Abstract
Streptomyces sp. TR1341 was isolated from the sputum of a man with a history of lung and kidney tuberculosis, recurrent respiratory infections, and COPD. It produces secondary metabolites associated with cytotoxicity and immune response modulation. In this study, we complement our previous results by identifying the genetic features associated with the production of these secondary metabolites and other characteristics that could benefit the strain during its colonization of human tissues (virulence factors, modification of the host immune response, or the production of siderophores). We performed a comparative phylogenetic analysis to identify the genetic features that are shared by environmental isolates and human respiratory pathogens. The results showed a high genomic similarity of Streptomyces sp. TR1341 to the plant-associated Streptomyces sp. endophyte_N2, inferring a soil origin of the strain. Putative virulence genes, such as mammalian cell entry (mce) genes were not detected in the TR1341’s genome. The presence of a type VII secretion system, distinct from the ones found in Mycobacterium species, suggests a different colonization strategy than the one used by other actinomycete lung pathogens. We identified a higher diversity of genes related to iron acquisition and demonstrated that the strain produces ferrioxamine B in vitro. These results indicate that TR1341 may have an advantage in colonizing environments that are low in iron, such as human tissue.
1. Introduction
Actinomycetes have become one of the most relevant groups of microorganisms since the discovery of antibiotics in the 1940s. They are gram-positive bacteria with high GC content, and comprise taxa known to be producers of a wide range of secondary metabolites. The Streptomyces genus is considered as the most relevant genus, since it has become the source of about two-thirds of all known natural antibiotics [1,2].
Even though streptomycetes are predominantly known as soil- and sediment-associated microorganisms, they have also been reported as free-living sea water bacteria [3,4], and have been found in extreme habitats [5,6,7,8] and as symbionts of plants [9], invertebrates [10,11,12,13], and turtles [14]. In most cases, their interaction with the host has been classified as mutualistic because streptomycetes protect the host against fungal or bacterial infections [15]. Nevertheless, some Streptomyces species cause diseases in plants through the production of phytotoxins and other specific metabolites. Streptomyces scabies, for instance, is the main causative agent of potato scab, which affects the roots and the tubers of the potato plant [16,17,18].
Due to their ability to produce antibacterials, antifungals, and immunomodulators as secondary metabolites, Streptomyces are generally viewed as a bioresource by humans. There are only a few Streptomyces species that are pathogenic under certain conditions: Streptomyces somalensis and Streptomyces sudanensis, which cause endemic actinomycetoma, for example [19,20].
Generally speaking, the main reservoir for human-associated Streptomyces strains is the soil. At the same time, contact with soil is an important factor in human health [21]. It is therefore not so farfetched to consider Streptomyces strains as potential control agents in the developing microbial communities as possible trainers of the human immune system [22,23]. Streptomyces species have been reported to inhabit healthy skin [24], the uterus [25], the gastrointestinal system [22], and the respiratory tract [26,27]. They are also suspected of mediating communication between the gut microbiota, the brain, and the lungs [28].
Albeit mostly soil dwellers, streptomycetes (and especially their spores) may be air-borne. Particularly in highly polluted areas with high particulate matter content, such as barns with hay storage, wet moldy houses, urban and industrial smog, or dusty air after soil and sand storm events [29,30,31]. Consequently, their spores can be inhaled easily and may start to germinate. It was shown that even at this development stage, Streptomyces coelicolor starts the production of secondary metabolites that not only control their growth but are also involved in signaling and communication with other organisms [32]. In vitro, Streptomyces was one of the main producers of cytotoxic and inflammatory compounds present in mold-infested houses [33]. Streptomyces californicus, along with Stachybotrys chartarum, are commonly isolated from water-damaged or mold-infested buildings together. It is hypothesized that these two organisms could be responsible for respiratory tract problems in patients living in water-damaged buildings [34]. So far, there are no reports of Streptomyces californicus isolation from lung tissue or sputum. However, its spores, in combination with Stachybotrys chartarum, induced an inflammatory response in macrophages of mice [35] and produced cytostatic compounds with immunotoxic properties [36]. Other clinical reports linked Streptomyces to pulmonary infections, abscesses of different organs, and bacteremias found mostly in immunocompromised patients and in a few immunocompetent individuals [37]. The limited data about the colonization of human tissues by non-pathogenic Streptomyces has recently been summarized by Herbrík et al. [27].
The human-colonization strategies of pathogenic actinomycetes are best studied in mycobacteria, which use mechanisms different to other bacterial pathogens. Host-adapted mycobacteria, such as Mycobacterium tuberculosis, do not employ virulence factors and toxins. Unlike other pathogenic bacteria, they have evolved specific strategies to outsmart the immune system and exploit the resources of the host [38]. A recently discovered mechanism used by actinomycetes for host immune evasion, modulation, and exploitation are the specialized Type VII secretion systems (T7SS), also called ESX [39]. Studied mostly in Mycobacterium but found also in Streptomyces, Corynebacterium, Nocardia, and Gordonia, ESX secretion systems allow the interaction of the infectious bacteria with the host immune system [40]. Mycobacterium tuberculosis has five paralogous T7S systems (ESX-1–ESX-5). ESX-1, together with horizontally acquired espACD loci, mediates the rupture of phagosomal membranes inside the host phagocytes. ESX-3 acts in metal ion homeostasis. ESX-5 induces caspase-independent cell death and facilitates the spread of the infection by modulating the innate immune response and inhibiting the production of cytokines in type I macrophages [41,42]. ESX-4 induces immunogenicity in mice characterized by significant lymphocyte proliferation and induction of TNF-α and IL-6. It is also the smallest and apparently the most ancestral ESX system in the genus Mycobacterium, with the other ESX systems evolving from it by gene duplication events and insertion. The putative function of ESX-2 remains unknown. So far, studies on Streptomyces scabies and Streptomyces coelicolor proved that their ESX system seems to be involved in sporulation and development rather than virulence [43,44].
Another important colonization mechanism is mediated by mammalian cell entry (MCE) proteins. They are a family of invasion proteins described in mycobacteria and present in some other Actinomycetales. MCE proteins allow nonpathogenic E. coli cells to enter and multiply within non-phagocytic cells [45]. The presence of mce genes in a genome does not guarantee the pathogenicity of the strain, but it highly correlates with it [45,46,47]. However, the deletion of the mce cluster in Streptomyces coelicolor A3(2) allowed for rapid germination of the spores inside the macrophage model Acanthamoeba polyphaga, but reduced the efficiency of colonization of the roots of Arabidopsis thaliana [46]. The enhanced virulence associated with a non-functioning mce operon was also observed by Shimono et al., who reported an increase in virulence in a mutant with a disruption in the mce-1 operon of M. tuberculosis [48]. Thus, these results suggest that if present before, the loss of the mce operon might increase the virulence of a given strain.
Once in the airways or lungs, microorganisms need a supply of essential trace elements, mostly iron, which in turn may unbalance the iron homeostasis of the host [49]. Invading microbes scavenge iron by secreting small molecules called siderophores. Siderophores have a large chemical structural diversity and are widely produced by streptomycetes [50]. They can chelate available iron ions, but also steal iron from iron-binding proteins produced by the human body, or by other members of the microbiome. This can lead to an antibiotic effect [51,52,53]. Siderophores can also be covalently linked to antibiotics forming sideromycins [54]. They are popular antibiotics because of the dual mechanism in which they function and most of them are produced by actinomycetes (albomycins, ferrimycins, danomycins, etc.) [51]. Other antibiotics produced by actinomycetes that also have metal biding characteristics are: streptomycin and novobiocin that bind Cu2+, erythromycin that binds Co2+, and tetracycline that binds Ca2+ and Fe2+ [55].
Streptomyces sp. TR1341 was isolated from the sputum of a senior man, who was living in the mining region of Příbram, Czechia, and had a history of lung and kidney tuberculosis (TB), recurrent respiratory infections, and chronic obstructive pulmonary disease (COPD). In our previous study we showed that TR1341’s antifungal, β-hemolytic, and cytolytic activities were due to the production of filipin III and fungichromin, and the antibacterial and cytotoxic activities were due to the production of actinomycin X2. In vitro experiments showed that Streptomyces sp. TR1341’s germinating spores reduce viability and activation of human macrophages. This could protect the strain upon its entrance into the human body. Co-culture of the strain with a human macrophage line further induces the cytotoxic activity [27].
In this study, we would like to address the following questions: (i) how does TR1341’s whole genome relate to other pathogenic actinomycetes? (ii) what mechanism of host evasion/colonization can we predict from TR1341 genome? (iii) does TR1341 possess genes or biosynthetic clusters that are commonly present in known pathogenic actinomycetes, but are absent in “environmental” strains?
In order to answer these questions, we performed a detailed analysis of the strain’s genome and compared it with 140 genomes of pathogenic and non-pathogenic actinomycetes from different environments. Here we show that Streptomyces sp. TR1341 has a potentially better adaptation strategy to the human tissue niche which involves the production of specific secondary metabolites, or other abovementioned colonization mechanisms. These mechanisms give TR1341 an edge over other environmental strain.
2. Materials and Methods
2.1. Streptomyces sp. TR13141: Isolation Method, Cultivation, and Genome Sequencing
Streptomyces sp. TR1341 was isolated from the sputum of an 81-year-old male patient with a complex clinical history including repeated infections of the respiratory tract. The sputum sample was subjected to selective cultivation for mycobacteria as described in Hebrík et al. [27]. The strain was kept in The National Reference Laboratory for Pathogenic Actinomycetes in the Local Hospital in Trutnov, Czech Republic, which kindly provided it the purposes of our research.
We routinely cultivated Streptomyces sp. TR1341 at 28 °C on M2 (malt extract 10 g, yeast extract 4 g, glucose 4 g, agar 20 g, distilled water 1 L). In order to investigate the effect of temperature on its growth, we also grew TR1314 at 37 °C, on M2 and tryptic soy agar (TSA) plates.
Details of the genome sequencing and assembly can be found in Hebrík et al. [27]. Briefly the library was prepared using the TruSeq PCR free LT library preparation kit (Illumina) and sequences with the Illumina MiSeq platform (Reagent kit v2, paired end, 300 bp). After polishing the reads with Trimmomatic 0.36 [56], the assembly was performed using FLASH [57] and SPAdes [58], and the quality of it was evaluated with QUAST [59] and Qualimap [60]. The draft genome (VSDL00000000.1) was annotated using Prokka v. 1.14.5 [61].
2.2. Morphological Characteristics and Microscopy
All the culture characteristics of the strain were determined through standard methods given in the International Streptomyces Project (ISP) [62]. The strain was streaked onto four different media: ISP2 (malt extract 10 g, yeast extract 4 g, dextrose 4 g, agar 20 g, distilled water 1 L), ISP3 (HiMedia; oat meal 20 g, FeSO4 × 7H2O 0.001 g, MnCl2 × 4H2O 0.001 g, ZnSO4 × 7H2O 0.001 g, agar 18 g, distilled water 1 L), M2 (malt extract 10 g, yeast extract 4 g, glucose 4 g, agar 20 g in 1 L of distilled water, pH 7.2) and MS (mannitol 20 g, soy flour 20 g, agar 20 g, tap water 1 L) [63]. The plates were incubated at 28 °C for 2 weeks, and colony morphology, signature characteristics of the spore coloration, and sporophore formation were observed.
The morphology of the sporophores produced by TR1341 that was grown on MS media for 14 days was observed with the scanning electron microscope JEOL 7401-FE (JEOL Ltd., Tokyo, Japan) at the accelerating voltage of 4 kV. The sample (aerial mycelium) was fixed and dehydrated in vapors of OsO4, then frozen and, after evaporation, coated with gold using a Sputter Coater (Baltec-SCD 050).
The spore surface was classified by the examination of carbon replicas of spores with the transmission electron microscope (TEM Jeol 1010) at the acceleration voltage of 80 kV with Mega View III camera (SIS).
2.3. Growth Curve and Carbon Utilization Assay
Strain TR1341 was pre-cultivated on M2 agar for 7 days at 28 °C and 37 °C. Because we wanted to use McFarland turbidity for standardizing of the inoculum, a suspension was prepared by vigorous vortexing of the mycelia and spore mixture in a small volume of sterile Luria-Bertani (LB) broth (tryptone 10 g, yeast extract 5 g, NaCl 10 g in 1 L of distilled water, pH 7.0) in a sterile microcentrifuge tube with glass beads. The supernatant was used to prepare a suspension of 0.5 McFarland turbidity (densitometer DEN-1, Biosan, Latvia). A total of 100 µL of suspension was inoculated into 25 mL of LB broth and cultivated at 28 °C and 37 °C in an orbital shaker (150 rpm, Multitron Standard, Infors AG, Bottmingen-Basel, Switzerland), in baffled 100 mL Erlenmeyer flasks supplemented with approximately 30 sterile glass beads (average 0.3 mm) to achieve a dispersed growth. OD600 measurements were carried out using a NanoDrop One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at regular intervals. The experiment was performed in triplicates. The same conditions were maintained for the determination of the growth rate of Streptomyces nodosus ssp. asukaensis ATCC 29757, which was chosen as a reference strain due to its homogeneous growth (no cell aggregates) in liquid media.
Carbon utilization was evaluated using sterile carbon sources and following the protocol given in the International Streptomyces Project (ISP) [62].
2.4. Siderophore Production and Identification
For the detection of siderophore production, a modified chromeazurol S (CAS) assay, using the dual agar plate method, was performed with malt extract agar as the cultivation medium and CAS agar as the detection medium [64,65]. Streptomyces sp. TR1341 was grown at 28 °C on MS agar plates to promote sporulation [63]. One loop of active spores was inoculated on the malt extract agar side at a 0.5 cm distance from the CAS agar side. The production of siderophores was indicated by the change of color from blue to red/orange in the CAS agar part. The zone was monitored and measured for two weeks, after which the area showing a color change, and thus containing the secreted siderophores (width ~ 2.0 cm), was cut. The siderophores were extracted by adding 2.5 mL of acetonitrile:water:formic acid solution (1:1:0.1 v/v/v) and shaking at 140 rpm 4 °C for 2.5 h. Experiments and extractions were performed in triplicates.
Siderophores were also extracted from freeze-dried biomass from 100 mL cultures (GYM medium, 150 rpm, 72 h, 28 °C) by adding 20 mL of acetonitrile:water:formic acid solution. These liquid assays were done by duplicates. Crude extracts were filtered with Chromafil® Regenerated cellulose syringe filters before analysis (pore size 0.45 μm, diam. 30 mm).
The filtered extracts were analysed using reversed-phase liquid chromatography with electrospray ionization and high-resolution mass spectrometry. A volume of 10 µL of the extract was injected first in the same phase precolumn (10 mm × 2.1 mm ID 5 µm particles), and then onto the analytical column (HypersilGold aQ, 50 mm length, 2.1 mm ID with 5 µm particles, Thermo Scientific, Waltham, MA, USA). A gradient of acetonitrile in water (acidified with 0.1% formic acid) was used for the separation of the analytes. The starting composition of the mobile phase was 100% of water at a flow of 0.35 µL·min−1 for 1 min, then the acetonitrile content was linearly increased to 25% in 4 min, 60% in 8 min, 100% in 10 min, followed with 2 min isocratic 100% acetonitrile.
The ion source conditions were as follows: vaporizer temperature 300 °C, capillary temperature 325 °C, sheath gas 40 arb. units, auxiliary gas 12 arb. Units, and spray ionization voltage 2800 and 3500 V for negative and positive ionizations modes, respectively. The QExactive HF (Thermo Fisher Scientific, Waltham, MA, USA) hybrid quadrupole/orbital trap high resolution mass spectrometer was used for detection in a combined full scan/data independent MS2 experiment.
One run was performed in positive ionization mode, the other in negative ionization mode. Full scan data were acquired in the range of 100–1300 m/z with an ion time of 100 ms. DIA was performed with an isolation window of 100 m/z for m/z 150, 250, 350, 450, 550, and 650 with an ion time of 70 ms, stepped collision energy 15, 30, and 50 NCE, and a 30,000 resolution of product scan. Data were processed and evaluated using Xcalibur and MassFrontier 7.1 software (ThermoScientific).
Full scan chromatograms were searched against an m/z list of known desferrioxiamines and ferrioxiamines published in literature [66,67]. A maximal difference from the theoretical mass of 5 ppm was set for this search.
2.5. Dataset
In addition to the genome of Streptomyces sp. TR1341, a total of 140 genomes were downloaded from the GenBank database (as of September 2020). The genomes were carefully selected based on genome quality and representativeness of a wide variety of isolation sources such as soil, rhizosphere, manure, water, air, marine invertebrates, and insects (Table S1). First, we selected the Streptomyces species clustering with Streptomyces sp. TR1341 on the 16S rRNA gene tree calculated in Hebrík et al. [27]. Next, we added well-studied organisms like Streptomyces albus DSM 41398, Streptomyces avermitilis MA-4680, and Streptomyces coelicolor A3(2). We also included Streptomyces albidoflavus NBRC 100770, Streptomyces daghestanicus NRRL B-2710, Streptomyces diastaticus subsp. ardesiacus NBRC 15402, Streptomyces koyangensis VK-A60T, Streptomyces flavogriseus ATCC 33331, Streptomyces seoulensis KCTC 9819, and Streptomyces xiamenensis MCCC 1A01550. Based on 16S rRNA gene analysis, these strains showed similarity to Streptomyces isolated from sputum samples collected in the same area of TR1341 (data not published yet). In order to investigate the potential role of TR1341 as a pathogenic agent, we included all the available genomes of human-associated Streptomyces species. We also selected 20 genomes of known human (lung-)pathogens having less than 200 contigs and affiliated to the following taxa: Actinomadura madurae, Gordonia, Mycobacterium, Nocardia asteroides, Rhodococcus, and Tsukamurella (Table S1).
We filtered our initial database by discarding all genomes with more than 300 contigs and then we evaluated genome completeness using BUSCO v. 3.0.2 [68]. Genomes that failed to satisfy the requirement of >90% completeness and overall quality of 50% were discarded. Exceptions were made when the low-quality assembly belonged to a pathogenic or reference species with no better assembly available (e.g., Streptomyces somaliensis DSM 40738).
All genomes were re-annotated using Prokka v. 1.14.5 [61] to avoid variability due to the annotation method in downstream analysis.
2.6. Orthologous Genes and Species Tree Inference
Groups of orthologous sequences were identified and phylogenetic inference was done using the abovementioned 141 genomes with the software OrthoFinder v. 2.4.0 [69]. OrthoFinder uses not only single copy orthologous genes, but also the full set of orthologous genes for phylogenetic inference. Due to the algorithm used for tree inference (STAG), the branch lengths correspond to the average number of substitutions per site across a large range of gene families [70]. The resulting tree is therefore a species tree that can be used directly in downstream analysis such as ancestral state reconstruction and calibration.
2.7. Biosynthetic Gene Clusters
Gene clusters were identified using antiSMASH v 5.1.2 set to the “relaxed” detection strictness option in order to detect complete clusters, as well as partial clusters, in which some genes might be missing in fragmented draft genomes [71]. For this analysis, we used a subset of genomes as reported in Table S2. All cluster predictions for TR1341 were further analyzed in PRISM [72].
The Antibiotic Resistant Target Seeker (ARTS 2.0) [73] was used to search for potential novel antibiotic targets in the genome of Streptomyces sp. TR1341.
2.8. Mammalian Cell Entry (mce) Genes and Type VII Secretion System (ESX)
Using the Batch CD-Search [74], we identified the main conserved protein domains of the mce genes: MlaE (PF02405.16) and MlaD (PF02470.20). In order to detect the type VII secretion system genes, we identified the conserved protein domain of five essential genes: WXG100 for esxA and esxB (PF06013.12), T7SS_ESX1_EccB for eccB (PF05108.13), FtsK_SpoIIIE for eccC (PF01580.18), YukD for eccD (PF08817.10), and EccE for eccE (PF11203.8). We downloaded the corresponding profiles from the Pfam database [75] and used them to search for mce and type VII secretion system genes in a subset of the selected genomes, using Hmmer v. 3.2 [76].
2.9. Genomic Comparison of Streptomyces sp. TR1341, Streptomyces sp. Endophyte_N2, and the Human Associated Streptomyces spp.
We ran two sets of comparisons: one set was done using the information produced by Orthofinder [69]; a second using the functional groups identified by RAST [77]. Shortly, we ran an analysis of amino acid sequence similarity between TR1341 and Streptomyces sp. Endophyte_N2, Streptomyces brasiliensis strain NRRL B-1626 (human associated, no clear pathogen), Streptomyces somaliensis DSM 40738 (human pathogen), and Streptomyces sp. KE1 (human associated, healthy skin).
Using the RAST annotation server, we identified genes associated with the following functional groups: antibiotic resistance, virulence factors, iron acquisition, acyl carrier proteins, phospholipases, glyoxylate, and mobile elements (phages and prophages).
To visualize the results of these comparisons, we constructed Venn diagrams using Venny 2.1.0 [78].
3. Results and Discussion
3.1. Streptomyces sp. TR1341: Morphological Characterization and Genome Features
Streptomyces sp. TR1341 was isolated from the sputum of an elderly man, who was treated for several respiratory tract-related diseases for decades. The strain produces aerial mycelium with spores that are smooth and form spiral sporophores (Figure S1).
We evaluated TR1341 growth rate at 28 °C and at 37 °C, and observed that TR1341 reaches the maximum OD faster at 37 °C (day 22) than at 28 °C (day 30). The reference strain, on the other hand, had a higher OD measurement at 28 °C (Figure S2).
When growing at 28 °C, TR1341 produced an orange pigment in the substrate mycelia on both M2 and TSA (color of substrate mycelium, ISP coding: Oc4r-Coo4b-4s/Grayish brownish orange), but the aerial spores developed completely only on M2. Cultivated at 37 °C on TSA, however, TR1341 does not produce an orange pigment, nor does it develop aerial mycelia. When grown at 37 °C on M2, the strain produces a small amount of orange pigment in the substrate mycelium and the typical pigmentation of aerial spores (ISP coding: GY5fe—Light grayish reddish brown) is partly inhibited (Figure S3).
The carbon utilization test (Table S2) was negative for L-arabinose, sucrose, rhamnose, raffinose, and cellulose. The results for D-xylose, I-inositol, and D-fructose were inconclusive. Interestingly, TR1341 can use both mannitol and glucose which is the positive control. Mannitol is commonly used in the treatment of patients with obstructive pulmonary disease. Inhaled mannitol (commercial name Bronchitol [79]) reduces the inflammation in the airways and improves mucociliary clearance [80,81]. Unfortunately, we lack information on the patient clinical history and were therefore unable to confirm a link between the use of mannitol as part of the patient’s treatment and its possible subsequent use as a carbon source by TR1341.
The draft genome of Streptomyces sp. TR1341 has a total length of 8.5 Mb and is divided into 170 contigs. It has a GC content of 71.8% and 7442 coding sequences, as reported in Table 1. The genome size and its GC content are comparable to the Streptomyces genomes selected for this study, which have an average genome size of 8.3 ± 1.5 Mb (Table S1). Among the selected Streptomyces, 16 have a genome shorter than 7 Mb. Two out of the three human-associated Streptomyces are part of this group: Streptomyces somaliensis DSM 40738 (5.2 Mb) and Streptomyces sp. KE1 (6.8 Mb). The other strains were isolated from marine habitats (6), plants (3), rhizospheres (3), and soil (2). Besides Streptomyces somaliensis DSM 40738, Streptomyces xiamensis is the only strain with a genome shorter than 6 Mb, despite the fact that it was isolated from mangrove sediments [82]. Another study shows that Streptomyces derived from marine sponges are remodeling their genomes by gene acquisition or loss in order to better adapt to different hosts [83]. When considering the plant pathogens of Streptomyces species, it appears that only a few of them have a reduced genome. For instance, the pathogens Streptomyces albidoflavus NBRC 13083 has a genome of 7 Mb (Table S1), whereas Streptomyces scabiei S58, Streptomyces turgidiscabies T45, and Streptomyces acidiscabies have much bigger genomes (around 10 Mb) [84].

Table 1.
Genomic features of Streptomyces sp. TR1341, and comparison with Streptomyces sp. endophyte_N2 and Streptomyces somaliensis DSM 40738.
Taken all together, it seems that Streptomyces strains keep a relatively large set of genes as a genome foundation and then they might add, silence or lose functions while colonizing a new environment and adapting to it. This flexibility allows them to survive in a wide range of environments. For instance, Streptomyces somaliensis DSM 40738 can live and survive either in the human body or the soil, despite having a reduced genome compared to the average Streptomyces genome size. Since these Streptomyces did not undergo a drastic genome reduction that forces them to steal the deleted function from the host (e.g., mycoplasma), we can conclude that these organisms, including TR1341, have a lifestyle that is different to that of obligate pathogens.
3.2. Orthologous Genes and Tree Inference
We identified 26,770 orthologous groups in the set of 141 strains. These were used to construct the tree and show the relationships between the strains. As expected, Streptomyces form a separate cluster from the other actinomycetes, with Actinomadura madurae being the closest taxon (Figure 1).
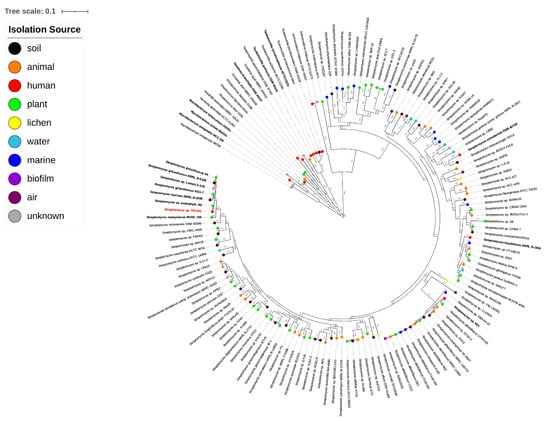
Figure 1.
Phylogenetic tree based on orthologous families calculated with Orthofinder. Branch lengths are indicated.
Streptomyces sp. TR1341 is located among the soil- and plant-derived strains, a large distance from other human-associated strains (Streptomyces somaliensis DSM 40738, Streptomyces brasiliensis NRRL B-1626, and Streptomyces sp. KE1) and as expected, far away from other pathogenic actinomycetes. The closest organism is Streptomyces sp. endophyte_N2 (Figure 1), with whom TR1341 shares more than 95% identity at amino acid level throughout the genome (Figure S4). This is also true for Streptomyces costaricanus DSM 41827, the closest clustering strain in the analysis made by Hebrík et al. [27]. Streptomyces sp. endophyte_N2 was isolated from Arabidopsis thaliana roots and has a broad antimicrobial activity [85]. It shares 5113 orthologous genes with TR1341 and has only 170 unique orthologous genes compared to the 293 that are present in TR1341 (Figure 2a). It seems that each of the four human-associated Streptomyces falls into a different cluster of the tree (Figure 1). This could reflect the fact that pathogenic and human-associated Streptomyces are principally opportunistic pathogens and therefore the closest related organisms in the tree are the ones from the primary habitat. For the moment, this remains a hypothesis that future studies could address with the addition of new publicly available genomic data of other Streptomyces isolated from human samples.
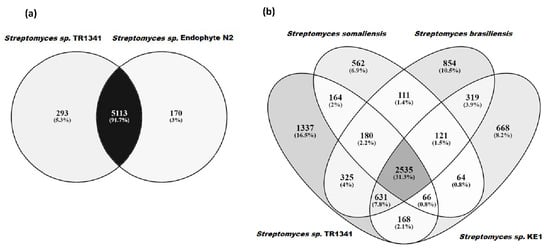
Figure 2.
Comparison of orthologous groups in (a) Streptomyces sp. TR1341 with the closely related Streptomyces sp. endophyte_N2, and (b) the human-associated Streptomyces species: Streptomyces somaliensis DSM 40738, Streptomyces brasiliensis NRRL B-1626, and Streptomyces sp. KE1.
For a deeper comparative genomics analysis, we arbitrarily chose 19 strains from our dataset: eight strains that closely cluster with TR1341 on the species tree (soil-derived and endophytes), the three human-associated strains of Streptomyces, three Mycobacterium strains, one Gordonia, and two Tsukamurella strains, all associated with lung tissue as human pathogens.
3.3. Biosynthetic Gene Clusters
The software antiSMASH was used to predict secondary metabolite gene clusters. Streptomyces sp. TR1341 harbors 42 clusters. Among these, 13 had more than 50% of the genes showing similarities to already known biosynthetic gene clusters (BGCs) (Table 2). A total of 26 clusters were confirmed using PRISM, whereas 16 and 8 clusters were predicted only by antiSMASH and PRISM, respectively (Table S3).

Table 2.
Biosynthetic gene clusters (BGCs) predicted in Streptomyces sp. TR1341 using antiSMASH [66].
The most represented category is non-ribosomal protein synthetases (NRPS), followed by polyketide synthases (PKS). In our previous study, we already proved that TR1341 is able to produce filipin and actinomycin X2 [27]. The filipin BGC is similar to the one characterized in Streptomyces avermitilis NRRL 8165 [86] and Streptomyces filipinensis DSM 40112 [87], from which it was first isolated as an antifungal agent [88]. It belongs to the group of polyene antibiotics and it is highly toxic for human cells. In TR1341, it is responsible for its β-hemolytic activity, providing the strain an advantage over fungal competitors and probably enhancing the release of nutrients from the host cells. Besides the loss of β-hemolytic activity, the deletion of the key PKS genes in the filipin cluster causes an increase in the production of actinomycin X2, a derivative of actinomycin D [27]. The actinomycin BGC of TR1341 shows similarity to the one of Streptomyces chrysomallus ATCC 11523 [89]. We were able to identify both compounds, actinomycin D and actinomycin X2, in the extracts of the submersed cultures (GYM media, samples 1 and 2). Like Streptomyces antibioticus IMRU 3720 [90], TR1341 has only one biosynthetic arm and the predicted substrates of the actinomycin synthetases (ACMS) are threonine-valine (ACMS II) and proline-glycine-valine (ACMS III). Moreover, in TR1341, the acmN gene, encoding a ferredoxin, is replaced by mobile elements, suggesting that this function might be carried out by a ferredoxin located outside the cluster. In the context of adaptation to the human body, the highly antibiotic and cytotoxic properties of actinomycin provide TR1341 with another tool to control gram-negative competitors and scavenge nutrients.
Other useful metabolites for the interaction of TR1341 with eukaryotic cells are hopene and rhizomide (Table 2). Hopene is the precursor for the synthesis of pentacyclic triterpenoids called hopanoids [91]. Besides their role in membrane stabilization [92], different studies demonstrated that they can act as anti-leukemia, anti-inflammatory, and anti-oxidation natural products [93,94]. Rhizomides have weak antitumor activity and protect against plant diseases like cucumber downy mildew. Their biosynthetic pathway has been characterized in Paraburkholderia rhizoxinica HKI 454 [95]. Even though three clusters of TR1341 show similarity to BGC for rhizomide A/B/C (Table 2), they might represent the same region. In fact, they are located on short contigs, which might have failed to assemble into a longer one due to the repetitions of the long NRPS gene involved in the synthesis of rhizomides (MIBiG accession BGC0001758).
The concentration of essential elements such as iron is strictly regulated inside the human body and the role of molecules such as siderophores is therefore essential. Streptomyces sp. TR1341 seems to be able to synthesize the siderophores mirubactin [96] and desferrioxamine [97] through a NRPS-dependent and -independent pathway, respectively (Table 2). We qualitatively confirmed the production of siderophores in vitro via the CAS agar test (Figure S5). In the crude extracts, we could not detect the siderophore mirubactin, but we were able to identify ferrioxamine B using reversed-phase liquid chromatography with electrospray ionization and high-resolution mass spectrometry (delta ppm from theoretical m/z −0.984 and isotope pattern corresponds to 1 Fe within the molecule). The structure was confirmed by finding a match with published MS/MS spectra fragments. In addition, we identified a homologue that has a very similar retention time but with a shorter chain (one CH2 less). Both compounds showed an identical low mass fragment while heavier fragments were specific for the given compound (Figure 3a). XIC at M+ peak areas of the extracts are reported in Figure 3b to document the semi-quantification of the produced compounds. The high reproducibility of the experiment and analysis is documented by the almost identical peak areas for the triplicates of the cultivation on dual CAS agar plates (MEA/CAS, TR1341/1–3), and the very similar peak areas for the duplicates of the submersed cultures (TR1431/1–2).
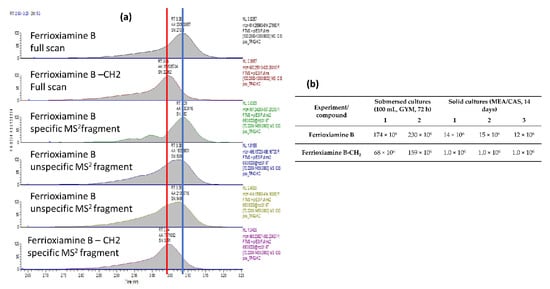
Figure 3.
Ferrioxiamine B and its -CH2 homologue detected in the extracts from the submersed cultures (1–2) and from the solid cultures (1–3). (a) Extracted XIC chromatograms of ferrioxiamine B and its -CH2 homologue. (b) Semiquantitative evaluation of ferrioxiamine B and its homologue.
In addition, one of TR1341’s clusters shows similarity to the BGC of diisonitrile antibiotic SF2768 identified in several Streptomyces species and other Actinobacteria. This molecule mediates the uptake of copper and acts as an antifungal and virulence factor [98]. In fact, besides their role in nutrient uptake, siderophores and other chelators act as antibiotic agents sequestering essential elements and preventing other microorganisms from access to these resources [99,100].
When looking at the BGC, in which at least 50% of the genes show similarity to a known cluster, we observed that most of TR1341’s BGCs are also present in the soil/plant-associated Streptomyces spp. belonging to its phylogenetic cluster (Figure 4). Streptomyces costaricanus DSM 41827, Streptomyces sp. endophyte_N2, and Streptomyces griseofuscus NG1-7 have almost the same predicted BGCs. For instance, previous studies reported the production of actinomycin and filipin in Streptomyces costaricanus SCSIO ZS0073 and Streptomyces sp. endophyte_N2, respectively [85,101]. The geosmin BGC is present in all selected Streptomyces spp. apart from Streptomyces somaliensis DSM 40738. On the other hand, the cluster for the osmoprotectant ectoine is widespread among Streptomyces [102] and it is the only BGC shared by TR1341, Mycobacterium smegmatis MC2-155, and the lung-associated actinomycetes such as Gordonia bronchialis DSM 43247, Tsukamurella pulmonis DSM 44142, and Tsukamurella tyrosinosolvens NCTC13231 (Figure 4). Only five of TR1341’s BGCs of TR1341 are present in the human-associated Streptomyces: desferrioxamine, diisonitrile antibiotic SF2768, geosmin, hopene, and melanin.
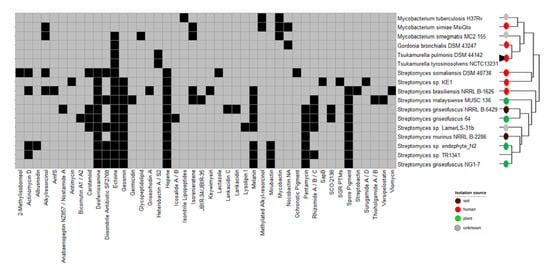
Figure 4.
Biosynthetic gene clusters (BGCs) predicted by antiSMASH with more than 50% of the genes showing similarity to a known BGC. Black squares indicate presence, while grey indicates absence. The isolation source of the organisms is shown with the colored circles at the tip of the branches.
In most cases, the number of predicted BGCs per organism correlates with the size of the genome: the larger the genome, the higher the number of predicted clusters (Figure 5, Table S4). It seems that the human-related Streptomyces strains are in an intermediate position between the lung-pathogen actinomycetes and the soil/plant-related Streptomyces. Indeed, smaller genomes are a sign of adaptation to a symbiotic/pathogenic lifestyle compared to community. For instance, the genome size of Streptomyces somaliensis DSM 40738 is comparable with that of the lung-pathogen actinomycetes, but it has more BGCs. These might still provide metabolites that are beneficial for survival in the soil before the colonization of human tissues via skin lesions. Therefore, the transition of Streptomyces somaliensis DSM 40738 to the human environment from the soil might have happened before the transition of other human-associated Streptomyces, such as TR1341. In any case, for S. somaliensis and TR1341, it seems that the human environment is just another possibility in terms of habitat and we do not have any indication of niche specificity.
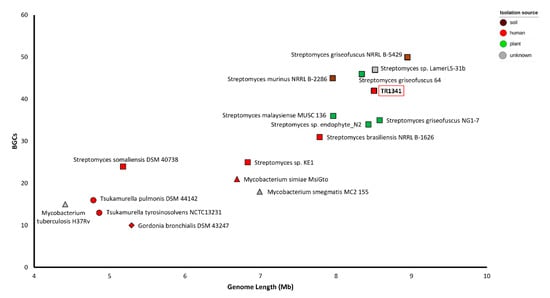
Figure 5.
Relationship between genome size (Mb) and number of biosynthetic gene clusters (BGCs). Squares: Streptomyces species; triangles: Mycobacterium species; circles: Tsukamurella species; diamond: Gordonia bronchialis DSM 43247.
To complement the antiSMASH analysis, the Antibiotic Resistant Target Seeker (ARTS) was used to search for potential novel antibiotic agents in the genome of Streptomyces sp. TR1341 [73]. A total of 587 genes (about 7% of the total) were identified as core/essential genes and 41 known resistance models were found. The major four functional categories of the core genes are: protein synthesis (18%), biosynthesis of cofactors, prosthetic groups, and carriers (12%), energy metabolism (12%), and unclassified (10%) (Table S5).
Among these, 14 genes satisfied at least three of the four criteria set by ARTS: duplication, proximity to BGC, phylogeny/horizontal gene transfer, and similarity to known resistance models. Regarding the proximity to a BGC, 24 clusters satisfy this criterium, having at least one core gene hit (Table S6). For instance, the BGC Region 20.1 corresponding to cluster 19 in ARTS contains eight core genes, but only 22% of the genes show similarity to the known BGC kinamycin. The genes showing similarity are not the main biosynthetic ones. In fact, the predicted PKS and NRPS genes seem to show similarity to the genes for the synthesis of other compounds like cystothiazole A and myxothiazol. In BGC Region 34.1 (cluster 27 in ARTS), we identified four genes and a resistance model related to a biotin-requiring enzyme, even though only 6% of the genes showed similarity to the formicamycins A-M BGC (Table 2, Table S6).
These examples show that even BGCs exhibiting a low percentage of similarity to previously characterized clusters are worth investigating. ARTS provides the information that enables the prioritization of research into unknown BGCs, which may lead to the discovery of new bioactive molecules.
3.4. Mammalian Cell Entry (mce) Genes and Type VII Secretion System (ESX)
We could not detect the presence of genes encoding the conserved domains MlaD and MlaE, which characterize the mce genes (Table 3) in the genome of Streptomyces sp. TR1341.

Table 3.
Summary of Mammalian Cell Entry (MCE) genes presence in the studied genomes. MleD (Pfam PF02470.20): conserved domain of the two ABC-transporter integral membrane proteins, yrbEA; MleE (Pfam PF02405.16): conserved domain of the six hypothetical proteins, mceABCDEF.
Among the selected Streptomyces strains, only Streptomyces sp. KE1 and Streptomyces brasiliensis NRRL B-1626 harbor one complete mce operon. They were both isolated from human samples but are not clearly associated with pathogenicity.
On the other hand, all the other actinomycetes harbor multiple mce operons, with Mycobacterium simiae MsiGto having the highest number (nine operons). Interestingly, the Streptomyces somaliensis DSM 40738 genome does not possess any mce operon, indicating that this strain uses a different virulence strategy than the typical strategy employed by actinobacterial lung pathogens. The fact that Streptomyces somaliensis DSM 40738 is not a lung pathogen but affects skin, soft tissues, and bones, probably relates to the differences that we observe in the infection strategies at this level (mce genes). A Streptomyces somaliensis DSM 40738 infection is characterized primarily by an inflammatory reaction [103], which is more related to the presence of type VII secretion systems (ESX) and not to mce-encoded proteins. Unlike Mycobacterium species, where all five ESX secretion systems are widespread and sometimes even duplicated [42,104], all the selected Streptomyces genomes, with the exception of Streptomyces sp. KE1, harbor at least one ESX system similar to the one characterized in Streptomyces scabies 87.22 and Streptomyces coelicolor A3(2) (Figure 6, Table S7).
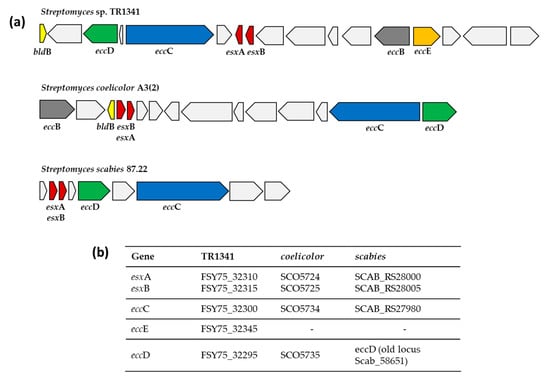
Figure 6.
Comparison of type VII secretion system (ESX) present in Streptomyces sp. TR1341 and the ones characterized in Streptomyces coelicolor A3(2) and Streptomyces scabies 87.22. (a) Schematic representation of the genomic region of the different type VII secretion systems. (b) Genomic locus tags of the main genes.
The fact that this ESX system has a different structure than the one present in pathogenic mycobacteria and that it is present in several Streptomyces isolated from different sources suggests that it indeed might be involved in developmental processes instead of virulence.
The complete ESX system in TR1341 is more similar to that of Streptomyces coelicolor A3(2) than to that of Streptomyces scabies 87.22 (Figure 6). In the plant pathogen Streptomyces scabies 87.22, esxA and esxB do not have the characteristic WXG100 protein family domain, but instead have an FNG-motif and FQA-motif, respectively. Moreover, a subtilisin-type serine protease distant from the ESX cluster might act as the transmembrane protein EccB [43,44].
Streptomyces sp. endophyte_N2, the Streptomyces griseofuscus strains (NG1-7, NRRL B-5429, 64), Streptomyces malaysiense MUSC 136, and Streptomyces brasiliensis NRRL B-1626 had a second copy of the system, where eccE was missing (Table S7). In Streptomyces sp. TR1341, Streptomyces murinus NRRL B-2286, and Streptomyces sp. LamerLS-31b, this second copy is lacking the two WXG100-Esx genes probably due to the fact that they are located near the edge of a contig. Other genes from the WXG100 family were found in other parts of the genomes. Gordonia bronchialis seems to have all the genes necessary for a functional ESX system, but they are scattered along the whole genome. Tsukamurella pulmonis DSM 44142 harbors two ESX systems, whereas in Tsukamurella tyrosinosolvens NCTC13231 we could detect all the domains located on different plasmids. The presence of ESX-encoding plasmids has been reported before in mycobacteria and is considered to play a major role in the spreading and diversification of type VII systems [105].
The lack of mce genes and a type VII secretion system that is distinct from the one found in pathogenic mycobacterium species suggest an alternative invasion and colonization strategy in TR1341, the nature of which is unfortunately still not clear.
3.5. Genomic Comparison of Streptomyces sp. TR1341, Streptomyces sp. Endophyte_N2, and the Human Associated Streptomyces spp.
To the best of our knowledge, only three genomes of human-associated Streptomyces are deposited in the GenBank database (September 2020). Streptomyces somaliensis DSM 40738 is the causative agent of actinomycetoma and shows the genetic adaptation of a pathogenic lifestyle (i.e., small genome size) [20]. Streptomyces brasiliensis NRRL B-1626 and Streptomyces sp. KE1 were isolated from “human disease” [106] and healthy human skin surface (SAMN03454257, unpublished), respectively. Streptomyces sp. TR1341 shares the highest number of orthologous gene families with Streptomyces brasiliensis NRRL B-1626 (3671), followed by Streptomyces sp. KE1 (3400) and Streptomyces somaliensis DSM 40738 (2945) (Figure 2b). These numbers reflect the distances showed in the phylogenetic tree in Figure 1.
In order to easily assign the identified orthologous genes to specific functional categories, we used RAST [77] to annotate the genomes of Streptomyces sp. endophyte_N2 and the three human-associated Streptomyces. Results of the comparison at protein level showed that TR1341 and Streptomyces sp. endophyte_N2 share 99% of sequence identity along most of the genome, with the exception of two small regions that have a similarity of around 60% (henceforth referred to as the red region) and 80% (the blue region) (Figure S6).
The high similarity at the amino acid level indicates that perhaps these two strains have originated from a similar environmental niche and so respond to similar evolutionary constrains [107]. The red region (Figure S7a) is a low similarity region in the central part of the chromosome and is composed of a few annotated genes and a group of hypothetical proteins. We cannot predict any specific biological activity from the group of annotated genes and antiSMASH failed to predict any BGCs in this region. However, these genes might be involved in a biosynthetic pathway, since most of them are synthetases or transferases, and there is a LysR transcriptional regulator.
The blue region is at the end of the left arm of the genome (Figure S7b) and it is unique to TR1341, not appearing in any of the other human-associated Streptomyces. Most of the genes present in TR1341 code for hypothetical proteins and those with an assigned function are annotated as mobile elements, phage/prophage-associated genes, or are involved in the mobilization of genetic material (recombinational DNA repair, DNA helicase). These kinds of genes are often found at the distal arms of the Streptomyces chromosome representations [108].
After, we performed a thorough analysis on the following genes and functional categories: antibiotic resistance and virulence, iron metabolisms, phospholipases, and glyoxylate pathway and mobile elements.
Regarding the antibiotic resistance genes and virulence factors, TR1341 has four genes, which are not found in the human-associated Streptomyces (Figure 7a, Table S8). The first of these genes is annotated as a putative internalin (RAST) from marine actinobacterium PHSC20C and Flavobacterium johnsoniae, or as hypothetical protein (Prokka, NCBI). Internalin is a surface protein, which is well-studied in Listeria monocytogenes and is involved in the invasion of mammalian cells via cadherins transmembrane proteins and Met receptors (hepatocyte growth factor receptor, HGFR) [109]. The putative internalin in TR1341 has very low amino acid similarity with the sequence of the internalin gene from Listeria, which could indicate a very different function in TR1341, or it could be that the differences in sequences are not translated to differences in 3D structure. However, at the moment, we have no way of testing any of these hypotheses. The amino acid sequence in TR1341 contains a DUF11 domain that has been associated with porin formation in Chlamydia trachomatis, and so it is regarded as important for the pathogenicity of the bacteria. The sequence DUF11 is widely distributed and well conserved among Streptomyces genomes of environmental origin (Figure S8). The second gene unique to TR1341 is the streptothricin acetyltransferase (Streptomyces lavendulae type); this gene provides resistance to streptothricin [110]. The other two unique genes belong to the family of MerR transcriptional regulators.
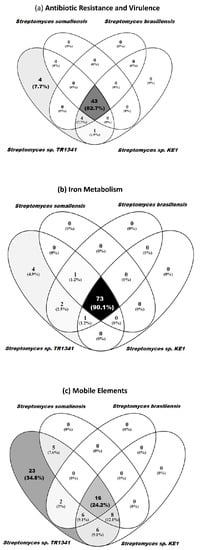
Figure 7.
Genes shared between Streptomyces sp. TR1341 and the human-associated Streptomyces species in the following RAST categories: (a) proteins associated with antibiotic resistance and virulence factors; (b) proteins associated with iron metabolism; (c) mobile elements (associated to secondary metabolism but no other subsystem).
Our previous study showed that TR1341 is resistant to penicillin, ampicillin, cephalosporin, quinolones, rifampicin, tetracycline, and trimethoprim/sulfamethoxazole [27]. Here, we classified the genes associated with these resistance profiles (Table S9). TR1341 has 13 genes associated with resistance to penicillin, ampicillin, and cephalosporin. Most of these genes are part of the class C beta-lactamases. Four genes are involved in the strain’s resistance to fluoroquinolones: two topoisomerase and two DNA gyrases. We found two genes for the strain’s resistance to rifampicin, three genes for the strain’s resistance to tetracycline, and three genes for the strain’s resistance to trimethoprim-sulfamethoxazole. These final three are ABC transporters.
In the RAST category for iron acquisition, there are four genes unique to TR1341 when compared to the other human-associated Streptomyces (Figure 7b, Table S10). These genes are unique forms of segments of the EfeUOB system; specifically, two unique forms of EfeU and EfeO. EfeU serve as integral membrane iron permeases, whereas the role of EfuO is not entirely known but it could act as an iron-binding or an electron-transfer component [111].
These results, coupled with the positive result of siderophore production assay and the presence of siderophore BGCs (Figure S5, Table 2), suggest that TR1341 has a wider variety of iron acquisition mechanisms compared to other strains in our analysis. This high efficiency could confer TR1341 with an advantage for colonizing low-iron environments such as the lungs.
When inside the human body, microbes are in contact with macrophages as a primary line of defense against infection. Studies showed that when captured by macrophages, the yeast Saccharomyces cerevisiae, the opportunistic fungal pathogen Candida albicans, and the lung pathogen Mycobacterium tuberculosis activate the glyoxylate shunt in order to use even-chain fatty acids as a carbon source [112,113,114]. Interestingly, in heterotrophic bacteria, the glyoxylate shunt plays an important role under iron deficient conditions [115]. The two enzymes of the glyoxylate shunt, isocitrate lyase and malate synthase, are present in Streptomyces sp. TR1341 and Streptomyces brasiliensis NRRL B-1626, but could not be found in the other selected genomes (Table S11).
Other enzymes that influence the interaction of microbes with eukaryotic cells are phospholipases and sphingomyelinases [116]. TR1341 harbors only phospholipases C and lysophospholipases, whereas the other Streptomyces strains also have phospholipases D and sphingomyelinases (Table S12). These enzymes are produced by both gram-positive and gram-negative bacteria and some can act as virulence factors. For instance, they hydrolyse membrane lipids that release nutrients essential for colonization, and in some cases they disrupt the immune response by affecting the homeostasis of the host’s signaling molecules [117,118].
It comes as no surprise that mobile elements are far more variable among the analyzed strains (Figure 7c, Table S13). There are nine sequences in this subsystem that are unique to TR1341. Most of them (seven) have ambiguous annotations (mobile element protein, phage related protein, phage protein). Only two have better understood functions and are annotated as the recombinational DNA repair protein RecT (prophage-associated) and as a transposase.
4. Conclusions
Our results show that TR1341 has an increased growth rate at 37 °C when compared to 28 °C. It produces the siderophore ferrioxamine B and can use mannitol as a carbon source. These physiological characteristics can be considered as an adaptation to the human niche.
The phylogenetic analysis shows that Streptomyces sp. TR1341 is quite distant from both human-associated and pathogenic strains, even though it was isolated from a sputum sample. We did not observe a genome reduction, which is usually associated with obligate pathogens. We could predict a large number of BGCs and we were able to identify several genes associated with the scavenging of nutrients (e.g., iron), which are not present in other human-associated Streptomyces. The various genes related to antibiotic resistance may be the result of the frequent antibiotic treatments that the patient underwent and could thus have been acquired after colonization. On the other hand, the diversity of ABC transporters present in TR1341 could point to an innate advantage of the strain related to antibiotic resistance. The lack of mce genes and the presence of a type VII secretion system distinct from the one employed by Mycobacterium tuberculosis species suggest a different colonization strategy.
All these results imply a greater metabolic plasticity of TR1341 when compared to other typical human-associated streptomycetes. A highly plastic metabolic capacity confers the microorganism with the ability to grow in many different environments, including the human lungs. This high potential for metabolic plasticity is also a characteristic of soil-associated bacteria when compared to specialized pathogens because soil-associated bacteria need to survive in highly unstable environments. On the other hand, specialized pathogens tend to discard the information that is not needed in their new and “stable” environment. Moreover, we detected more gene variants in TR1341 (e.g., multiple slightly different copies of the same gene) than in the other pathogenic strains. This could be an adaptive response to a constantly changing environment, and could also point to the fact that TR1341 is not a specialized pathogen but an opportunistic one. To sum up, it seems that TR1341 was a soil-dwelling strain that accidentally entered the lungs of the patient and had enough genetic mechanisms at its disposal to allow it to not only survive, but to thrive inside the patient’s lungs.
This thorough genomic analysis can be used as a road map for future experiments that will improve understanding of the specific colonization strategies of the respiratory tract by TR1341 and further advance our knowledge of the role of streptomycetes in the human body.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081547/s1, Figure S1: Electron microscopy images of Streptomyces sp. TR1341, Figure S2: The effect of temperature on the growth characteristics of Streptomyces sp. TR1341, Figure S3: Comparison of Streptomyces sp. TR1341 with Streptomyces costaricanus DSM 41827 and Streptomyces sp. endophyte_N2, Figure S4: Siderophore production assay, Figure S5: Comparison of Streptomyces sp. TR1341 with Streptomyces sp. endophyte_N2 and human-associated Streptomyces at amino acid level, Figure S6: Comparison of Streptomyces sp. TR1341 with Streptomyces sp. endophyte_N2 and human-associated Streptomyces at amino acid level. Bidirectional best hit calculated in RAST. TR1341 vs (a) Streptomyces sp. endophyte_N2, (b) Streptomyces brasiliensis NRRL B-1626, (c) Streptomyces somaliensis DSM 40738, (d) Streptomyces sp. KE1. Red arrow indicates a region (red region)) of the genome that has low identity between TR1341 and Streptomyces sp. endophyte_N2. Blue arrows indicate a region (blue region) that seems to be unique in TR1341 when compared with human-associated Streptomyces, Figure S7: Dissimilar genomic regions of Streptomyces sp. TR1341 with Streptomyces sp. endophyte_N2 and human-associated Streptomyces, Figure S8: Distribution and similarity of genes with DUFF11 conserved domain among Streptomyces genomes available in the NCBI database, Table S1: List of genomes used in this study, Table S2: Carbon source utilization test, Table S3: BGCs of Streptomyces sp. TR1341 predicted by antiSMASH and PRISM, Table S4: Biosynthetic gene clusters (BGC) predicted by antiSMASH, Table S5: Core/essential genes of Streptomyces sp. TR1341 identified in ARTS, Table S6: BGCs of Streptomyces sp. TR1341 predicted in ARTS, Table S7: ESX systems, Table S8: Virulence and antibiotic resistance genes annotated in RAST, Table S9: Specific antibiotic resistance genes in Streptomyces sp. TR1341, Table S10: Iron-acquisition-related genes annotated in RAST, Table S11: Genes involved in the glyoxylate metabolism identified in RAST, Table S12: Phospholipases and sphingomyelinases identified in RAST, Table S13: Mobile elements annotated in RAST.
Author Contributions
Conceptualization, A.C.L., E.C., K.P. and A.C.; methodology, A.C.L., E.C., A.C. and R.G.; formal analysis, A.C.L. and E.C.; investigation, A.C.L., E.C., R.G., L.K. and F.L.; data curation, A.C.L. and E.C.; writing—original draft preparation, A.C.L. and E.C.; writing—review and editing, F.L., K.P., R.G. and A.C.; visualization, A.C.L. and E.C.; project administration, K.P. and A.C.; funding acquisition, K.P. and A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Czech Health Research Council project No. 17-30091A.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
We acknowledge the core facility Laboratory of Electron Microscopy, Biology Centre CAS supported by the MEYS CR (LM2018129 Czech-BioImaging) and ERDF (No. CZ.02.1.01/0.0/0.0/16_013/0001775); Josef Scharfen, Jr. (Regional Hospital Trutnov, a. s., National Reference Laboratory for Pathogenic Actinomycetes, Trutnov, Czechia) for providing TR1341 to our laboratory; Petr Ježek (District Hospital in Příbram, Czechia) for the isolation of strain; Václav Krištůfek for sharing the experience; and Martina Petrlíková, Petra Beníšková, and Nikola Fuková for technical help with laboratory analyses, strain cultivation, maintenance, and long-term storage. Natalie Bergman is thanked for English corrections of the manuscript. Tereza Chroňáková is thanked for help with graphical abstract design.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; OXford University Press, Inc.: Oxford, UK, 2007; ISBN-13 978-0-19-515066-7. [Google Scholar]
- Müller, R.; Wink, J. Future potential for anti-infectives from bacteria—How to exploit biodiversity and genomic potential. Int. J. Med. Microbiol. 2014, 304, 3–13. [Google Scholar] [CrossRef]
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Chen, M.; Chai, W.; Song, T.; Ma, M.; Lian, X.Y.; Zhang, Z. Anti-glioma Natural Products Downregulating Tumor Glycolytic Enzymes from Marine Actinomycete Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef]
- Kämpfer, P. The Family Streptomycetaceae, Part I: Taxonomy. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 538–604. [Google Scholar]
- Amin, A.; Ahmed, I.; Khalid, N.; Osman, G.; Khan, I.U.; Xiao, M.; Li, W.J. Streptomyces caldifontis sp. nov., isolated from a hot water spring of Tatta Pani, Kotli, Pakistan. Antonie Leeuwenhoek 2017, 110, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Řeháková, K.; Chroňáková, A.; Krištůfek, V.; Kuchtová, B.; Čapková, K.; Scharfen, J.; Čapek, P.; Doležal, J. Bacterial community of cushion plant Thylacospermum ceaspitosum on elevational gradient in the Himalayan cold desert. Front. Microbiol. 2015, 6, 304. [Google Scholar] [CrossRef] [PubMed]
- Chroňáková, A.; Krištůfek, V.; Tichý, M.; Elhottová, D. Biodiversity of Streptomycetes isolated from a succession sequence at a post-mining site and their evidence in Miocene lacustrine sediment. Microbiol. Res. 2010, 165, 594–608. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M.; Göttler, W.; Herzner, G.; Strohm, E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr. Biol. 2005, 15, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Haeder, S.; Wirth, R.; Herz, H.; Spiteller, D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4742–4746. [Google Scholar] [CrossRef]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.M.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef]
- Kim, D.R.; Cho, G.; Jeon, C.W.; Weller, D.M.; Thomashow, L.S.; Paulitz, T.C.; Kwak, Y.S. A mutualistic interaction between Streptomyces bacteria, strawberry plants and pollinating bees. Nat. Commun. 2019, 10, 4802. [Google Scholar] [CrossRef]
- Grubbs, K.J.; Surup, F.; Biedermann, P.H.W.; McDonald, B.R.; Klassen, J.L.; Carlson, C.M.; Clardy, J.; Currie, C.R. Cycloheximide-Producing Streptomyces Associated with Xyleborinus saxesenii and Xyleborus affinis Fungus-Farming Ambrosia Beetles. Front. Microbiol. 2020, 11, 2207. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento-Ramírez, J.M.; Van Der Voort, M.; Raaijmakers, J.M.; Diéguez-Uribeondo, J. Unravelling the microbiome of eggs of the endangered sea turtle Eretmochelys imbricata identifies bacteria with activity against the emerging pathogen Fusarium falciforme. PLoS ONE 2014, 9, e95206. [Google Scholar] [CrossRef]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Sawada, H.; Tanaka, F.; Matsuda, I. Phylogenetic analysis of Streptomyces spp. causing potato scab based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 1996, 46, 476–479. [Google Scholar] [CrossRef]
- Li, Y.; Liu, J.; Díaz-Cruz, G.; Cheng, Z.; Bignell, D.R.D. Virulence mechanisms of plant-pathogenic Streptomyces species: An updated review. Microbiology 2019, 165, 1025–1040. [Google Scholar] [CrossRef]
- Khalil, M.; Lerat, S.; Beaudoin, N.; Beaulieu, C. The Plant Pathogenic Bacterium Streptomyces scabies Degrades the Aromatic Components of Potato Periderm via the β-Ketoadipate Pathway. Front. Microbiol. 2019, 10, 2795. [Google Scholar] [CrossRef]
- Quintana, E.T.; Wierzbicka, K.; Mackiewicz, P.; Osman, A.; Fahal, A.H.; Hamid, M.E.; Zakrzewska-Czerwinska, J.; Maldonado, L.A.; Goodfellow, M. Streptomyces sudanensis sp. nov., a new pathogen isolated from patients with actinomycetoma. Antonie Leeuwenhoek 2008, 93, 305–313. [Google Scholar] [CrossRef]
- Kirby, R.; Sangal, V.; Tucker, N.P.; Zakrzewska-Czerwińska, J.; Wierzbicka, K.; Herron, P.R.; Chu, C.J.; Chandra, G.; Fahal, A.H.; Goodfellow, M.; et al. Draft genome sequence of the human pathogen Streptomyces somaliensis, a significant cause of actinomycetoma. J. Bacteriol. 2012, 194, 3544–3545. [Google Scholar] [CrossRef] [PubMed]
- Sing, D.; Sing, C.F. Impact of direct soil exposures from airborne dust and geophagy on human health. Int. J. Environ. Res. Public Health 2010, 7, 1205–1223. [Google Scholar] [CrossRef]
- Bolourian, A.; Mojtahedi, Z. Streptomyces, shared microbiome member of soil and gut, as “old friends” against colon cancer. FEMS Microbiol. Ecol. 2018, 94, fiy120. [Google Scholar] [CrossRef]
- Bolourian, A.; Mojtahedi, Z. Immunosuppressants produced by Streptomyces: Evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ. Microbiol. Rep. 2018, 10, 123–126. [Google Scholar] [CrossRef]
- Gallo, R.L.; Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012, 12, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Nariya, S.; Harris, J.M.; Lynch, S.V.; Choy, D.F.; Arron, J.R.; Boushey, H. The airway microbiome in patients with severe asthma: Associations with disease features and severity. J. Allergy Clin. Immunol. 2015, 136, 874–884. [Google Scholar] [CrossRef]
- Herbrík, A.; Corretto, E.; Chroňáková, A.; Langhansová, H.; Petrásková, P.; Hrdý, J.; Čihák, M.; Krištůfek, V.; Bobek, J.; Petříček, M.; et al. A Human Lung-Associated Streptomyces sp. TR1341 Produces Various Secondary Metabolites Responsible for Virulence, Cytotoxicity and Modulation of Immune Response. Front. Microbiol. 2020, 10, 3028. [Google Scholar] [CrossRef] [PubMed]
- Engevik, M.A.; Versalovic, J. Biochemical Features of Beneficial Microbes: Foundations for Therapeutic Microbiology. In Bugs as Drugs; American Society of Microbiology: Washington, DC, USA, 2017; pp. 3–47. [Google Scholar]
- Siddiqui, S.; Anderson, V.L.; Hilligoss, D.M.; Abinun, M.; Kuijpers, T.W.; Masur, H.; Witebsky, F.G.; Shea, Y.R.; Gallin, J.I.; Malech, H.L.; et al. Fulminant mulch pneumonitis: An emergency presentation of chronic granulomatous disease. Clin. Infect. Dis. 2007, 45, 673–681. [Google Scholar] [CrossRef] [PubMed]
- Lertcanawanichakul, M.; Chawawisit, K. Identification of Streptomyces spp. isolated from air samples and its cytotoxicity of anti-MRSA bioactive compounds. Biocatal. Agric. Biotechnol. 2019, 20, 101236. [Google Scholar] [CrossRef]
- Kettleson, E.; Kumar, S.; Reponen, T.; Vesper, S.; Méheust, D.; Grinshpun, S.A.; Adhikari, A. Stenotrophomonas, Mycobacterium, and Streptomyces in home dust and air: Associations with moldiness and other home/family characteristics. Indoor Air 2013, 23, 387–396. [Google Scholar] [CrossRef]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofronová, O.; Petrícková, K.; Bobek, J. Secondary metabolites produced during the germination of Streptomyces coelicolor. Front. Microbiol. 2017, 8, 2495. [Google Scholar] [CrossRef]
- Huttunen, K.; Hyvärinen, A.; Nevalainen, A.; Komulainen, H.; Hirvonen, M.R. Production of proinflammatory mediators by indoor air bacteria and fungal spores in mouse and human cell lines. Environ. Health Perspect. 2003, 111, 85–92. [Google Scholar] [CrossRef]
- Jussila, J.; Komulainen, H.; Huttunen, K.; Roponen, M.; Hälinen, A.; Hyvärinen, A.; Kosma, V.-M.; Pelkonen, J.; Hirvonen, M.-R. Inflammatory Responses in Mice after Intratracheal Instillation of Spores of Streptomyces californicus Isolated from Indoor Air of a Moldy Building. Toxicol. Appl. Pharmacol. 2001, 171, 61–69. [Google Scholar] [CrossRef]
- Penttinen, P.; Huttunen, K.; Pelkonen, J.; Hirvonen, M.R. The proportions of Streptomyces californicus and Stachybotrys chartarum in simultaneous exposure affect inflammatory responses in mouse RAW264.7 macrophages. Inhal. Toxicol. 2005, 17, 79–85. [Google Scholar] [CrossRef]
- Penttinen, P.; Pelkonen, J.; Huttunen, K.; Hirvonen, M.-R.R. Co-cultivation of Streptomyces californicus and Stachybotrys chartarum stimulates the production of cytostatic compound(s) with immunotoxic properties. Toxicol. Appl. Pharmacol. 2006, 217, 342–351. [Google Scholar] [CrossRef]
- Yacoub, A.T.; Velez, A.P.; Khwaja, S.I.; Sandin, R.L.; Greene, J. Streptomyces pneumonia in an immunocompromised patient: A case report and a review of literature. Infect. Dis. Clin. Pract. 2014, 22, e113–e115. [Google Scholar] [CrossRef]
- Cambier, C.J.; Falkow, S.; Ramakrishnan, L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 2014, 159, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Bitter, W.; Houben, E.N.G.; Bottai, D.; Brodin, P.; Brown, E.J.; Cox, J.S.; Derbyshire, K.; Fortune, S.M.; Gao, L.-Y.; Liu, J.; et al. Systematic Genetic Nomenclature for Type VII Secretion Systems. PLoS Pathog. 2009, 5, e1000507. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, M.; Constantinidou, C.; Palmer, T.; Pallen, M.J. The Enigmatic Esx Proteins: Looking Beyond Mycobacteria. Trends Microbiol. 2017, 25, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Gröschel, M.I.; Sayes, F.; Simeone, R.; Majlessi, L.; Brosch, R. ESX secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016, 14, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Forrellad, M.A.; Klepp, L.I.; Gioffré, A.; García, J.S.; Morbidoni, H.R.; de la Paz Santangelo, M.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef]
- Fyans, J.K.; Bignell, D.; Loria, R.; Toth, I.; Palmer, T. The ESX/type VII secretion system modulates development, but not virulence, of the plant pathogen Streptomyces scabies. Mol. Plant Pathol. 2013, 14, 119–130. [Google Scholar] [CrossRef]
- Roman, S.A.S.; Facey, P.D.; Fernandez-Martinez, L.; Rodriguez, C.; Vallin, C.; Del Sol, R.; Dyson, P. A heterodimer of EsxA and EsxB is involved in sporulation and is secreted by a type VII secretion system in Streptomyces coelicolor. Microbiology 2010, 156, 1719–1729. [Google Scholar] [CrossRef] [PubMed]
- Haile, Y.; Caugant, D.A.; Bjune, G.; Wiker, H.G. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol. Med. Microbiol. 2002, 33, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.C.; Seipke, R.F.; Prieto, P.; Willemse, J.; Van Wezel, G.P.; Hutchings, M.I.; Hoskisson, P.A. Mammalian cell entry genes in Streptomyces may provide clues to the evolution of bacterial virulence. Sci. Rep. 2013, 3, 1109. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Riley, L.W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 2007, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Shimono, N.; Morici, L.; Casali, N.; Cantrell, S.; Sidders, B.; Ehrt, S.; Riley, L.W. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. USA 2003, 100, 15918–15923. [Google Scholar] [CrossRef]
- Zhang, V.; Nemeth, E.; Kim, A. Iron in lung pathology. Pharmaceuticals 2019, 12, 30. [Google Scholar] [CrossRef]
- Terra, L.; Dyson, P.; Ratcliffe, N.; Castro, H.C.; Vicente, A.C.P. Biotechnological Potential of Streptomyces Siderophores as New Antibiotics. Curr. Med. Chem. 2021, 28, 1407–1421. [Google Scholar] [CrossRef]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Kronstad, J.W.; Caza, M. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front. Cell. Infect. Microbiol. 2013, 4, 80. [Google Scholar]
- Parrow, N.L.; Fleming, R.E.; Minnick, M.F. Sequestration and scavenging of iron in infection. Infect. Immun. 2013, 81, 3503–3514. [Google Scholar] [CrossRef]
- Braun, V.; Pramanik, A.; Gwinner, T.; Köberle, M.; Bohn, E. Sideromycins: Tools and antibiotics. Biometals 2009, 22, 3–13. [Google Scholar] [CrossRef]
- Wang, W.; Qiu, Z.; Tan, H.; Cao, L. Siderophore production by actinobacteria. Biometals 2014, 27, 623–631. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibel-ski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Kieser, T. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000; ISBN 9780708406236. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Milagres, A.M.F.; Machuca, A.; Napoleão, D. Detection of siderophore production from several fungi and bacteria by a modification of chrome azurol S (CAS) agar plate assay. J. Microbiol. Methods 1999, 37, 1–6. [Google Scholar] [CrossRef]
- Sidebottom, A.M.; Karty, J.A.; Carlson, E.E. Accurate Mass MS/MS/MS Analysis of Siderophores Ferrioxamine B and E1 by Collision-Induced Dissociation Electrospray Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 1899–1902. [Google Scholar] [CrossRef]
- Senges, C.H.R.; Al-Dilaimi, A.; Marchbank, D.H.; Wibberg, D.; Winkler, A.; Haltli, B.; Nowrousian, M.; Kalinowski, J.; Kerr, R.G.; Bandow, J.E. The secreted metabolome of Streptomyces chartreusis and implications for bacterial chemistry. Proc. Natl. Acad. Sci. USA 2018, 115, 2490–2495. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simao, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. STAG: Species Tree Inference from All Genes. bioRxiv 2018, 267914. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 2020, 11, 6058. [Google Scholar] [CrossRef] [PubMed]
- Mungan, M.D.; Alanjary, M.; Blin, K.; Weber, T.; Medema, M.H.; Ziemert, N. ARTS 2.0: Feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Res. 2020, 48, W546–W552. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. Available online: http://bioinfogp.cnnb.csic.es/tools/venny/index.html (accessed on 9 September 2020).
- Chiesi USA, Inc. Bronchitol ® Inhaled Dry Powder Mannitol (DPM) for Adult Patients with Cystic Fibrosis; Chiesi USA, Inc.: Cary, NC, USA, 2019. [Google Scholar]
- Mitri, C.; Xu, Z.; Bardin, P.; Corvol, H.; Touqui, L.; Tabary, O. Novel Anti-Inflammatory Approaches for Cystic Fibrosis Lung Disease: Identification of Molecular Targets and Design of Innovative Therapies. Front. Pharmacol. 2020, 11, 1096. [Google Scholar] [CrossRef] [PubMed]
- Nevitt, S.J.; Thornton, J.; Murray, C.S.; Dwyer, T. Inhaled mannitol for cystic fibrosis. Cochrane Database Syst. Rev. 2020, 2020, CD008649. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.J.; Wang, J.H.; Bu, X.L.; Yu, H.L.; Li, P.; Ou, H.Y.; He, Y.; Di Xu, F.; Hu, X.Y.; Zhu, X.M.; et al. Deciphering the streamlined genome of Streptomyces xiamenensis 318 as the producer of the anti-fibrotic drug candidate xiamenmycin. Sci. Rep. 2016, 6, 18977. [Google Scholar] [CrossRef] [PubMed]
- Ian, E.; Malko, D.B.; Sekurova, O.N.; Bredholt, H.; Rückert, C.; Borisova, M.E.; Albersmeier, A.; Kalinowski, J.; Gelfand, M.S.; Zotchev, S.B. Genomics of Sponge-Associated Streptomyces spp. Closely Related to Streptomyces albus J1074: Insights into Marine Adaptation and Secondary Metabolite Biosynthesis Potential. PLoS ONE 2014, 9, e96719. [Google Scholar] [CrossRef] [PubMed]
- Tomihama, T.; Nishi, Y.; Sakai, M.; Ikenaga, M.; Okubo, T.; Ikeda, S. Draft genome sequences of Streptomyces scabiei S58, Streptomyces turgidiscabies T45, and Streptomyces acidiscabies a10, the pathogens of potato common scab, isolated in Japan. Genome Announc. 2016, 4, e00062-16. [Google Scholar] [CrossRef]
- Worsley, S.F.; Newitt, J.; Rassbach, J.; Batey, S.F.D.; Holmes, N.A.; Murrell, J.C.; Wilkinson, B.; Hutchings, M.I. Streptomyces endophytes promote host health and enhance growth across plant species. Appl. Environ. Microbiol. 2020, 86, e01053-20. [Google Scholar] [CrossRef]
- Vicente, C.M.; Santos-Aberturas, J.; Payero, T.D.; Barreales, E.G.; de Pedro, A.; Aparicio, J.F. PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 2014, 98, 9311–9324. [Google Scholar] [CrossRef] [PubMed]
- Payero, T.D.; Vicente, C.M.; Rumbero, Á.; Barreales, E.G.; Santos-Aberturas, J.; de Pedro, A.; Aparicio, J.F. Functional analysis of filipin tailoring genes from Streptomyces filipinensis reveals alternative routes in filipin III biosynthesis and yields bioactive derivatives. Microb. Cell Factories 2015, 14, 114. [Google Scholar] [CrossRef]
- Whitfield, G.B.; Brock, T.D.; Ammann, A.; Gottlieb, D.; Carter, H.E. Filipin, an Antifungal Antibiotic: Isolation and Properties. J. Am. Chem. Soc. 1955, 77, 4799–4801. [Google Scholar] [CrossRef]
- Keller, U.; Lang, M.; Crnovcic, I.; Pfennig, F.; Schauwecker, F. The actinomycin biosynthetic gene cluster of Streptomyces chrysomallus: A genetic hall of mirrors for synthesis of a molecule with mirror symmetry. J. Bacteriol. 2010, 192, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Crnovčić, I.; Rückert, C.; Semsary, S.; Lang, M.; Kalinowski, J.; Keller, U. Genetic interrelations in the actinomycin biosynthetic gene clusters of Streptomyces antibioticus IMRU 3720 and Streptomyces chrysomallus ATCC11523, producers of actinomycin X and actinomycin C. Adv. Appl. Bioinform. Chem. 2017, 10, 29–46. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.S.; Pearson, A.; Sáenz, J.P.; Marx, C.J. Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis. Org. Geochem. 2010, 41, 1075–1081. [Google Scholar] [CrossRef]
- Doughty, D.M.; Dieterle, M.; Sessions, A.L.; Fischer, W.W.; Newman, D.K. Probing the Subcellular Localization of Hopanoid Lipids in Bacteria Using NanoSIMS. PLoS ONE 2014, 9, e84455. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; WASHIO, T.; SATO, M.; SUZUKI, Y. Cytotoxic Effects of Several Hopanoids on Mouse Leukemia L1210 and P388 Cells. Biol. Pharm. Bull. 1995, 18, 421–423. [Google Scholar] [CrossRef][Green Version]
- Moreau, R.A.; Hicks, K.B. Bacteriohopanetetrol and Related Compounds Useful for Modulation of Lipoxygenase Activity and Anti-Inflammatory Applications. U.S. Patent No US6177415B1, 23 January 2001. [Google Scholar]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L.; et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar] [CrossRef]
- Giessen, T.W.; Franke, K.B.; Knappe, T.A.; Kraas, F.I.; Bosello, M.; Xie, X.; Linne, U.; Marahiel, M.A. Isolation, structure elucidation, and biosynthesis of an unusual hydroxamic acid ester-containing siderophore from Actinosynnema mirum. J. Nat. Prod. 2012, 75, 905–914. [Google Scholar] [CrossRef]
- Barona-Gómez, F.; Wong, U.; Giannakopulos, A.E.; Derrick, P.J.; Challis, G.L. Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 2004, 126, 16282–16283. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, M.; Zhang, Q.; Zhang, X.; Yang, P.; Liu, Z.; Deng, Y.; Zhu, Y.; Huang, X.; Han, L.; et al. Diisonitrile Natural Product SF2768 Functions as a Chalkophore That Mediates Copper Acquisition in Streptomyces thioluteus. ACS Chem. Biol. 2017, 12, 3067–3075. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- van Zyl, W.F.; Deane, S.M.; Dicks, L.M.T. Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 2020, 12, 1831339. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jia, Y.; Xie, Y.; Zhang, C.; Ma, J.; Sun, C.; Ju, J. Identification of the actinomycin D biosynthetic pathway from marine-derived Streptomyces costaricanus SCSIO ZS0073. Mar. Drugs 2019, 17, 240. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Soltani, B.M.; Nekouei, M.K.; Jouzani, G.S.; Mirzaei, H.H.; Sadeghizadeh, M. Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 2014, 169, 699–708. [Google Scholar] [CrossRef]
- Hassan, A.M.E.; Fahal, A.H.; Ahmed, A.O.; Ismail, A.; Veress, B. The immunopathology of actinomycetoma lesions caused by Streptomyces somaliensis. Trans. R. Soc. Trop. Med. Hyg. 2001, 95, 89–92. [Google Scholar] [CrossRef]
- Shah, S.; Briken, V. Modular Organization of the ESX-5 Secretion System in Mycobacterium tuberculosis. Front. Cell. Infect. Microbiol. 2016, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Dumas, E.; Boritsch, E.C.; Vandenbogaert, M.; De La Vega, R.C.R.; Thiberge, J.M.; Caro, V.; Gaillard, J.L.; Heym, B.; Girard-Misguich, F.; Brosch, R.; et al. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol. Evol. 2016, 8, 387–402. [Google Scholar] [CrossRef]
- Ju, K.S.; Gao, J.; Doroghazi, J.R.; Wang, K.K.A.; Thibodeaux, C.J.; Li, S.; Metzger, E.; Fudala, J.; Su, J.; Zhang, J.K.; et al. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc. Natl. Acad. Sci. USA 2015, 112, 12175–12180. [Google Scholar] [CrossRef]
- Koonin, E.V.; Galperin, M.Y. Sequence-Evolution-Function: Computational Approaches in Comparative Genomics. In Sequence-Evolution-Function: Computational Approaches in Comparative Genomics; Springer: Boston, MA, USA, 2003; ISBN 1-40207-274-0. [Google Scholar]
- Karoonuthaisiri, N.; Weaver, D.; Huang, J.; Cohen, S.N.; Kao, C.M. Regional organization of gene expression in Streptomyces coelicolor. Gene 2005, 353, 53–66. [Google Scholar] [CrossRef]
- Gaillard, J.-L.; Berche, P.; Frehei, C.; Gouin, E.; Cossartt, P. Entry of L. monocytogenes into Cells Is Mediated by Internalin, a Repeat Protein Reminiscent of Surface Antigens from Gram-Positive Cocci. Cell 1991, 65, 1127–1141. [Google Scholar] [CrossRef]
- Kobayashi, T.; Uozomi, T.; Beppu, T. Cloning and characterization of the streptothricin-resistance gene which encodes streptothricin acetyltransferase from Streptomyces lavendulae. J. Antibiot. 1986, 39, 688–693. [Google Scholar] [CrossRef]
- Rajasekaran, M.B.; Nilapwar, S.; Andrews, S.C.; Watson, K.A. EfeO-cupredoxins: Major new members of the cupredoxin superfamily with roles in bacterial iron transport. Biometals 2010, 23, 1. [Google Scholar] [CrossRef]
- Chew, S.Y.; Chee, W.J.Y.; Than, L.T.L. The glyoxylate cycle and alternative carbon metabolism as metabolic adaptation strategies of Candida glabrata: Perspectives from Candida albicans and Saccharomyces cerevisiae. J. Biomed. Sci. 2019, 26, 52. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Puckett, S.; Trujillo, C.; Wang, Z.; Eoh, H.; Ioerger, T.R.; Krieger, I.; Sacchettini, J.; Schnappinger, D.; Rhee, K.Y.; Ehrt, S. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2017, 114, E2225–E2232. [Google Scholar] [CrossRef] [PubMed]
- Koedooder, C.; Guéneuguès, A.; Van Geersdaële, R.; Vergé, V.; Bouget, F.-Y.; Labreuche, Y.; Obernosterer, I.; Blain, S. The Role of the Glyoxylate Shunt in the Acclimation to Iron Limitation in Marine Heterotrophic Bacteria. Front. Mar. Sci. 2018, 5, 435. [Google Scholar] [CrossRef]
- Flores-Díaz, M.; Monturiol-Gross, L.; Naylor, C.; Alape-Girón, A.; Flieger, A. Bacterial Sphingomyelinases and Phospholipases as Virulence Factors. Microbiol. Mol. Biol. Rev. 2016, 80, 597–628. [Google Scholar] [CrossRef]
- van der Meer-Janssen, Y.P.M.; van Galen, J.; Batenburg, J.J.; Helms, J.B. Lipids in host-pathogen interactions: Pathogens exploit the complexity of the host cell lipidome. Prog. Lipid Res. 2010, 49, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Songer, J.G. Bacterial phospholipases and their role in virulence. Trends Microbiol. 1997, 5, 156–161. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).