Engineering the Steroid Hydroxylating System from Cochliobolus lunatus in Mycolicibacterium smegmatis
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Strains, Oligonucleotides and Culture Growth
2.3. Construction of the M. smegmatis Heterologous Strains
2.4. Steroid Biotransformation Assay
2.5. Steroid Extraction and Analyses
| Time (min) | %A | %B | %C |
| 0 | 50 | 50 | 0 |
| 5 | 50 | 50 | 0 |
| 15 | 20 | 71 | 9 |
| 20 | 4 | 87 | 9 |
| 40 | 0 | 85 | 15 |
| 41 | 0 | 85 | 15 |
| 42 | 50 | 50 | 0 |
| 52 | 50 | 50 | 0 |
| Time (min) | %A | %B | %C |
| 0 | 50 | 50 | 0 |
| 5 | 50 | 50 | 0 |
| 15 | 20 | 71 | 9 |
| 20 | 0 | 91 | 9 |
| 40 | 0 | 70 | 30 |
| 41 | 0 | 85 | 15 |
| 42 | 50 | 50 | 0 |
| 52 | 50 | 50 | 0 |
2.6. NMR Spectra
3. Results and Discussion
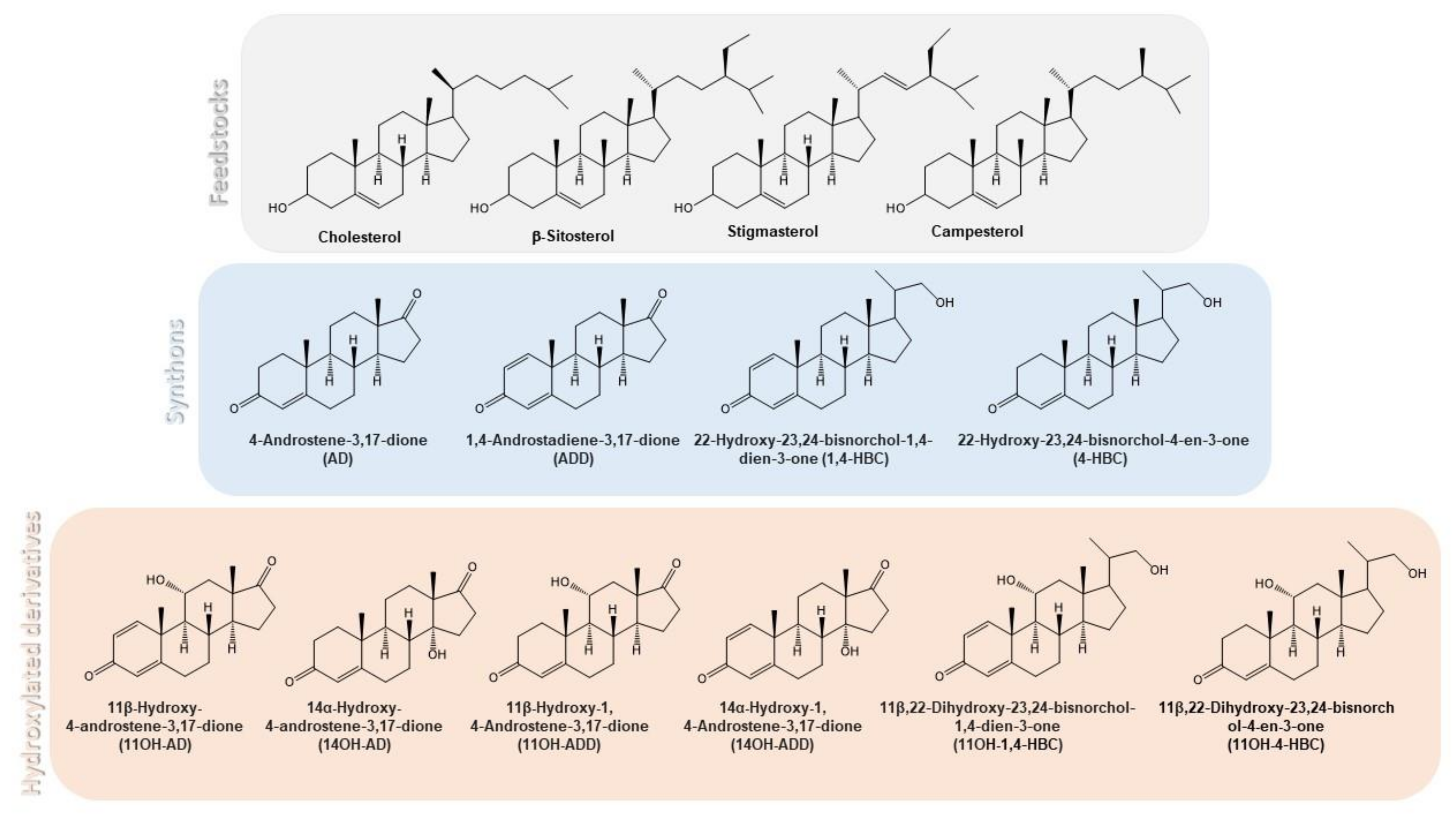
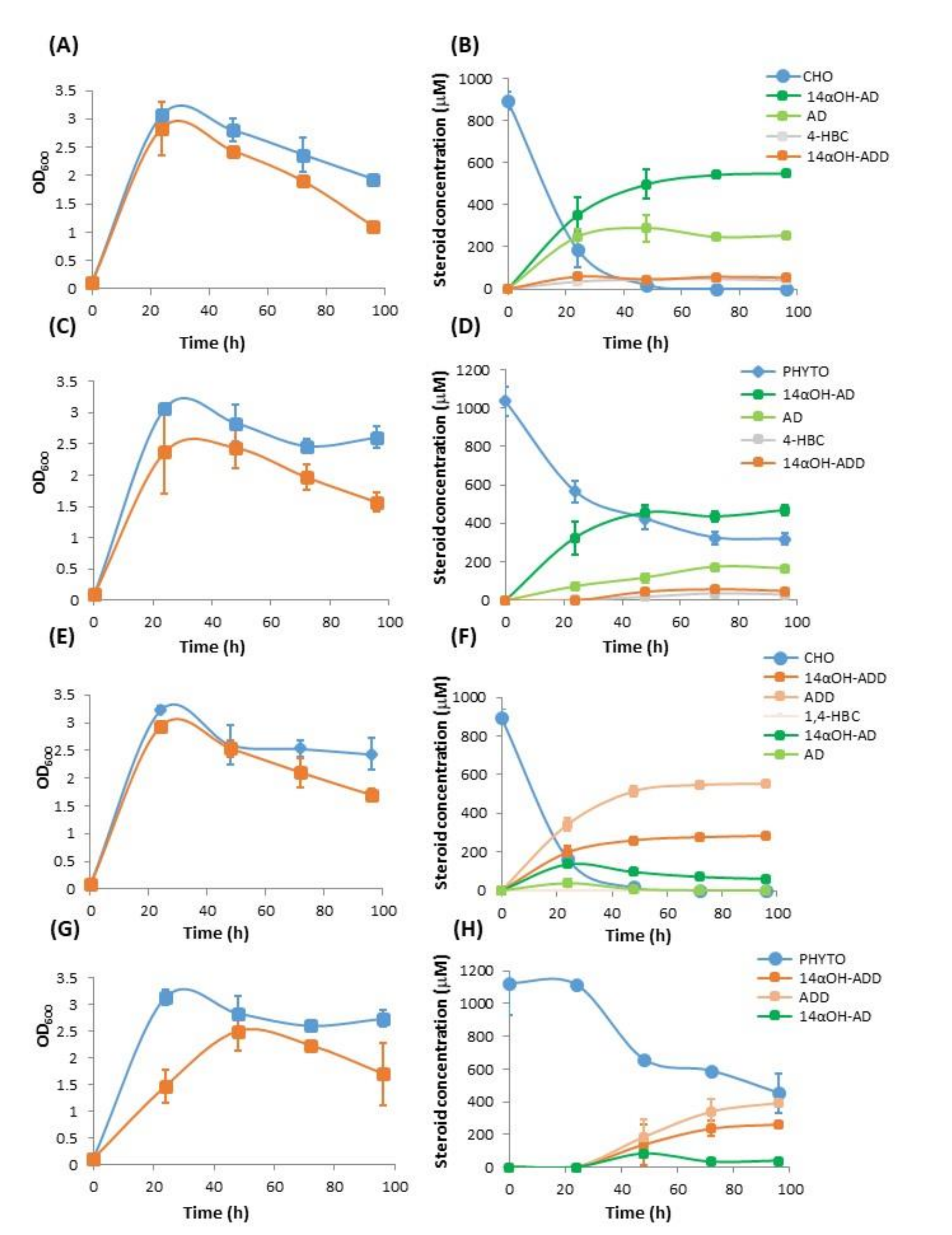
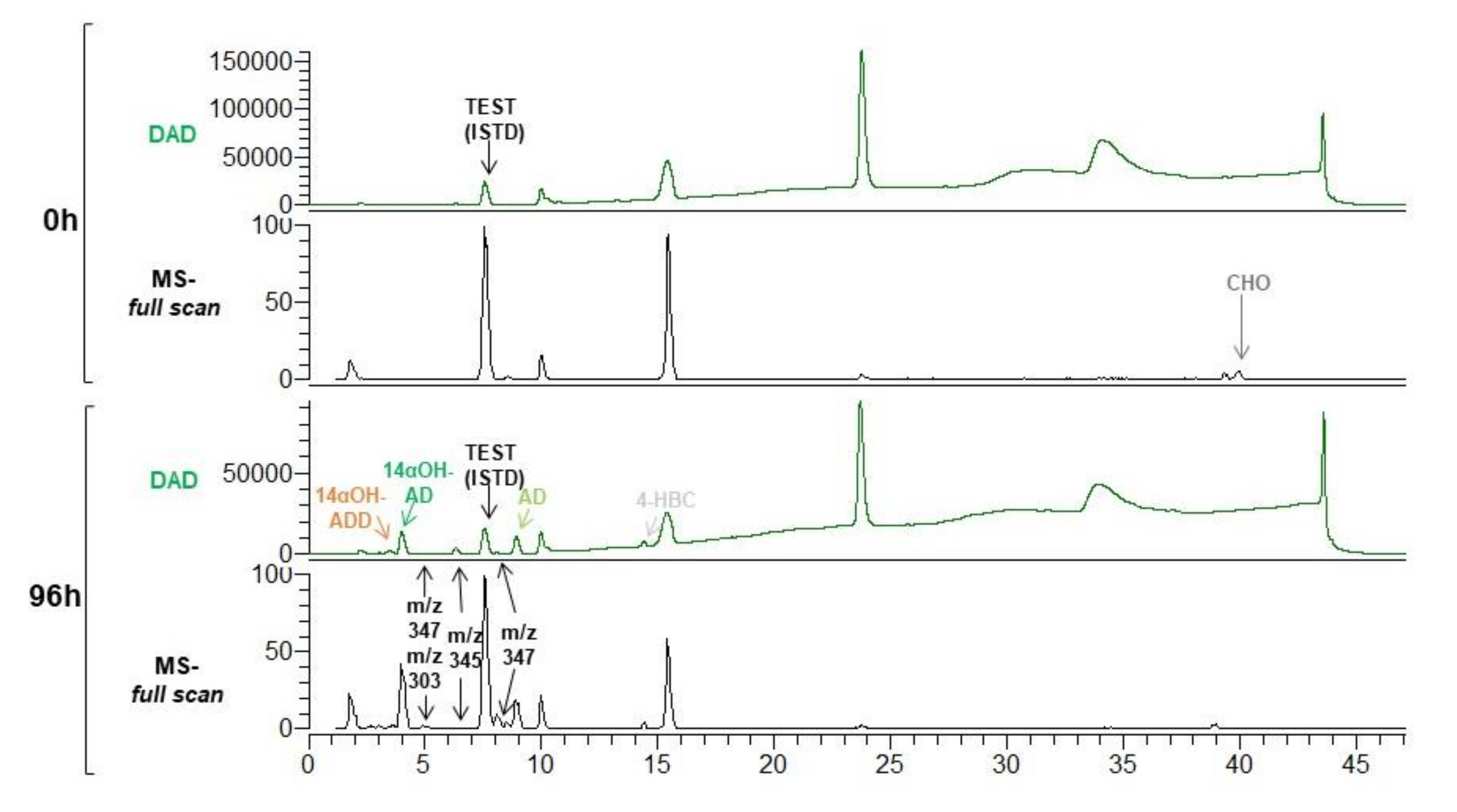
3.1. Production of 14αOH-AD in M. Smegmatis from Sterols in Single Fermentation Step
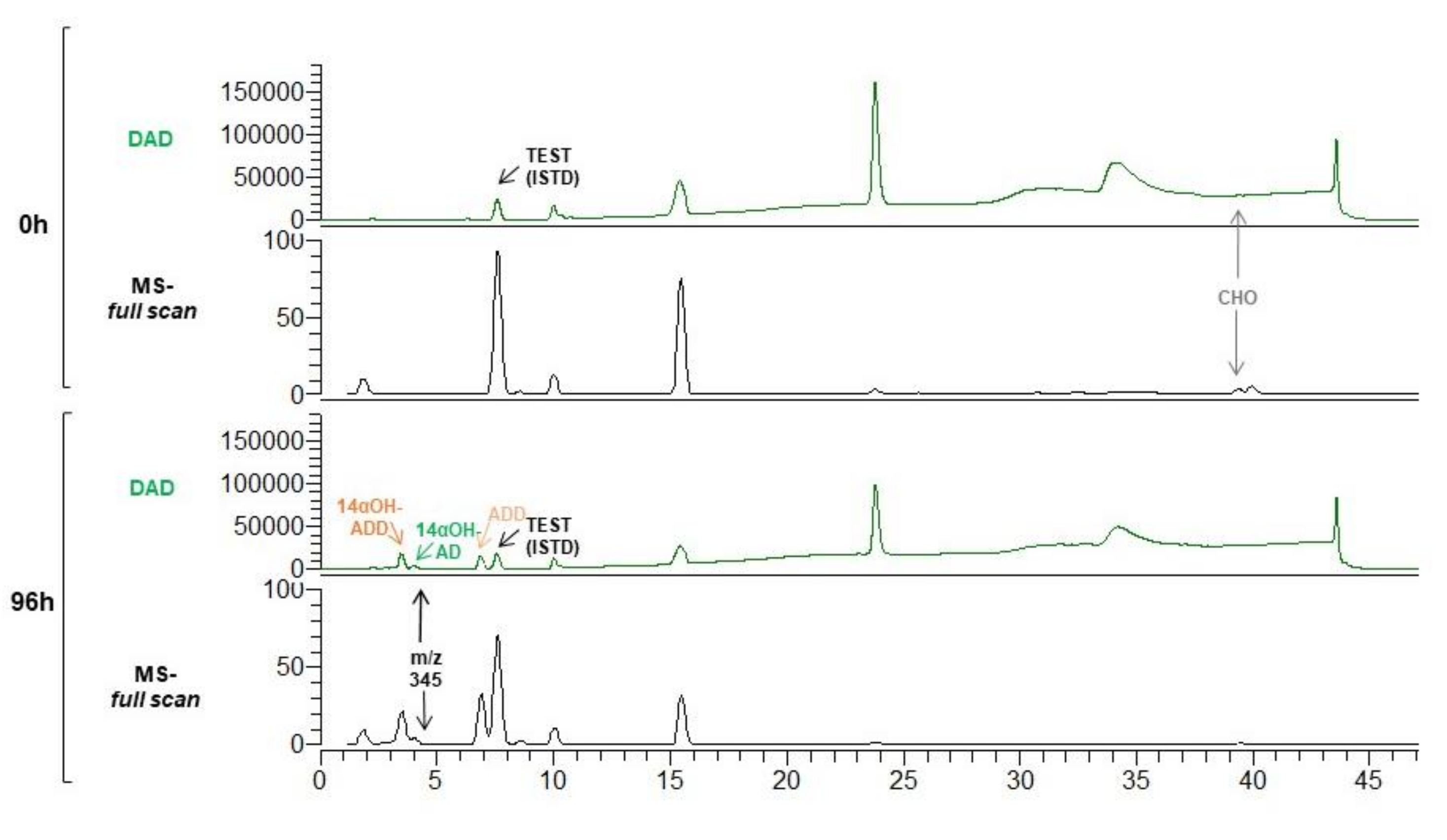
3.2. Production of 14αOH-ADD in M. smegmatis from Sterols in Single Fermentation Step
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Fernández-Cabezón, L.; Galán, B.; García, J.L. New insights on steroid biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- Lednicer, D. Steroid Chemistry at a Glance; John Wiley and Sons: New York, NY, USA, 2011; Volume 10. [Google Scholar]
- Donova, M.V.; Egorova, O.V. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012, 94, 1423–1447. [Google Scholar] [CrossRef] [PubMed]
- Szaleniec, M.; Wojtkiewicz, A.M.; Bernhardt, R.; Borowski, T.; Donova, M.V. Bacterial steroid hydroxylases: Enzyme classes, their functions and comparison of their catalytic mechanisms. Appl. Microbiol. Biotechnol. 2018, 102, 8153–8171. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.; Rižner, T.L. Steroid-transforming enzymes in fungi. J. Steroid Biochem. Mol. Biol. 2012, 129, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V. Steroid bioconversions. Methods Mol. Biol. 2017, 1645, 1–13. [Google Scholar]
- Bernhardt, R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006, 124, 128–145. [Google Scholar] [CrossRef]
- Crešnar, B.; Petrič, Š. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta 2011, 1814, 29–35. [Google Scholar] [CrossRef]
- Mahato, S.B.; Garai, S. Advances in microbial steroid biotransformation. Steroids 1997, 62, 332–345. [Google Scholar] [CrossRef]
- Schiffer, L.; Anderko, S.; Hobler, A.; Hannemann, F.; Kagawa, N.; Bernhardt, R. A recombinant CYP11B1 dependent Escherichia coli biocatalyst for selective cortisol production and optimization towards a preparative scale. Microb. Cell Fact. 2015, 14, 25. [Google Scholar] [CrossRef]
- Vita, M.; Smith, K.; Rozman, D.; Komel, R. Progesterone metabolism by the filamentous fungus Cochliobolus lunatus. J. Steroid Biochem. Mol. Biol. 1994, 49, 87–92. [Google Scholar] [CrossRef]
- Paraszkiewicz, K.; Długon, J. Cortexolone 11β-hydroxylation in protoplasts of Curvularia lunata. J. Biotechnol. 1998, 65, 217–224. [Google Scholar] [CrossRef]
- Lu, W.; Du, L.; Wang, M.; Jia, X.; Wen, J.; Huang, Y.; Guo, Y.; Gong, W.; Bao, H.; Yang, J.; et al. Optimization of hydrocortisone production by Curvularia lunata. Appl. Biochem. Biotechnol. 2007, 142, 17–28. [Google Scholar] [CrossRef]
- Felpeto-Santero, C.; Galán, B.; Luengo, J.M.; Fernández-Cañon, J.M.; Del Cerro, C.; Medrano, F.J.; García, J.L. Identification and expression of the 11β-steroid hydroxylase from Cochliobolus lunatus in Corynebacterium glutamicum. Microb. Biotechnol. 2019, 12, 856–868. [Google Scholar] [CrossRef]
- Fernández-Cabezón, L.; Galán, B.; García, J.L. Engineering Mycobacterium smegmatis for testosterone production. Microb. Biotechnol. 2017, 10, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Felpeto-Santero, C.; Galán, B.; García, J.L. Production of 11α-hydroxysteroids from sterols in a single fermentation step by Mycolicibacterium smegmatis. Microb. Biotechnol. 2021. [Google Scholar] [CrossRef]
- Schmitz, D.; Zapp, J.; Bernhardt, R. Steroid conversion with CYP106A2—Production of pharmaceutically interesting DHEA metabolites. Microb. Cell Fact. 2014, 13, 81. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ichinose, H.; Hatakeyama, M.; Yamauchi, Y. Sequence modifications and heterologous expression of eukaryotic cytochromes P450 in Escherichia coli. J. Biosci. Bioeng. 2015, 120, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hull, C.M.; Warrilow, A.G.S.; Rolley, N.J.; Price, C.L.; Donnison, I.S.; Kelly, D.E.; Kelly, S.L. Biotechnology for biofuels co-production of 11α-hydroxyprogesterone and ethanol using recombinant yeast expressing fungal steroid hydroxylases. Biotechnol. Biofuels 2017, 10, 226. [Google Scholar] [CrossRef]
- Uno, T.; Yanase, T.; Imaishi, H. Functional characterization of CYP52G3 from Aspergillus oryzae and its application for bioconversion and synthesis of hydroxyl flavanone and steroids. Biotechnol. Appl. Biochem. 2017, 64, 385–391. [Google Scholar] [CrossRef]
- Wang, R.; Sui, P.; Hou, X.; Cao, T.; Jia, L.; Lu, F.; Singh, S.; Wang, Z.; Liu, X. Cloning and identification of a novel steroid 11α-hydroxylase gene from Absidia coerulea. J. Steroid Biochem. Mol. Biol. 2017, 171, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Chen, X.; Feng, J.; Bao, Y.; Wang, Y. A fungal P450 enzyme from Thanatephorus cucumeris. Appl. Environ. Microbiol. 2018, 84, e00503-18. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Xi, Y.; Dai, Z.; Bi, C.; Chen, X.; Fan, F.; Zhang, X. Production of 14α-hydroxysteroids by a recombinant Saccharomyces cerevisiae biocatalyst expressing of a fungal steroid 14α-hydroxylation system. Appl. Microbiol. Biotechnol. 2019, 103, 8363–8374. [Google Scholar] [CrossRef]
- Barnes, H.J.; Arlotto, M.P.; Waterman, M.R. Expression and enzymatic activity of recombinant cytochrome P450 17α-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1991, 88, 5597–5601. [Google Scholar] [CrossRef]
- Gonzalez, F.J.; Korzekwa, K.R. Cytochromes P450 expression systems. Annu. Rev. Pharmacol. Toxicol. 1995, 35, 369–390. [Google Scholar] [CrossRef]
- Bureik, M.; Lisurek, M.; Bernhardt, R. The human steroid hydroxylase CYP11B1 and CYP11B2. Biol. Chem. 2002, 383, 1537–1551. [Google Scholar] [CrossRef]
- Szczebara, F.M.; Chandelier, C.; Villeret, C.; Masurel, A.; Bourot, S.; Duport, C.; Blanchard, S.; Groisillier, A.; Testet, E.; Costaglioli, P.; et al. Total biosynthesis of hydrocortisone from a simple carbon source in yeast. Nat. Biotechnol. 2003, 21, 143–149. [Google Scholar] [CrossRef]
- Drǎgan, C.A.; Zearo, S.; Hannemann, F.; Bernhardt, R.; Bureik, M. Efficient conversion of 11-deoxycortisol to cortisol (hydrocortisone) by recombinant fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 2005, 5, 621–625. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hannemann, F.; Virus, C.; Bernhardt, R. Design of an Escherichia coli system for whole cell mediated steroid synthesis and molecular evolution of steroid hydroxylases. J. Biotechnol. 2006, 124, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Hakki, T.; Zearo, S.; Drǎgan, C.A.; Bureik, M.; Bernhardt, R. Coexpression of redox partners increases the hydrocortisone (cortisol) production efficiency in CYP11B1 expressing fission yeast Schizosaccharomyces pombe. J. Biotechnol. 2008, 133, 351–359. [Google Scholar] [CrossRef]
- Bernhardt, R.; Cresnar, B.; Hakki, T.; Petric, S. Cytochrome P450 from Rhizopus oryzae and Uses Thereof. WO2011042143, 5 October 2009. [Google Scholar]
- Fernandes, P.; Cruz, A.; Angelova, B.; Pinheiro, H.M.; Cabral, J.M.S. Microbial conversion of steroid compounds: Recent developments. Enzyme Microb. Technol. 2003, 32, 688–705. [Google Scholar] [CrossRef]
- García, J.L.; Uhía, I.; Galán, B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb. Biotechnol. 2012, 5, 679–699. [Google Scholar] [CrossRef]
- Galán, B.; Martínez, I.; Bahillo, E.; de la Fuente, J.L.; Barredo, J.L.; Fernández-Cabezón, L.; García, J.L. Mycobacterium smegmatis is a suitable cell factory for the production of steroidic synthons. Microb. Biotechnol. 2017, 10, 138–150. [Google Scholar] [CrossRef]
- Janig, G.; Pfeil, D.; Muller-Frohne, M.; Riemer, H.; Henning, M.; Schwarze, W.; Ruckpaul, K. Steroid 11 beta-hydroxylation by a fungal microsomal cytochrome P450. J. Steroid Biochem. Mol. Biol. 1992, 43, 1117–1123. [Google Scholar] [CrossRef]
- Suzuki, K.; Sanga, K.I.; Chikaoka, Y.; Itagaki, E. Purification and properties of cytochrome P-450 (P-450lun) catalyzing steroid 11β-hydroxylation in Curvularia lunata. Biochim. Biophys. Acta Protein Struct. Mol. 1993, 1203, 215–223. [Google Scholar] [CrossRef]
- Shuvalova, S.D.; Gabinskaya, K.N.; Popova, E.V.; Savinova, T.S.; Andryushina, V.A. Hydroxylation of androst-4-ene-3,17-dione with the aid of Curvularia lunata fungus. Pharm. Chem. J. 2001, 35, 279–281. [Google Scholar] [CrossRef]
- Andryushina, V.A.; Voishvillo, N.E.; Druzhinina, A.V.; Stytsenko, T.S.; Yaderets, V.V.; Petrosyan, M.A.; Zeinalov, O.A. 14α-Hydroxylation of steroids by mycelium of the mold fungus Curvularia lunata (VKPM F-981) to produce precursors for synthesizing new steroidal drugs. Pharm. Chem. J. 2013, 47, 103–108. [Google Scholar] [CrossRef]
- Andryushina, V.A.; Iaderets, V.V.; Stytsenko, T.S.; Druzhinina, A.V.; Voĭshvillo, N.E. Effect of the steroid molecule structure on the direction of its hydroxylation by the fungus Curvularia lunata. Prikl. Biokhim. Mikrobiol. 2013, 49, 382–390. [Google Scholar] [CrossRef]
- Hu, S.H.; Genain, G.; Azerad, R. Microbial transformation of steroids: Contribution to 14α-hydroxylations. Steroids 1995, 60, 337–352. [Google Scholar] [CrossRef]
- Ahmad, S.; Henderson, K.; Dumsday, G.; Zachariou, M. Microbial biotransformations: Stereoselective synthesis of pharmaceutical drug precursors. Australas. Biotechnol. 2001, 11, 26–28. [Google Scholar]
- Snapper, S.B.; Melton, R.E.; Mustafa, S.; Kieser, T.; Jacobs, W.R.J. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990, 4, 1911–1919. [Google Scholar] [CrossRef]
- Stover, C.K.; Cruz, V.F.D.I.; Fuerst, T.R.; Burlein, J.E.; Benson, L.A.; Bennett, L.T.; Bansal, G.P.; Young, J.F.; Lee, M.H.; Hatfull, G.F.; et al. New use of BCG for recombinant vaccines. Nature 1991, 351, 8763–8767. [Google Scholar] [CrossRef] [PubMed]
- García, J.L.; Uhía, I.; Galán, B. Selective Mycobacterium smegmatis mc2 155 Recombinant Mutants and Use Thereof for Producing 1,4-Androstadiene-3,17-Dione or 4-Androstene-3,17-Dione from Natural Sterols. EP112472A1, 27 March 2019. [Google Scholar]
- Kollerov, V.V.; Shutov, A.A.; Fokina, V.V.; Sukhodol’skaia, G.V.; Gulevskaia, S.A.; Donova, M.V. Bioconversion of C19- and C21-steroids with parent and mutant strains of Curvularia lunata. Prikl. Biokhim. Mikrobiol. 2010, 46, 212–220. [Google Scholar] [CrossRef] [PubMed]

| Strain | Description | Reference |
|---|---|---|
| Mycolicibacterium smegmatis mc2 155 | ept−1, mutant mc26 | [42] |
| Mycolicibacterium smegmatis mc2 155 MS6039 | M. smegmatis mc2 155 ΔMSMEG_6039 | [34] |
| Mycolicibacterium smegmatis mc2 155 MS6039-5941 | M. smegmatis mc2 155 ΔMSMEG_6039-ΔMSMEG_5941 | [34] |
| Escherichia coli DH10B | F−, mcrA, Δ(mrr-hsdRMS-mcrBC), f80ΔlacZDM15 ΔlacX74, deoR, recA1, endA1, araD139, Δ(ara-leu)7697, galU, galK, rpsL, nupG, λ− | Invitrogen |
| Plasmid | Description | Reference |
| pMV261 | KmR, Mycobacterium expression vector, PHsp60 | [43] |
| pMVFAN | KmR, synthetic operon FAN into pMV261 | This work |
| Oligonucleotide | Sequence | Application |
| pMV4260 F | TTGCCGTCACCCGGTGACC | pMV261 sequencing |
| pMV4486 R | ATCACCGCGGCCATGATGG | pMV261 sequencing |
| 64795 EcoRIF | CCGGAATTCTGACCTGAGAGAAAGGGAGTGATAAATGGCACAACTCGACACGC | CPR64795 amplification |
| 64795 XbaIR | GCTCTAGATTATCATGACCAGACGTCTTCCTG | CPR64795 amplification |
| 103168 BglXbaMunF | GAAGATCTTCTAGACAATTGTGACCTGAGAGAAAGGGAGTGATAAATGGATCCCCAGACTGTCG | CYP103168 amplification |
| 103168 XhoR | CCGCTCGAGTTACTACACTACCACTCTCTTGAAAGC | CYP103168 amplification |
| 64795 F2 | AATCAGCATTGCTGGCTCC | FAN operon sequencing |
| 64795 F3 | CTCCAACTTCAAGCTTCCTTCG | FAN operon sequencing |
| 64795 F4 | AATACGTCGCTTTCGGTCTCG | FAN operon sequencing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felpeto-Santero, C.; Galán, B.; García, J.L. Engineering the Steroid Hydroxylating System from Cochliobolus lunatus in Mycolicibacterium smegmatis. Microorganisms 2021, 9, 1499. https://doi.org/10.3390/microorganisms9071499
Felpeto-Santero C, Galán B, García JL. Engineering the Steroid Hydroxylating System from Cochliobolus lunatus in Mycolicibacterium smegmatis. Microorganisms. 2021; 9(7):1499. https://doi.org/10.3390/microorganisms9071499
Chicago/Turabian StyleFelpeto-Santero, Carmen, Beatriz Galán, and José Luis García. 2021. "Engineering the Steroid Hydroxylating System from Cochliobolus lunatus in Mycolicibacterium smegmatis" Microorganisms 9, no. 7: 1499. https://doi.org/10.3390/microorganisms9071499
APA StyleFelpeto-Santero, C., Galán, B., & García, J. L. (2021). Engineering the Steroid Hydroxylating System from Cochliobolus lunatus in Mycolicibacterium smegmatis. Microorganisms, 9(7), 1499. https://doi.org/10.3390/microorganisms9071499





