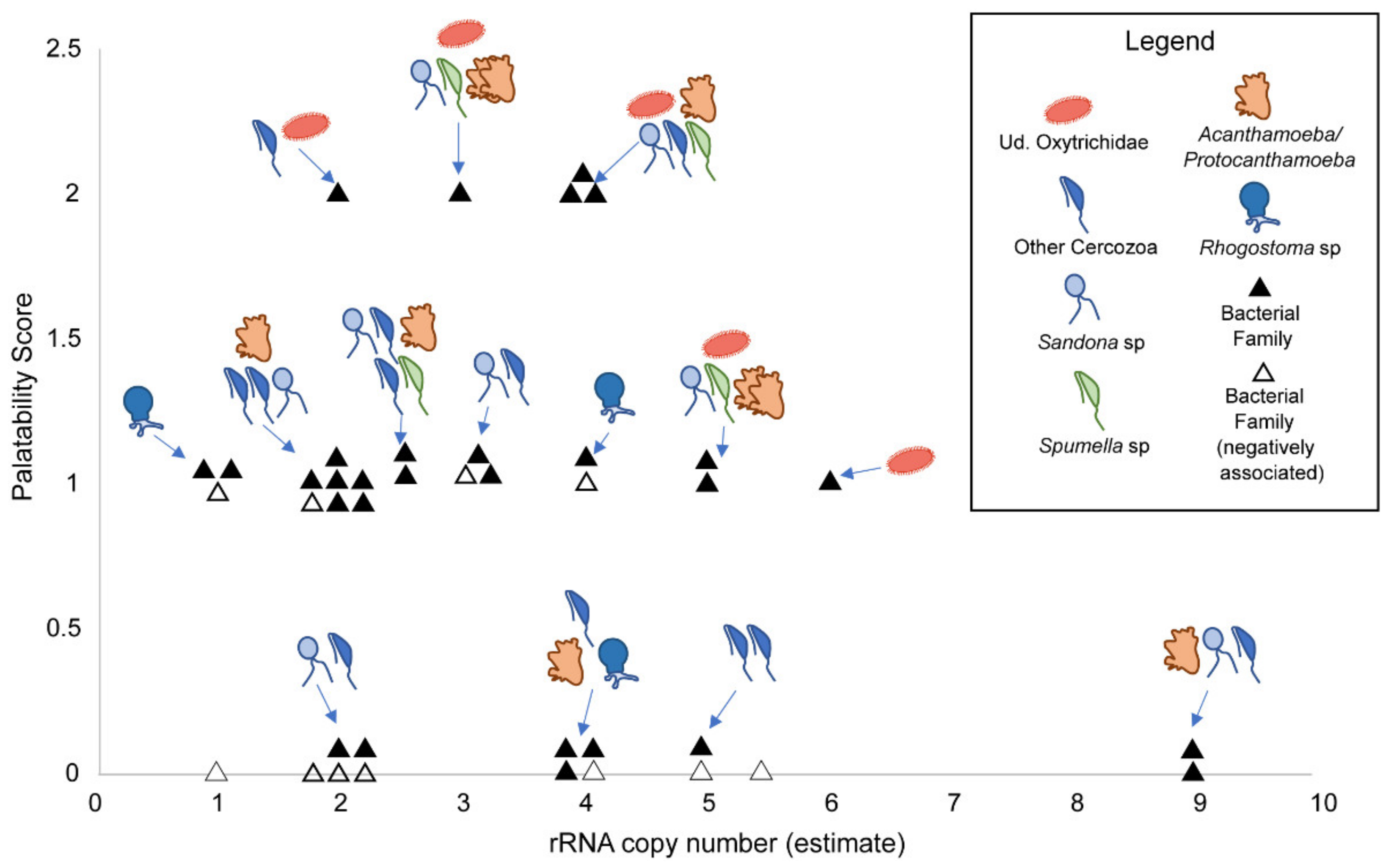
3.3. Associations with Eukaryotes
3.4. Associations with Bacteria
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wardle, D.A. The influence of biotic interactions on soil biodiversity. Ecol. Lett. 2006, 9, 870–886. [Google Scholar] [CrossRef] [PubMed]
- Heemsbergen, D.A.; Berg, M.P.; Loreau, M.; van Hal, J.R.; Faber, J.H.; Verhoef, H.A. Biodiversity effects on soil processes explained by interspecific functional dissimilarity. Science 2004, 306, 1019–1020. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.A.D.; Adl, S.; Bonkowski, M.; Dunthorn, M.; Ekelund, F.; Fernandez, L.D.; Jousset, A.; Krashevska, V.; Singer, D.; et al. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 2018, 42, 293–323. [Google Scholar] [CrossRef]
- Seppey, C.V.W.; Singer, D.; Dumack, K.; Fournier, B.; Belbahri, L.; Mitchell, E.A.D.; Lara, E. Distribution patterns of soil microbial eukaryotes suggests widespread algivory by phagotrophic protists as an alternative pathway for nutrient cycling. Soil Biol. Biochem. 2017, 112, 68–76. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Leff, J.W.; Barberan, A.; Bates, S.T.; Betley, J.; Crowther, T.W.; Kelly, E.F.; Oldfield, E.E.; Shaw, E.A.; Steenbock, C.; et al. Biogeographic patterns in below-ground diversity in New York City’s Central Park are similar to those observed globally. Proc. Biol. Sci. 2014, 281. [Google Scholar] [CrossRef]
- Wilkinson, D.M.; Creevy, A.L.; Valentine, J. The past, present and future of soil protist ecology. Acta Protozool. 2012, 51, 189–199. [Google Scholar] [CrossRef]
- Crotty, F.V.; Adl, S.M.; Blackshaw, R.P.; Murray, P.J. Protozoan pulses unveil their pivotal position within the soil food web. Microb. Ecol. 2012, 63, 905–918. [Google Scholar] [CrossRef]
- Saleem, M.; Fetzer, I.; Dormann, C.F.; Harms, H.; Chatzinotas, A. Predator richness increases the effect of prey diversity on prey yield. Nat. Commun. 2012, 3, 1305. [Google Scholar] [CrossRef] [PubMed]
- Erktan, A.; Or, D.; Scheu, S. The physical structure of soil: Determinant and consequence of trophic interactions. Soil Biol. Biochem. 2020, 148, 107876. [Google Scholar] [CrossRef]
- Geisen, S. The bacterial-fungal energy channel concept challenged by enormous functional versatility of soil protists. Soil Biol. Biochem. 2016, 102, 22–25. [Google Scholar] [CrossRef]
- Thakur, M.P.; Geisen, S. Trophic regulations of the soil microbiome. Trends Microbiol. 2019, 27, 771–780. [Google Scholar] [CrossRef]
- Neidig, N.; Jousset, A.; Nunes, F.; Bonkowski, M.; Paul, R.J.; Scheu, S. Interference between bacterial feeding nematodes and amoebae relies on innate and inducible mutual toxicity. Funct. Ecol. 2010, 24, 1133–1138. [Google Scholar] [CrossRef]
- Trap, J.; Bonkowski, M.; Plassard, C.; Villenave, C.; Blanchart, E. Ecological importance of soil bacterivores for ecosystem functions. Plant Soil 2016, 398, 1–24. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.; Wilkinson, D.; Adl, S.; Bonkowski, M.; Brown, M.; Fiore-Donno, A.M.; Heger, T.; Jassey, V.; Krashevska, V.; et al. Soil protistology rebooted: 30 fundamental questions to start with. Soil Biol. Biochem. 2017, 111, 94–103. [Google Scholar] [CrossRef]
- Glucksman, E.; Bell, T.; Griffiths, R.I.; Bass, D. Closely related protist strains have different grazing impacts on natural bacterial communities. Environ. Microbiol. 2010, 12, 3105–3113. [Google Scholar] [CrossRef]
- Adams, B.J.; Bardgett, R.D.; Ayres, E.; Wall, D.H.; Aislabie, J.; Bamforth, S.; Bargagli, R.; Cary, C.; Cavacini, P.; Connell, L.; et al. Diversity and distribution of Victoria Land biota. Soil Biol. Biochem. 2006, 38, 3003–3018. [Google Scholar] [CrossRef]
- Allesina, S.; Bodini, A.; Pascual, M. Functional links and robustness in food webs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Karlsson, I.; Geisen, S.; Kowalchuk, G.; Jousset, A. Protists: Puppet masters of the rhizosphere microbiome. Trends Plant Sci. 2019, 24, 165–176. [Google Scholar] [CrossRef]
- Wu, T.; Ayres, E.; Li, G.; Bardgett, R.D.; Wall, D.H.; Garey, J.R. Molecular profiling of soil animal diversity in natural ecosystems: Incongruence of molecular and morphological results. Soil Biol. Biochem. 2009, 41, 849–857. [Google Scholar] [CrossRef]
- Decaëns, T. Macroecological patterns in soil communities. Glob. Ecol. Biogeogr. 2010, 19, 287–302. [Google Scholar] [CrossRef]
- Bardgett, R.D. Causes and consequences of biological diversity in soil. Zoology 2002, 105, 367–374. [Google Scholar] [CrossRef]
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Bamforth, S.S.; Wall, D.H.; Virginia, R.A. Distribution and diversity of soil protozoa in the McMurdo Dry Valleys of Antarctica. Polar Biol. 2005, 28, 756–762. [Google Scholar] [CrossRef]
- Thompson, A.R.; Geisen, S.; Adams, B.J. Shotgun metagenomics reveal a diverse assemblage of protists in a model Antarctic soil ecosystem. Environ. Microbiol. 2020, 22, 4620–4632. [Google Scholar] [CrossRef]
- Burton-Johnson, A.; Black, M.; Fretwell, P.T.; Kaluza-Gilbert, J. An automated methodology for differentiating rock from snow, clouds and sea in Antarctica from Landsat 8 imagery: A new rock outcrop map and area estimation for the entire Antarctic continent. Cryosphere 2016, 10, 1665–1677. [Google Scholar] [CrossRef]
- Levy, J. How big are the McMurdo Dry Valleys? Estimating ice-free area using Landsat image data. Antarct. Sci. 2012, 25, 119–120. [Google Scholar] [CrossRef]
- Cary, S.C.; McDonald, I.R.; Barrett, J.E.; Cowan, D.A. On the rocks: The microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 2010, 8, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.E.; Virginia, R.A.; Hopkins, D.W.; Aislabie, J.; Bargagli, R.; Bockheim, J.G.; Campbell, I.B.; Lyons, W.B.; Moorhead, D.L.; Nkem, J.N.; et al. Terrestrial ecosystem processes of Victoria Land, Antarctica. Soil Biol. Biochem. 2006, 38, 3019–3034. [Google Scholar] [CrossRef]
- Wall, D.H. Global change tipping points: Above- and below-ground biotic interactions in a low diversity ecosystem. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 2291–2306. [Google Scholar] [CrossRef]
- Burkins, M.B.; Virginia, R.A.; Wall, D.H. Organic carbon cycling in Taylor Valley, Antarctica: Quantifying soil reservoirs and soil respiration. Glob. Chang. Biol. 2001, 7, 113–125. [Google Scholar] [CrossRef]
- Virginia, R.A.; Wall, D.H. How soils structure communities in the Antarctic Dry Valleys. Bioscience 1999, 49, 973–983. [Google Scholar] [CrossRef]
- Barrett, J.E.; Virginia, R.A.; Wall, D.H.; Parsons, A.H.; Powers, L.E.; Burkins, M.B. Variation in biochemistry and soil biodiversity across spatial scales in a polar desert ecosystem. Ecology 2004, 85, 3105–3118. [Google Scholar] [CrossRef]
- Wall, D.H. Biodiversity and ecosystem functioning in terrestrial habitats of Antarctica. Antarct. Sci. 2005, 17, 523–531. [Google Scholar] [CrossRef]
- Barrett, J.E.; Virginia, R.A.; Wall, D.H.; Cary, S.C.; Adams, B.J.; Hacker, A.L.; Aislabie, J.M. Co-variation in soil biodiversity and biogeochemistry in northern and southern Victoria Land, Antarctica. Antarct. Sci. 2006, 18, 1–14. [Google Scholar] [CrossRef]
- Lee, C.K.; Laughlin, D.C.; Bottos, E.M.; Caruso, T.; Joy, K.; Barrett, J.E.; Brabyn, L.; Nielsen, U.N.; Adams, B.J.; Wall, D.H.; et al. Biotic interactions are an unexpected yet critical control on the complexity of an abiotically driven polar ecosystem. Commun. Biol. 2019, 2, 62. [Google Scholar] [CrossRef]
- Caruso, T.; Hogg, I.D.; Nielsen, U.N.; Bottos, E.M.; Lee, C.K.; Hopkins, D.W.; Cary, S.C.; Barrett, J.E.; Green, T.G.A.; Storey, B.C.; et al. Nematodes in a polar desert reveal the relative role of biotic interactions in the coexistence of soil animals. Commun. Biol. 2019, 2, 63. [Google Scholar] [CrossRef]
- Lekfeldt, J.D.S.; Rønn, R. A common soil flagellate (Cercomonas sp.) grows slowly when feeding on the bacterium Rhodococcus fascians in isolation, but does not discriminate against it in a mixed culture with Sphingopyxis witflariensis. FEMS Microbiol. Ecol. 2008, 65, 113–124. [Google Scholar] [CrossRef][Green Version]
- Foissner, W. Faunistics, taxonomy and ecology of moss and soil ciliates (Protozoa, Ciliophora) from Antarctica, with description of new species, including Pleuroplitoides smithi gen. n., sp. n. Acta Protozool. 1996, 35, 95–123. [Google Scholar]
- Corno, G.; Jurgens, K. Structural and functional patterns of bacterial communities in response to protist predation along an experimental productivity gradient. Environ. Microbiol. 2008, 10, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
- Bjørnlund, L.; Rønn, R. ‘David and Goliath’ of the soil food web—Flagellates that kill nematodes. Soil Biol. Biochem. 2008, 40, 2032–2039. [Google Scholar] [CrossRef]
- Rønn, R.; Vestergård, M.; Ekelund, F. Interactions between bacteria, protozoa and nematodes in soil. Acta Protozool. 2012, 51, 223–235. [Google Scholar] [CrossRef]
- Bjørnlund, L.; Mørk, S.; Vestergård, M.; Rønn, R. Trophic interactions between rhizosphere bacteria and bacterial feeders influenced by phosphate and aphids in barley. Biol. Fertil. Soils 2006, 43, 1–11. [Google Scholar] [CrossRef]
- Rønn, R.; McCaig, A.E.; Griffiths, B.S.; Prosser, J.I. Impact of protozoan grazing on bacterial community structure in soil microcosms. Appl. Environ. Microbiol. 2002, 68, 6094–6105. [Google Scholar] [CrossRef]
- Murase, J.; Noll, M.; Frenzel, P. Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl. Environ. Microbiol. 2006, 72, 5436–5444. [Google Scholar] [CrossRef] [PubMed]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef]
- Courtright, E.M.; Wall, D.H.; Virginia, R.A. Determining habitat suitability for soil invertebrates in an extreme environment: The McMurdo Dry Valleys, Antarctica. Antarct. Sci. 2001, 13, 9–17. [Google Scholar] [CrossRef]
- Shaw, E.A.; Adams, B.; Barrett, J.; Lyons, W.; Virginia, R.; Wall, D. Stable C and N isotope ratios reveal soil food web structure and identify the nematode Eudorylaimus antarcticus as an omnivore–predator in Taylor Valley, Antarctica. Polar Biol. 2018, 41, 1013–1018. [Google Scholar] [CrossRef]
- Raoult, D.; Boyer, M. Amoebae as Genitors and Reservoirs of Giant Viruses. Intervirology 2010, 53, 321–329. [Google Scholar] [CrossRef]
- Hamels, I.; Mussche, H.; Sabbe, K.; Muylaert, K.; Vyverman, W. Evidence for constant and highly specific active food selection by benthic ciliates in mixed diatoms assemblages. Limnol. Oceanogr. 2004, 49, 58–68. [Google Scholar] [CrossRef]
- Velasco-Castrillón, A.; Gibson, J.A.E.; Stevens, M.I. A review of current Antarctic limno-terrestrial microfauna. Polar Biol. 2014, 37, 1517–1531. [Google Scholar] [CrossRef]
- Velasco-Castrillón, A.; McInnes, S.J.; Schultz, M.B.; Arróniz-Crespo, M.; D’Haese, C.A.; Gibson, J.A.E.; Adams, B.J.; Page, T.J.; Austin, A.D.; Cooper, S.J.B.; et al. Mitochondrial DNA analyses reveal widespread tardigrade diversity in Antarctica. Invertebr. Syst. 2015, 29, 578–590. [Google Scholar] [CrossRef]
- Schulz-Bohm, K.; Geisen, S.; Wubs, E.R.; Song, C.; de Boer, W.; Garbeva, P. The prey’s scent—Volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J. 2017, 11, 817–820. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, C.; Mouillot, D.; Kulbicki, M.; Gravel, D. Extensions of Island Biogeography Theory predict the scaling of functional trait composition with habitat area and isolation. Ecol. Lett. 2017, 20, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Aanderud, Z.T.; Saurey, S.; Ball, B.A.; Wall, D.H.; Barrett, J.E.; Muscarella, M.E.; Griffin, N.A.; Virginia, R.A.; Adams, B.J. Frontiers in Ecology and the Environment. Front. Microbiol. 2018, 9, 1401. [Google Scholar] [CrossRef]
- Knox, M.A.; Wall, D.H.; Virginia, R.A.; Vandegehuchte, M.L.; Gil, I.S.; Adams, B.J. Impact of diurnal freeze–thaw cycles on the soil nematode Scottnema lindsayae in Taylor Valley, Antarctica. Polar Biol. 2015, 39, 583–592. [Google Scholar] [CrossRef]
- Veech, J.A. A probabilistic model for analysing species co-occurrence. Glob. Ecol. Biogeogr. 2013, 22, 252–260. [Google Scholar] [CrossRef]
- Veech, J.A. The pairwise approach to analysing species co-occurrence. J. Biogeogr. 2014, 41, 1029–1035. [Google Scholar] [CrossRef]
- Trivellone, V.; Bougeard, S.; Giavi, S.; Krebs, P.; Balseiro, D.; Dray, S.; Moretti, M. Factors shaping community assemblages and species co-occurrence of different trophic levels. Ecol. Evol. 2017, 7, 4745–4754. [Google Scholar] [CrossRef]
- Goberna, M.; Montesinos-Navarro, A.; Valiente-Banuet, A.; Colin, Y.; Gómez-Fernández, A.; Donat, S.; Navarro-Cano, J.A.; Verdú, M. Incorporating phylogenetic metrics to microbial co-occurrence networks based on amplicon sequences to discern community assembly processes. Mol. Ecol. Resour. 2019, 19, 1552–1564. [Google Scholar] [CrossRef]
- Thurman, L.L.; Barner, A.K.; Garcia, T.S.; Chestnut, T. Testing the link between species interactions and species co-occurrence in a trophic network. Ecography 2019, 42, 1658–1670. [Google Scholar] [CrossRef]
- Freilich, M.A.; Wieters, E.; Broitman, B.R.; Marquet, P.A.; Navarrete, S.A. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities? Ecology 2018, 99, 690–699. [Google Scholar] [CrossRef]
- Blanchet, F.G.; Cazelles, K.; Gravel, D. Co-occurrence is not evidence of ecological interactions. Ecol. Lett. 2020, 23, 1050–1063. [Google Scholar] [CrossRef]
- Peres-Neto, P.R.; Legendre, P.; Dray, S.; Borcard, D. Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology 2006, 87, 2614–2625. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hartmann, M.; Eriksson, K.M.; Pal, C.; Thorell, K.; Larsson, D.G.; Nilsson, R.H. METAXA2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 2015, 15, 1403–1414. [Google Scholar] [CrossRef]
- Eddy, S.R. HMMER: Biosequence Analysis Using Profile Hidden Markov Models. Available online: http://hmmer.org/ (accessed on 29 June 2021).
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Sayers, E.W.; Beck, J.; Bolton, E.E.; Bourexis, D.; Brister, J.R.; Canese, K.; Comeau, D.C.; Funk, K.; Kim, S.; Klimke, W.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef]
- Guo, J.; Cole, J.R.; Zhang, Q.; Brown, C.T.; Tiedje, J.M. Microbial community analysis with ribosomal gene fragments from shotgun metagenomes. Appl. Environ. Microbiol. 2016, 82, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.B.; Wallroth, M.; Jonsson, V.; Kristiansson, E. Comparison of normalization methods for the analysis of metagenomic gene abundance data. BMC Genom. 2018, 19, 274. [Google Scholar] [CrossRef]
- Wilke, A.; Bischof, J.; Harrison, T.; Brettin, T.; D’Souza, M.; Gerlach, W.; Matthews, H.; Paczian, T.; Wilkening, J.; Glass, E.M.; et al. A RESTful API for accessing microbial community data for MG-RAST. PLoS Comput. Biol. 2015, 11, e1004008. [Google Scholar] [CrossRef]
- Bates, S.T.; Clemente, J.C.; Flores, G.E.; Walters, W.A.; Parfrey, L.W.; Knight, R.; Fierer, N. Global biogeography of highly diverse protistan communities in soil. ISME J. 2013, 7, 652–659. [Google Scholar] [CrossRef]
- Petz, W. Ecology of the active soil microfauna (protozoa, metazoa) of Wilkes Land Antarctica. Polar Biol. 1997, 18, 33–44. [Google Scholar] [CrossRef]
- Griffith, D.M.; Veech, J.A.; Marsh, C.J. cooccur: Probabilistic species co-occurrence analysis in R. J. Stat. Softw. 2016, 69, 1–17. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph software package for complex network research. InterJ. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology in R, 2nd ed.; Borcard, D., Gillet, F., Legendre, P., Eds.; Springer: New York, NY, USA, 2018. [Google Scholar]
- Mevik, B.-H. The pls package: Principal component and partial least squares regression in R. J. Stat. Softw. 2007, 18, 1–23. [Google Scholar] [CrossRef]
- Wold, S.; Eriksson, L. Statistical Validation of QSAR Results. In Methods and Principles in Medicinal Chemistry; de Waterbeemd, H.V., Ed.; VCH: Weinheim, Germany, 1995; pp. 309–338. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.4-1. 2016. Available online: https://CRAN.R-project.org/package=vegan (accessed on 29 June 2021).
- Blois, J.L.; Gotelli, N.J.; Behrensmeyer, A.K.; Faith, J.T.; Lyons, S.K.; Williams, J.W.; Amatangelo, K.L.; Bercovici, A.; Du, A.; Eronen, J.T.; et al. A framework for evaluating the influence of climate, dispersal limitation, and biotic interactions using fossil pollen associations across the late Quaternary. Ecography 2014, 37, 1095–1108. [Google Scholar] [CrossRef]
- R Core Development Team. R: A Language and Environment for Statistical Computing; R foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Gupta, R.S. Origin of diderm (Gram-negative) bacteria: Antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes. Antonie Van Leeuwenhoek 2011, 100, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B. (Ed.) Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Son, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Roller, B.R.K.; Stoddard, S.F.; Schmidt, T.M. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 2016, 1, 16160. [Google Scholar] [CrossRef]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.; Schmidt, T.M. rrnDB: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015, 43, D593–D598. [Google Scholar] [CrossRef]
- Berg, M.P.; Bengtsson, J. Temporal and spatial variability in soil food web structure. Oikos 2007, 116, 1789–1804. [Google Scholar] [CrossRef]
- Adams, B.; Wall, D.; Virginia, R.; Broos, E.; Knox, M. Ecological Biogeography of the Terrestrial Nematodes of Victoria Land, Antarctica. ZooKeys 2014, 419, 29–71. [Google Scholar] [CrossRef] [PubMed]
- Katsu-Kimura, Y.; Nakaya, F.; Baba, S.A.; Mogami, Y. Substantial energy expenditure for locomotion in ciliates verified by means of simultaneous measurement of oxygen consumption rate and swimming speed. J. Exp. Biol. 2009, 212, 1819–1824. [Google Scholar] [CrossRef]
- Howe, A.T.; Bass, D.; Chao, E.E.; Cavalier-Smith, T. New genera, species, and improved phylogeny of Glissomonadida (Cercozoa). Protist 2011, 162, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Venter, P.C.; Nitsche, F.; Arndt, H. The hidden diversity of flagellated protists in soil. Protist 2018, 169, 432–449. [Google Scholar] [CrossRef]
- Newsham, K.K.; Rolf, J.; Pearce, D.A.; Strachan, R.J. Differing preferences of Antarctic soil nematodes for microbial prey. Eur. J. Soil Bio. 2004, 40, 1–8. [Google Scholar] [CrossRef]
- Majdi, N.; Traunspurger, W.; Fueser, H.; Gansfort, B.; Laffaille, P.; Maire, A. Effects of a broad range of experimental temperatures on the population growth and body-size of five species of free-living nematodes. J. Therm. Biol. 2019, 80, 21–36. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, A.R.; Roth-Monzón, A.J.; Aanderud, Z.T.; Adams, B.J. Phagotrophic Protists and Their Associates: Evidence for Preferential Grazing in an Abiotically Driven Soil Ecosystem. Microorganisms 2021, 9, 1555. https://doi.org/10.3390/microorganisms9081555
Thompson AR, Roth-Monzón AJ, Aanderud ZT, Adams BJ. Phagotrophic Protists and Their Associates: Evidence for Preferential Grazing in an Abiotically Driven Soil Ecosystem. Microorganisms. 2021; 9(8):1555. https://doi.org/10.3390/microorganisms9081555
Chicago/Turabian StyleThompson, Andrew R., Andrea J. Roth-Monzón, Zachary T. Aanderud, and Byron J. Adams. 2021. "Phagotrophic Protists and Their Associates: Evidence for Preferential Grazing in an Abiotically Driven Soil Ecosystem" Microorganisms 9, no. 8: 1555. https://doi.org/10.3390/microorganisms9081555
APA StyleThompson, A. R., Roth-Monzón, A. J., Aanderud, Z. T., & Adams, B. J. (2021). Phagotrophic Protists and Their Associates: Evidence for Preferential Grazing in an Abiotically Driven Soil Ecosystem. Microorganisms, 9(8), 1555. https://doi.org/10.3390/microorganisms9081555





