Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms
Abstract
:1. Introduction
2. Materials and Methods
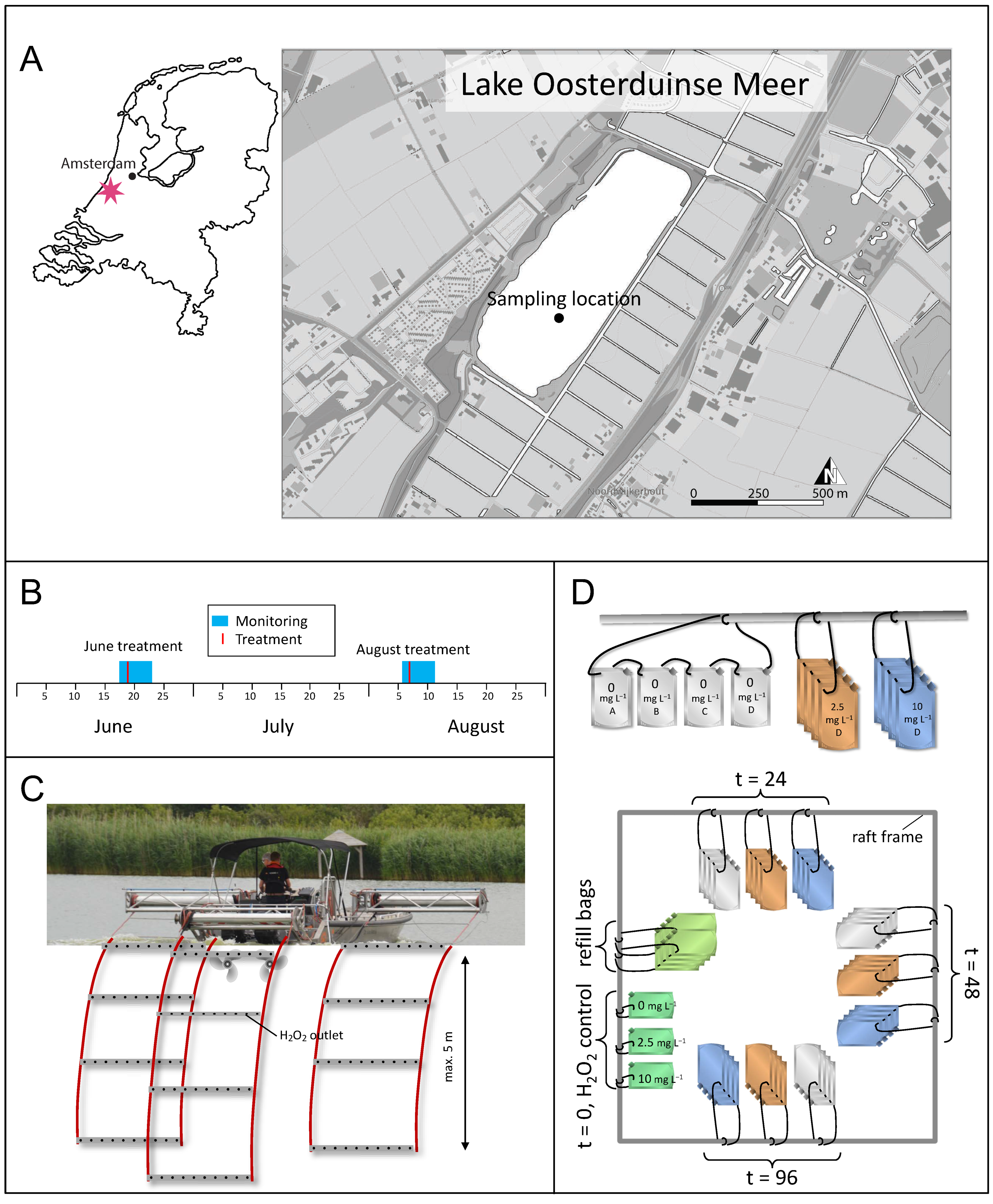
2.1. Lake, H2O2 Treatment and Sampling Information
2.2. Incubation Experiments
2.3. H2O2 Measurements
2.4. Quantification of Dissolved Inorganic Nutrients
2.5. Microscopic Analysis of Phytoplankton
2.6. Flow Cytometric Analysis of Prokaryotes
2.7. DNA Extraction
2.8. 16S rRNA Gene Amplicon Sequencing and Data Analysis
3. Results
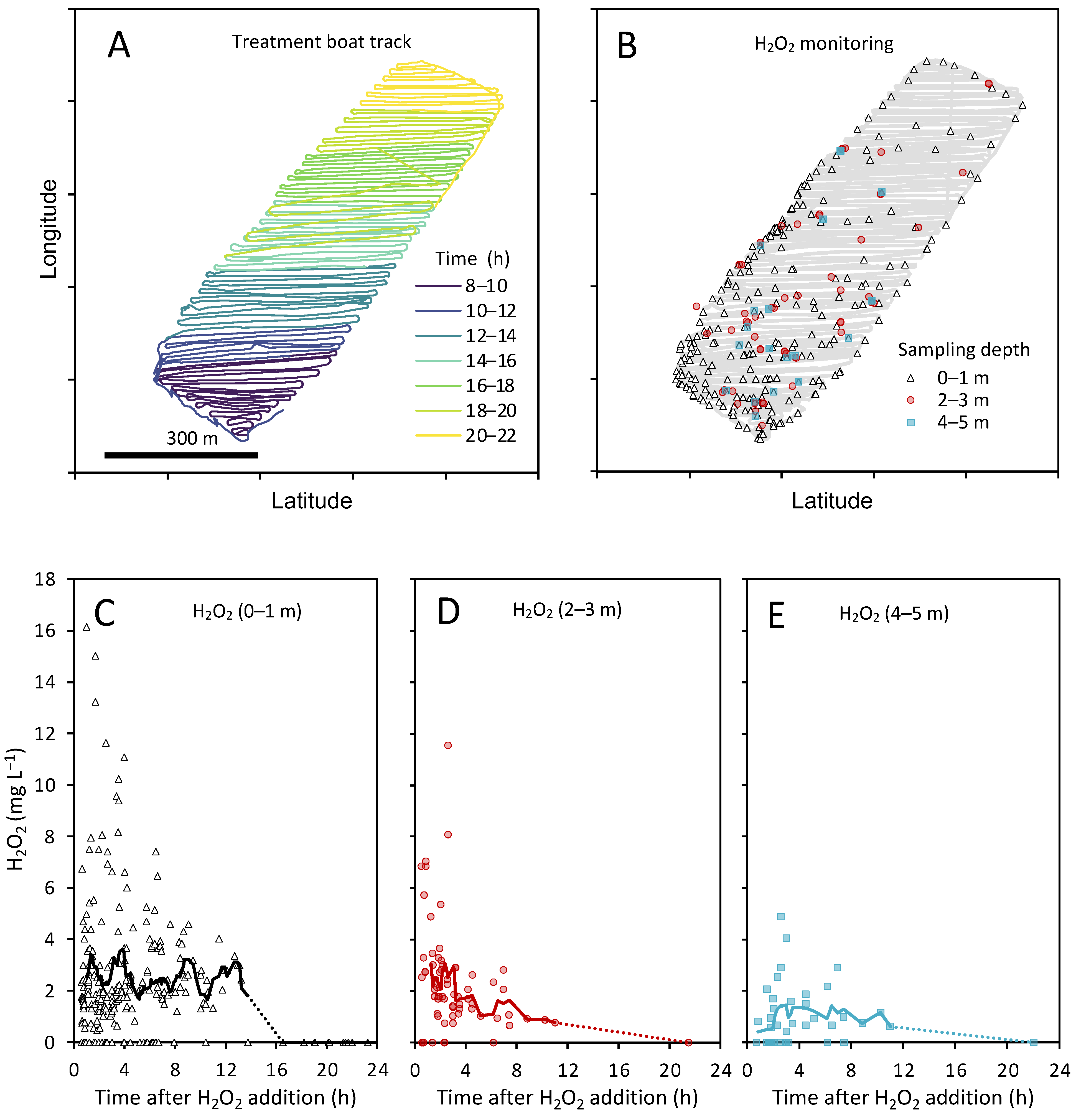
3.1. H2O2 Concentrations during the Treatment
3.2. Environmental Data during the Treatment
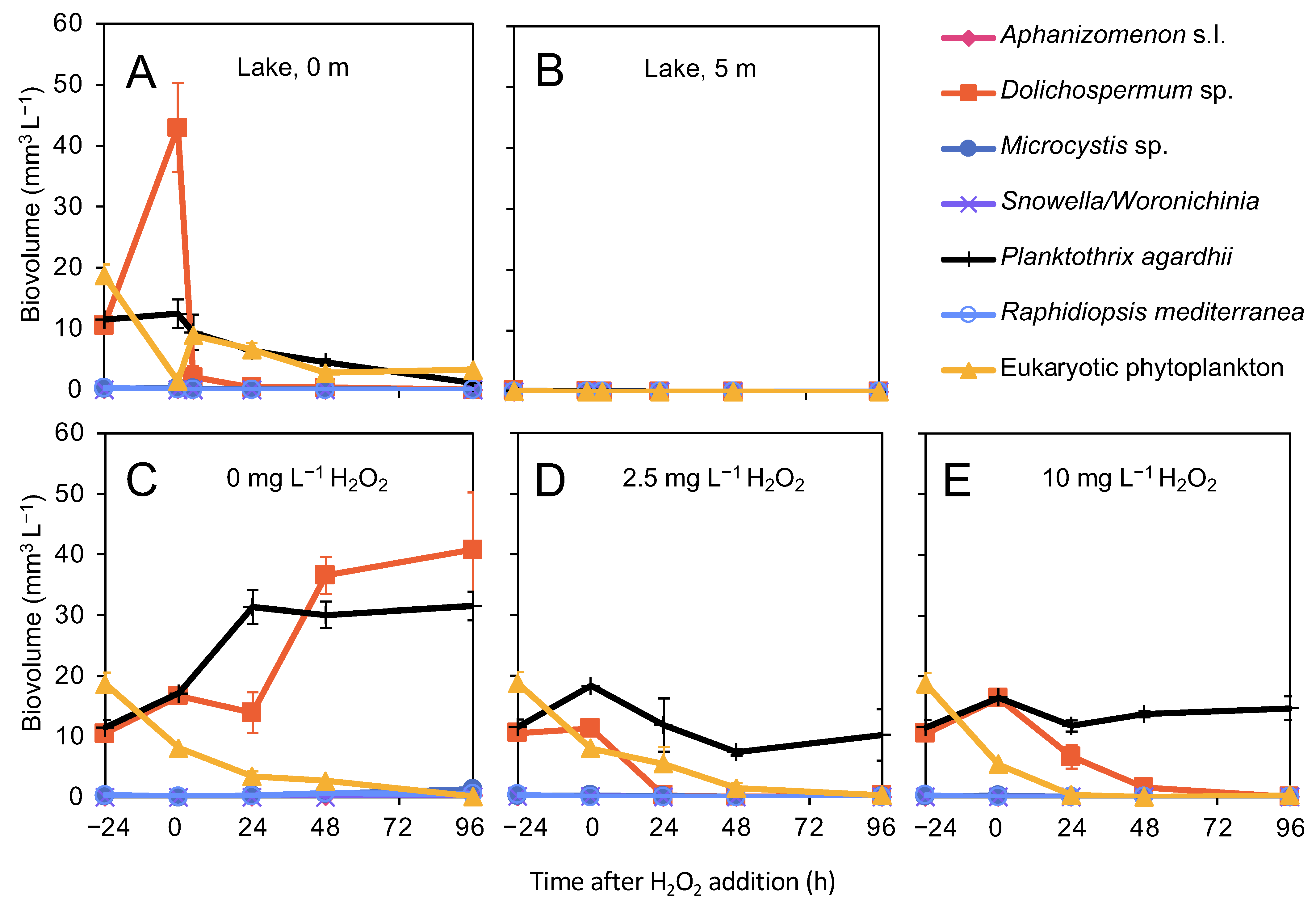
3.3. Effects of H2O2 on Phytoplankton
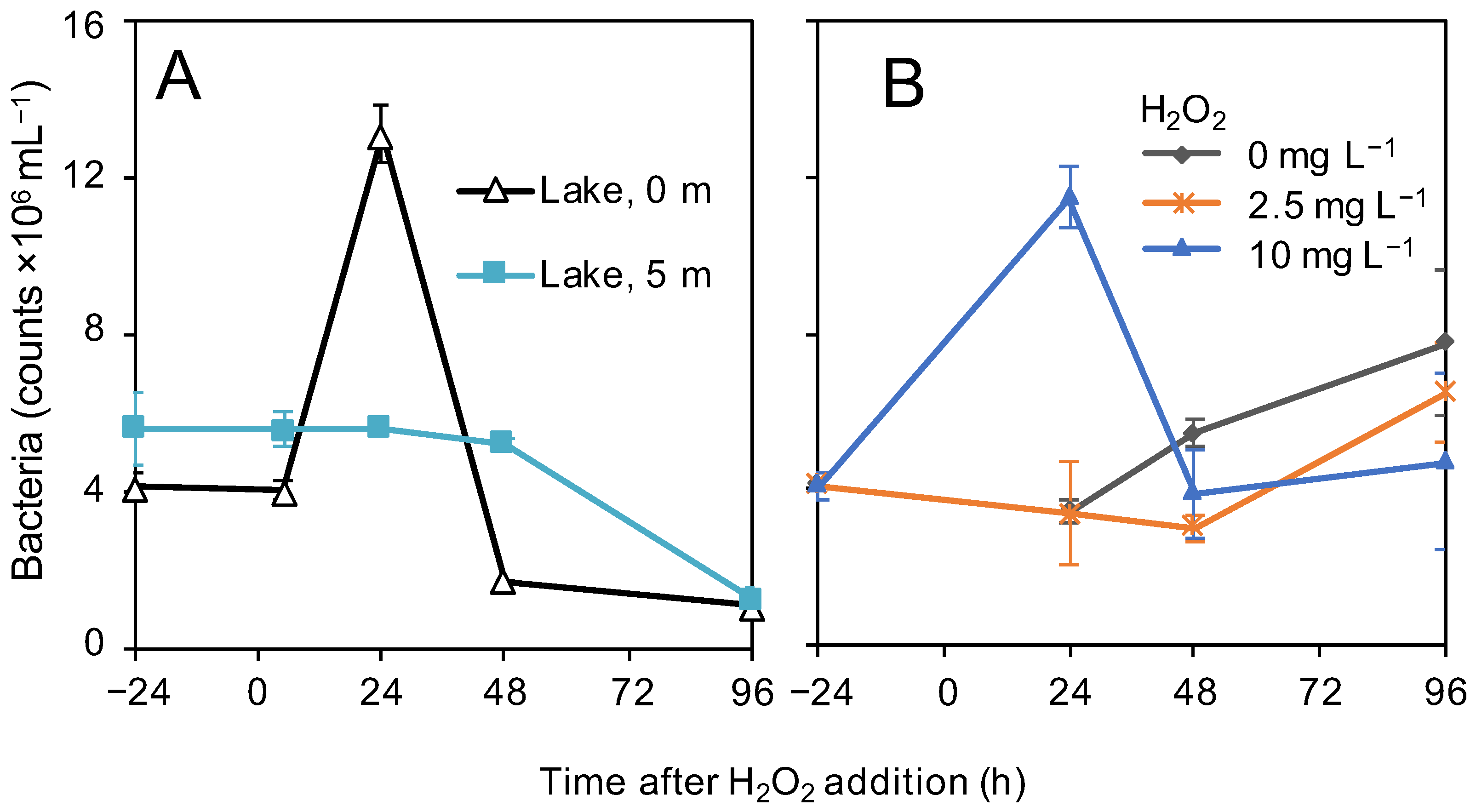
3.4. Bacterial Abundances
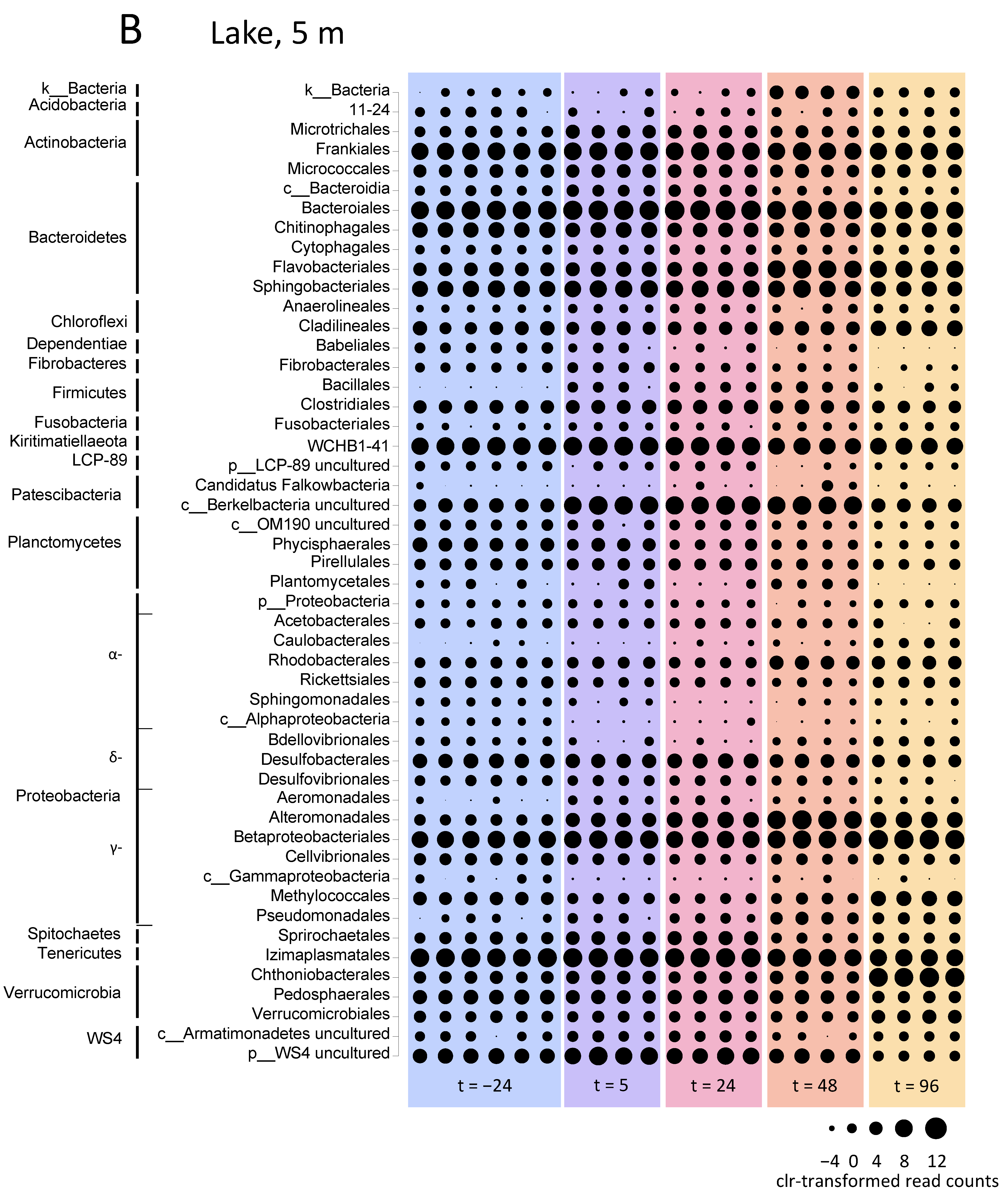
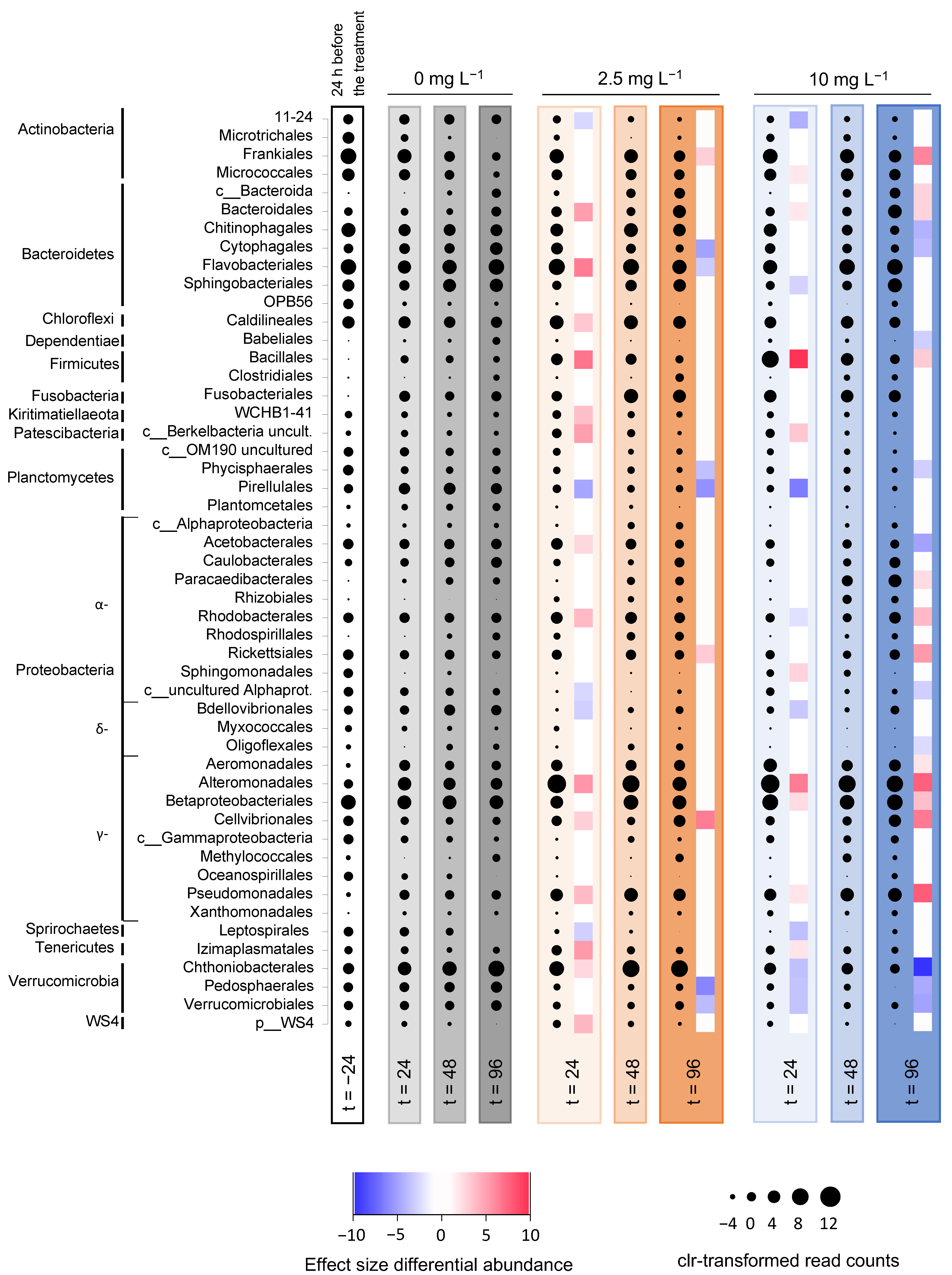
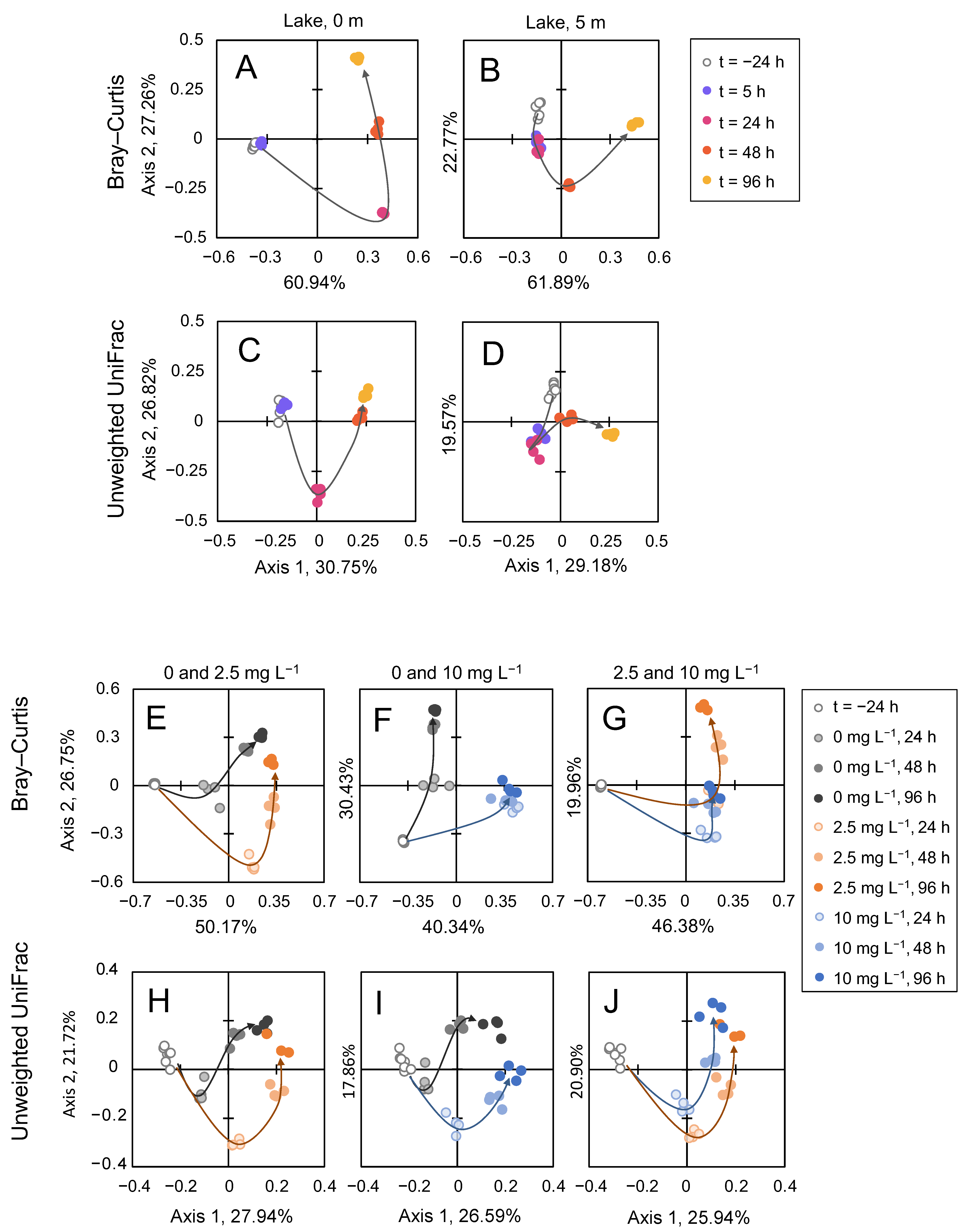
3.5. Microbial Community Analysis
3.5.1. Microbial Community Composition in the Lake
3.5.2. Microbial Community Composition in the Incubation Experiments
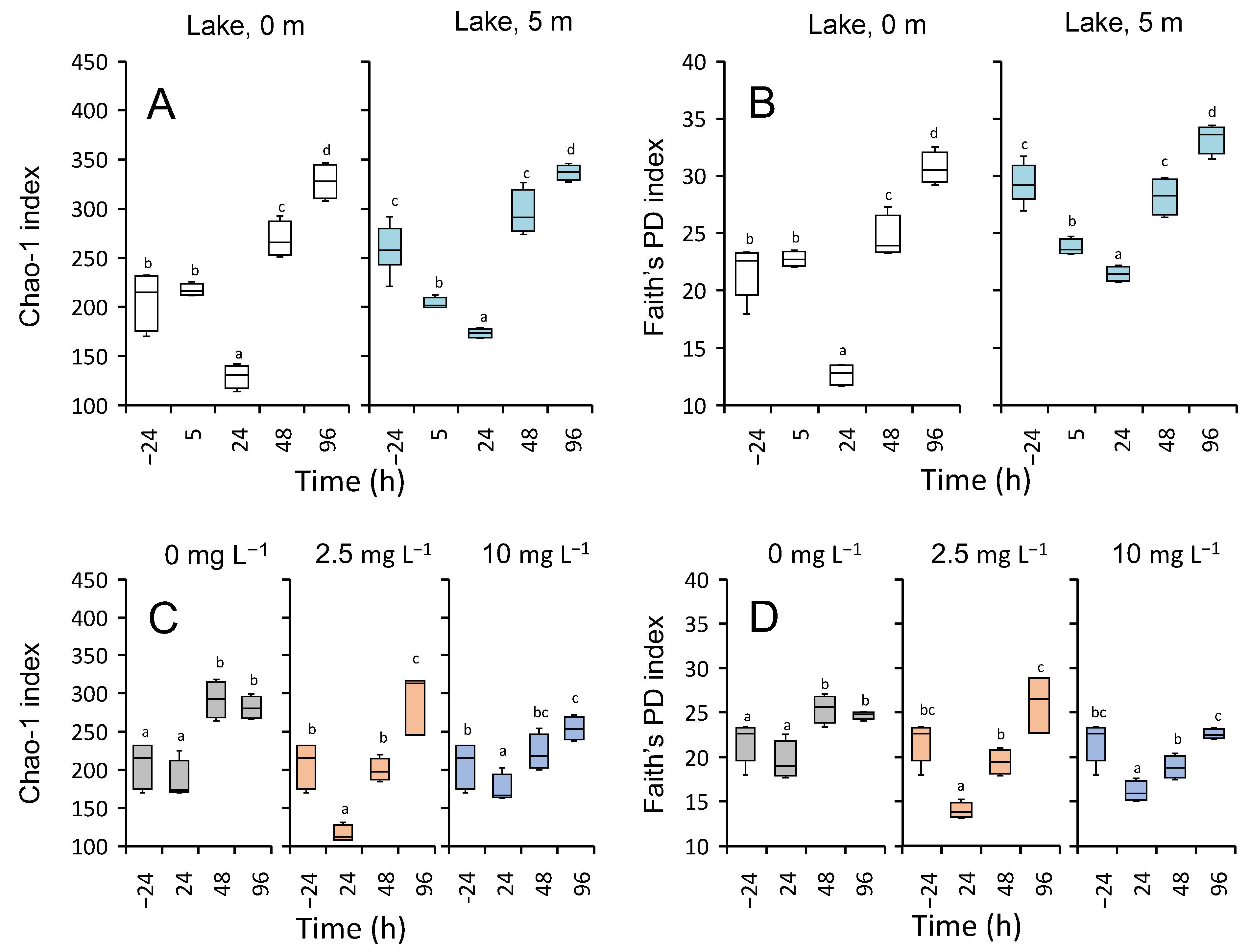
3.5.3. Microbial Diversity
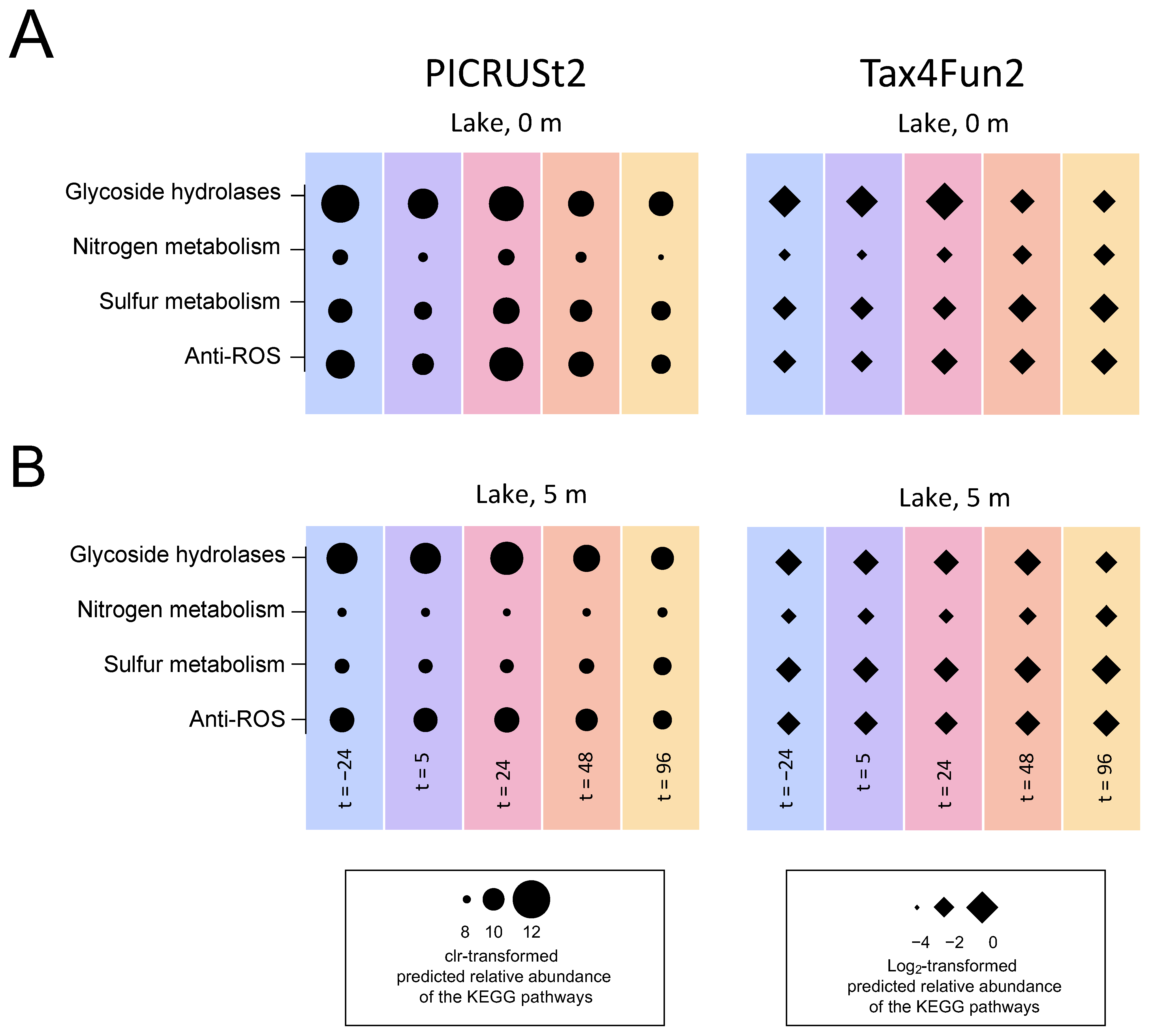
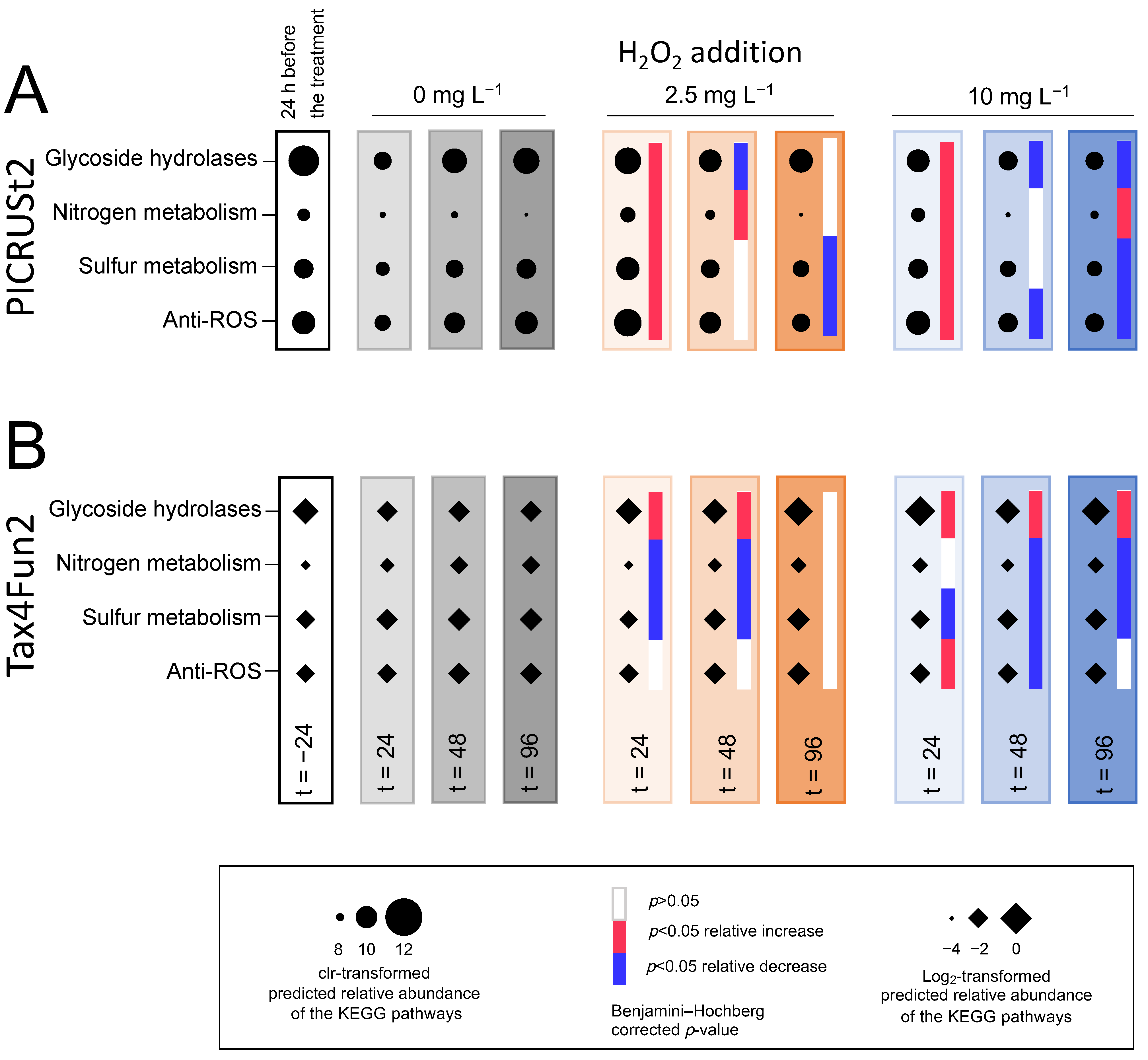
3.5.4. Functional Prediction
3.6. Comparison of the June and August Treatments
4. Discussion
4.1. Effect of H2O2 on the Phytoplankton Community
4.2. Bacterial Response to the H2O2 Treatment
4.3. Effect of H2O2 on Microbial Community Composition
4.4. Community Resilience after the Treatment with H2O2
4.5. Microbial Functions Show Resistance after the Treatment with H2O2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Michalak, A.M.; Anderson, E.J.; Beletsky, D.; Boland, S.; Bosch, N.S.; Bridgeman, T.B.; Chaffin, J.D.; Cho, K.; Confesor, R.; Daloglu, I.; et al. Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6448–6452. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, D.P.; Wood, S.A.; Dietrich, D.R.; Puddick, J. Costs of harmful blooms of freshwater cyanobacteria. In Cyanobacteria: An Economic Perspective; Naveen, K.S., Rai, A.K., Stal, L.J., Eds.; Wiley Online Library: Chichester, UK, 2014; pp. 247–256. [Google Scholar]
- Sanseverino, I.; Conduto, D.; Pozzoli, L.; Dobricic, S.; Lettieri, T. Algal Bloom and Its Economic Impact. Eur. Comm. Jt. Res. Cent. Inst. Environ. Sustain. 2016, EUR 27905 EN, 1–48. [Google Scholar] [CrossRef]
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Carmichael, W.W. Health effects of toxin-producing cyanobacteria: “The CyanoHABs”. Hum. Ecol. Risk Assess. (HERA) 2001, 7, 1393–1407. [Google Scholar] [CrossRef]
- Codd, G.A.; Morrison, L.F.; Metcalf, J.S. Cyanobacterial toxins: Risk management for health protection. Toxicol. Appl. Pharmacol. 2005, 203, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Poste, A.E.; Hecky, R.E.; Guildford, S.J. Evaluating microcystin exposure risk through fish consumption. Environ. Sci. Technol. 2011, 45, 5806–5811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bukaveckas, P.A.; Lesutienė, J.; Gasiūnaitė, Z.R.; Ložys, L.; Olenina, I.; Pilkaitytė, R.; Pūtys, Ž.; Tassone, S.; Wood, J. Microcystin in aquatic food webs of the Baltic and Chesapeake Bay Regions. Estuar. Coast. Shelf Sci. 2017, 191, 50–59. [Google Scholar] [CrossRef] [Green Version]
- Visser, P.M.; Ibelings, B.W.; Bormans, M.; Huisman, J. Artificial mixing to control cyanobacterial blooms: A review. Aquat. Ecol. 2016, 50, 423–441. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.C.; Michalak, A.M.; Pahlevan, N. Widespread global increase in intense lake phytoplankton blooms since the 1980s. Nature 2019, 574, 667–670. [Google Scholar] [CrossRef]
- O’Neil, J.M.; Davis, T.W.; Burford, M.A.; Gobler, C.J. The rise of harmful cyanobacteria blooms: The potential roles of eutrophication and climate change. Harmful Algae 2012, 14, 313–334. [Google Scholar] [CrossRef]
- Paerl, H.W.; Huisman, J. Climate: Blooms like it hot. Science 2008, 320, 57–58. [Google Scholar] [CrossRef] [Green Version]
- Jeppesen, E.; Søndergaard, M.; Jensen, J.P.; Havens, K.E.; Anneville, O.; Carvalho, L.; Coveney, M.F.; Deneke, R.; Dokulil, M.T.; Foy, B.; et al. Lake responses to reduced nutrient loading: An analysis of contemporary long-term data from 35 case studies. Freshw. Biol. 2005, 50, 1747–1771. [Google Scholar] [CrossRef]
- Fastner, J.; Abella, S.; Litt, A.; Morabito, G.; Vörös, L.; Pálffy, K.; Straile, D.; Kümmerlin, R.; Matthews, D.; Phillips, M.G.; et al. Combating cyanobacterial proliferation by avoiding or treating inflows with high P load: Experiences from eight case studies. Aquat. Ecol. 2016, 50, 367–383. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, D.P.; Salmaso, N.; Paerl, H.W. Mitigating harmful cyanobacterial blooms: Strategies for control of nitrogen and phosphorus loads. Aquat. Ecol. 2016, 50, 351–366. [Google Scholar] [CrossRef]
- Søndergaard, M.; Jensen, J.P.; Jeppesen, E. Role of sediment and internal loading of phosphorus in shallow lakes. Hydrobiologia 2003, 506–509, 135–145. [Google Scholar] [CrossRef]
- Ibelings, B.W.; Bormans, M.; Fastner, J.; Visser, P.M. Cyanocost Special Issue on Cyanobacterial Blooms: Synopsis—A critical review of the management options for their prevention, control and mitigation. Aquat. Ecol. 2016, 50, 595–605. [Google Scholar] [CrossRef]
- Paerl, H.W. Mitigating toxic planktonic cyanobacterial blooms in aquatic ecosystems facing increasing anthropogenic and climatic pressures. Toxins 2018, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Barroin, G.; Feuillade, M. Hydrogen peroxide as a potential algicide for Oscillatoria rubescens D.C. Water Res. 1986, 20, 619–623. [Google Scholar] [CrossRef]
- Drábková, M.; Admiraal, W.; Maršálek, B. Combined exposure to hydrogen peroxide and light-selective effects on cyanobacteria, green algae, and diatoms. Environ. Sci. Technol. 2007, 41, 309–314. [Google Scholar] [CrossRef]
- Kumar Sinha, A.; Eggleton, M.A.; Lochmann, R.T. An environmentally friendly approach for mitigating cyanobacterial bloom and their toxins in hypereutrophic ponds: Potentiality of a newly developed granular hydrogen peroxide-based compound. Sci. Total Environ. 2018, 637–638, 524–537. [Google Scholar] [CrossRef]
- Lin, L.; Shan, K.; Xiong, Q.; Zhou, Q.; Li, L.; Gan, N.; Song, L. The ecological risks of hydrogen peroxide as a cyanocide: Its effect on the community structure of bacterioplankton. J. Oceanol. Limnol. 2018, 36, 2231–2242. [Google Scholar] [CrossRef]
- Yang, Z.; Buley, R.P.; Fernandez-Figueroa, E.G.; Barros, M.U.G.; Rajendran, S.; Wilson, A.E. Hydrogen peroxide treatment promotes chlorophytes over toxic cyanobacteria in a hyper-eutrophic aquaculture pond. Environ. Pollut. 2018, 240, 590–598. [Google Scholar] [CrossRef]
- Wang, B.; Song, Q.; Long, J.; Song, G.; Mi, W.; Bi, Y. Optimization method for Microcystis bloom mitigation by hydrogen peroxide and its stimulative effects on growth of chlorophytes. Chemosphere 2019, 228, 503–512. [Google Scholar] [CrossRef]
- Weenink, E.F.J.; Matthijs, H.C.P.; Schuurmans, J.M.; Piel, T.; Herk, M.J.; Sigon, C.A.M.; Visser, P.M.; Huisman, J. Interspecific protection against oxidative stress: Green algae protect harmful cyanobacteria against hydrogen peroxide. Environ. Microbiol. 2021, 23, 2404–2419. [Google Scholar] [CrossRef]
- Lusty, M.W.; Gobler, C.J. The efficacy of hydrogen peroxide in mitigating cyanobacterial blooms and altering microbial communities across four lakes in NY, USA. Toxins 2020, 12, 428. [Google Scholar] [CrossRef]
- Matthijs, H.C.P.; Jančula, D.; Visser, P.M.; Maršálek, B. Existing and emerging cyanocidal compounds: New perspectives for cyanobacterial bloom mitigation. Aquat. Ecol. 2016, 50, 443–460. [Google Scholar] [CrossRef] [Green Version]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts: I. Mechanism of the reduction of oxygen and other Hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 601–639. [Google Scholar] [CrossRef] [PubMed]
- Helman, Y.; Tchernov, D.; Reinhold, L.; Shibata, M.; Ogawa, T.; Schwarz, R.; Ohad, I.; Kaplan, A. Genes encoding A-type flavoproteins are essential for photoreduction of O2 in Cyanobacteria. Curr. Biol. 2003, 13, 230–235. [Google Scholar] [CrossRef] [Green Version]
- Allahverdiyeva, Y.; Isojärvi, J.; Zhang, P.; Aro, E.M. Cyanobacterial oxygenic photosynthesis is protected by flavodiiron proteins. Life 2015, 5, 716–743. [Google Scholar] [CrossRef] [PubMed]
- Drábková, M.; Matthijs, H.C.P.; Admiraal, W.; Maršálek, B. Selective effects of H2O2 on cyanobacterial photosynthesis. Photosynthetica 2007, 45, 363–369. [Google Scholar] [CrossRef]
- Blot, N.; Mella-Flores, D.; Six, C.; le Corguillé, G.; Boutte, C.; Peyrat, A.; Monnier, A.; Ratin, M.; Gourvil, P.; Campbell, D.A.; et al. Light history influences the response of the marine cyanobacterium Synechococcus sp. WH7803 to oxidative stress. Plant Physiol. 2011, 156, 1934–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikula, P.; Zezulka, S.; Jančula, D.; Maršálek, B. Metabolic activity and membrane integrity changes in Microcystis aeruginosa: New findings on hydrogen peroxide toxicity in cyanobacteria. Eur. J. Phycol. 2012, 47, 195–206. [Google Scholar] [CrossRef]
- Lürling, M.; Meng, D.; Faassen, E.J. Effects of hydrogen peroxide and ultrasound on biomass reduction and toxin release in the cyanobacterium, Microcystis aeruginosa. Toxins 2014, 6, 3260–3280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadimitriou, T.; Kormas, K.; Dionysiou, D.D.; Laspidou, C. Using H2O2 Treatments for the degradation of cyanobacteria and microcystins in a shallow hypertrophic reservoir. Environ. Sci. Pollut. Res. 2016, 23, 21523–21535. [Google Scholar] [CrossRef]
- Daniel, E.; Weiss, G.; Murik, O.; Sukenik, A.; Lieman-Hurwitz, J.; Kaplan, A. The response of Microcystis aeruginosa strain MGK to a single or two consecutive H2O2 applications. Environ. Microbiol. Rep. 2019, 11, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Piel, T.; Sandrini, G.; White, E.; Xu, T.; Schuurmans, J.M.; Huisman, J.; Visser, P.M. Suppressing cyanobacteria with hydrogen peroxide is more effective at high light intensities. Toxins 2020, 12, 18. [Google Scholar] [CrossRef] [Green Version]
- Sandrini, G.; Piel, T.; Xu, T.; White, E.; Qin, H.; Slot, P.C.; Huisman, J.; Visser, P.M. Sensitivity to hydrogen peroxide of the bloom-forming cyanobacterium Microcystis PCC 7806 depends on nutrient availability. Harmful Algae 2020, 99, 101916. [Google Scholar] [CrossRef]
- Keliri, E.; Paraskeva, C.; Sofokleous, A.; Sukenik, A.; Dziga, D.; Chernova, E.; Brient, L.; Antoniou, M.G. Occurrence of a single-species cyanobacterial bloom in a lake in Cyprus: Monitoring and treatment with hydrogen peroxide-releasing granules. Environ. Sci. Eur. 2021, 33, 31. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Chen, K.; Shi, X.; Yang, G. Using hydrogen peroxide to control cyanobacterial blooms: A mesocosm study focused on the effects of algal density in Lake Chaohu, China. Environ. Pollut. 2021, 272, 115923. [Google Scholar] [CrossRef]
- Santos, A.A.; Guedes, D.O.; Barros, M.U.G.; Oliveira, S.; Pacheco, A.B.F.; Azevedo, S.M.F.O.; Magalhães, V.F.; Pestana, C.J.; Edwards, C.; Lawton, L.A.; et al. Effect of hydrogen peroxide on natural phytoplankton and bacterioplankton in a drinking water reservoir: Mesocosm-scale study. Water Res. 2021, 197, 117069. [Google Scholar] [CrossRef]
- Matthijs, H.C.P.; Visser, P.M.; Reeze, B.; Meeuse, J.; Slot, P.C.; Wijn, G.; Talens, R.; Huisman, J. Selective suppression of harmful cyanobacteria in an entire lake with hydrogen peroxide. Water Res. 2012, 46, 1460–1472. [Google Scholar] [CrossRef]
- Barrington, D.J.; Reichwaldt, E.S.; Ghadouani, A. The use of hydrogen peroxide to remove cyanobacteria and microcystins from waste stabilization ponds and hypereutrophic systems. Ecol. Eng. 2013, 50, 86–94. [Google Scholar] [CrossRef]
- Burson, A.; Matthijs, H.C.P.; de Bruijne, W.; Talens, R.; Hoogenboom, R.; Gerssen, A.; Visser, P.M.; Stomp, M.; Steur, K.; van Scheppingen, Y.; et al. Termination of a toxic Alexandrium bloom with hydrogen peroxide. Harmful Algae 2014, 31, 125–135. [Google Scholar] [CrossRef]
- Lück, E.; Jager, M. Antimicrobial Food Additives: Characteristics, Uses, Effects; Springer: Berlin, Germany, 1997; Volume 2. [Google Scholar]
- Bjarnsholt, T.; Jensen, P.Ø.; Burmølle, M.; Hentzer, M.; Haagensen, J.A.J.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 2005, 151, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Sultana, S.T.; Atci, E.; Babauta, J.T.; Falghoush, A.M.; Snekvik, K.R.; Call, D.R.; Beyenal, H. Electrochemical scaffold generates localized, low concentration of hydrogen peroxide that inhibits bacterial pathogens and biofilms. Sci. Rep. 2015, 5, 14908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohwada, T.; Shirakawa, Y.; Kusumoto, M.; Masuda, H.; Sato, T. Susceptibility to hydrogen peroxide and catalase activity of root nodule bacteria. Biosci. Biotechnol. Biochem. 1999, 63, 457–462. [Google Scholar] [CrossRef]
- Zhao, Y.; Nickels, L.M.; Wang, H.; Ling, J.; Zhong, Z.; Zhu, J. OxyR-regulated catalase activity is critical for oxidative stress resistance, nodulation and nitrogen fixation in Azorhizobium caulinodans. FEMS Microbiol. Lett. 2016, 363, fnw130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strus, M.; Brzychczy-Włoch, M.; Gosiewski, T.; Kochan, P.; Heczko, P.B. The in vitro effect of hydrogen peroxide on vaginal microbial communities. FEMS Immunol. Med. Microbiol. 2006, 48, 56–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Kreth, J. The role of hydrogen peroxide in environmental adaptation of oral microbial communities. Oxid. Med. Cell. Longev. 2012, 2012, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xenopoulos, M.A.; Bird, D.F. Effect of acute exposure to hydrogen peroxide on the production of phytoplankton and bacterioplankton in a mesohumic lake. Photochem. Photobiol. 1997, 66, 471–478. [Google Scholar] [CrossRef]
- Kakosová, E.; Hrabák, P.; Černík, M.; Novotný, V.; Czinnerová, M.; Trögl, J.; Popelka, J.; Kuráň, P.; Zoubková, L.; Vrtoch, Ľ. Effect of various chemical oxidation agents on soil microbial communities. Chem. Eng. J. 2017, 314, 257–265. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef] [Green Version]
- Linz, A.M.; He, S.; Stevens, S.L.R.; Anantharaman, K.; Rohwer, R.R.; Malmstrom, R.R.; Bertilsson, S.; McMahon, K.D. Freshwater carbon and nutrient cycles revealed through reconstructed population genomes. PeerJ 2018, 6, e6075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.P.; Lin, C.T.; Chang, C.M.; Wu, S.H.; Lo, L.C. Nitrophenylboronic acids as highly chemoselective probes to detect hydrogen peroxide in foods and agricultural products. J. Agric. Food Chem. 2011, 59, 11403–11406. [Google Scholar] [CrossRef] [PubMed]
- Utermöhl, H. Zur Vervollkommnung Der Quantitativen Phytoplankton-Methodik: Mit 1 Tabelle Und 15 Abbildungen Im Text Und Auf 1 Tafel. Int. Ver. Theor. Angew. Limnol. Mitt. 1958, 9, 1–38. [Google Scholar] [CrossRef]
- Hillebrand, H.; Dürselen, C.-D.; Kirschtel, D.; Pollingher, U.; Zohary, T. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 1999, 35, 403–424. [Google Scholar] [CrossRef]
- Marie, D.; Partensky, F.; Vaulot, D.; Brussaard, C. Enumeration of phytoplankton, bacteria, and viruses in marine samples. Curr. Protoc. Cytom. 1999, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Chelius, M.K.; Triplett, E.W. The diversity of Archaea and Bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Hamady, M.; Walker, J.J.; Harris, J.K.; Gold, N.J.; Knight, R. Error-correcting barcoded primers allow hundreds of samples to be pyrosequenced in multiplex. Nat. Methods 2008, 5, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Knight, R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009, 19, 1141–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [Green Version]
- Janssen, S.; Mcdonald, D.; Gonzalez, A.; Navas-Molina, J.A.; Jiang, L.; Xu, Z.Z.; Winker, K.; Kado, D.M.; Orwoll, E.; Manary, M.; et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 2018, 3, e00021. [Google Scholar] [CrossRef] [Green Version]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [Green Version]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Q.; Luan, Y.; Sun, F. Variance Adjusted Weighted UniFrac: A powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinform. 2011, 12, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Bittinger, K.; Charlson, E.S.; Hoffmann, C.; Lewis, J.; Wu, G.D.; Collman, R.G.; Bushman, F.D.; Li, H. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Vázquez-Baeza, Y.; Koslicki, D.; McClelland, J.; Reeve, N.; Xu, Z.; Gonzalez, A.; Knight, R. Striped UniFrac: Enabling microbiome analysis at unprecedented scale. Nat. Methods 2018, 15, 847–848. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Reid, J.N.S.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the analysis of high-throughput sequencing datasets: Characterizing RNA-Seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef] [Green Version]
- Guevara Campoverde, N.C.; Hassenrück, C.; Buttigieg, P.L.; Gärdes, A. Characterization of bacterioplankton communities and quantification of organic carbon pools off the Galapagos Archipelago under contrasting environmental conditions. PeerJ 2018, 6, e5984. [Google Scholar] [CrossRef]
- Fernandes, A.D.; Vu, M.T.H.Q.; Edward, L.-M.; Macklaim, J.M.; Gloor, G.B. A reproducible effect size is more useful than an irreproducible hypothesis test to analyze high throughput sequencing datasets. arXiv 2018, arXiv:1809.02623. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2: Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome 2020, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Lürling, M.; Mucci, M.; Waajen, G. Removal of positively buoyant Planktothrix rubescens in lake restoration. Toxins 2020, 12, 700. [Google Scholar] [CrossRef] [PubMed]
- Schuurmans, J.M.; Brinkmann, B.W.; Makower, A.K.; Dittmann, E.; Huisman, J.; Matthijs, H.C.P. Microcystin interferes with defense against high oxidative stress in harmful cyanobacteria. Harmful Algae 2018, 78, 47–55. [Google Scholar] [CrossRef]
- Heaney, S.I.; Furnass, T.I. Laboratory models of diel vertical migration in the dinoflagellate Ceratium hirundinella. Freshw. Biol. 1980, 10, 163–170. [Google Scholar] [CrossRef]
- Arndt, H. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates): A review. Hydrobiologia 1993, 255/256, 231–246. [Google Scholar] [CrossRef]
- Agasild, H.; Nõges, T. Cladoceran and rotifer grazing on bacteria and phytoplankton in two shallow eutrophic lakes: In situ measurement with fluorescent microspheres. J. Plankton Res. 2005, 27, 1155–1174. [Google Scholar] [CrossRef] [Green Version]
- Hwang, S.-J.; Heath, R.T. Zooplankton bacterivory at coastal and offshore sites of Lake Erie. J. Plankton Res. 1999, 21, 699–719. [Google Scholar] [CrossRef]
- Eiler, A.; Bertilsson, S. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 2004, 6, 1228–1243. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Razzano, M.; Mou, X. Cyanobacterial blooms alter the relative importance of neutral and selective processes in assembling freshwater bacterioplankton community. Sci. Total Environ. 2020, 706, 135724. [Google Scholar] [CrossRef]
- Shade, A.; Read, J.S.; Youngblut, N.D.; Fierer, N.; Knight, R.; Kratz, T.K.; Lottig, N.R.; Roden, E.E.; Stanley, E.H.; Stombaugh, J.; et al. Lake microbial communities are resilient after a whole-ecosystem disturbance. ISME J. 2012, 6, 2153–2167. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Berghoff, B.A.; Stratmann, V.; Grossart, H.P.; Glaeser, J. Contrasting effects of singlet oxygen and hydrogen peroxide on bacterial community composition in a humic lake. PLoS ONE 2014, 9, e92518. [Google Scholar] [CrossRef]
- Woodhouse, J.N.; Kinsela, A.S.; Collins, R.N.; Bowling, L.C.; Honeyman, G.L.; Holliday, J.K.; Neilan, B.A. Microbial communities reflect temporal changes in cyanobacterial composition in a shallow ephemeral freshwater lake. ISME J. 2016, 10, 1337–1351. [Google Scholar] [CrossRef]
- Diao, M.; Sinnige, R.; Kalbitz, K.; Huisman, J.; Muyzer, G. Succession of bacterial communities in a seasonally stratified lake with an anoxic and sulfidic hypolimnion. Front. Microbiol. 2017, 8, 2511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Steinman, A.D.; Tang, X.; Xue, Q.; Zhao, Y.; Xie, L. Response of bacterial communities to cyanobacterial harmful algal blooms in Lake Taihu, China. Harmful Algae 2017, 68, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Poretsky, R.; Rodriguez, -R.L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.M.; Lin, C.Y.; Sheu, S.Y. Investigating antimicrobial activity in Rheinheimera sp. due to hydrogen peroxide generated by L-lysine oxidase activity. Enzym. Microb. Technol. 2010, 46, 487–493. [Google Scholar] [CrossRef]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genom. 2005, 6, 115. [Google Scholar] [CrossRef] [Green Version]
- Kiley, P.J.; Kaplan, S. Molecular genetics of photosynthetic membrane biosynthesis in Rhodobacter sphaeroides. Microbiol. Rev. 1988, 52, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Tabita, F.R. The biochemistry and metabolic regulation of carbon metabolism and CO2 fixation in purple bacteria. In Anoxygenic Photosynthetic Bacteria. Advances in Photosynthesis ad Respiration; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995; pp. 885–914. [Google Scholar]
- Satoh, T.; Hoshino, Y.; Kitamura, H. Rhodopseudomonas sphaeroides forma sp. denitrificans, a denitrifying strain as a subspecies of Rhodopseudomonas sphaeroides. Arch. Microbiol. 1976, 108, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, P.; Chen, X.; Hugenholtz, P.; Janssen, P.H. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 2004, 70, 5875–5881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, E.; Schmidt, M.L.; Berry, M.A.; Biddanda, B.A.; Burtner, A.; Johengen, T.H.; Palladino, D.; Denef, V.J. Verrucomicrobia are prevalent in north-temperate freshwater lakes and display class-level preferences between lake habitats. PLoS ONE 2018, 13, e0195112. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Lundin, D.; Labrenz, M.; Jürgens, K.; Zheng, Z.; Aspeborg, H.; Andersson, A.F. Metagenomic de novo assembly of an aquatic representative of the Verrucomicrobial class Spartobacteria. mBio 2013, 4, e00569. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Stevens, S.L.R.; Chan, L.-K.; Bertilsson, S.; Glavina del Rio, T.; Tringe, S.G.; Malmstrom, R.R.; McMahon, K.D. Ecophysiology of freshwater Verrucomicrobia inferred from metagenome-assembled genomes. mSphere 2017, 2, e00277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allison, S.D.; Martiny, J.B.H. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teeling, H.; Fuchs, B.M.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Berry, M.A.; Davis, T.W.; Cory, R.M.; Duhaime, M.B.; Johengen, T.H.; Kling, G.W.; Marino, J.A.; den Uyl, P.A.; Gossiaux, D.; Dick, G.J.; et al. Cyanobacterial harmful algal blooms are a biological disturbance to western Lake Erie bacterial communities. Environ. Microbiol. 2017, 19, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Bender, E.A.; Case, T.J.; Gilpin, M.E. Perturbation experiments in community ecology: Theory and practice. Ecology 1984, 65, 1–13. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H.; et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [Green Version]
- Shade, A.; Read, J.S.; Welkie, D.G.; Kratz, T.K.; Wu, C.H.; McMahon, K.D. Resistance, resilience and recovery: Aquatic bacterial dynamics after water column disturbance. Environ. Microbiol. 2011, 13, 2752–2767. [Google Scholar] [CrossRef]
- Baho, D.L.; Peter, H.; Tranvik, L.J. Resistance and resilience of microbial communities: Temporal and spatial insurance against perturbations. Environ. Microbiol. 2012, 14, 2283–2292. [Google Scholar] [CrossRef] [Green Version]
- Daehler, C.C.; Strong, D.R. Status, prediction and prevention of introduced cordgrass Spartina spp. invasions in Pacific estuaries, USA. Biol. Conserv. 1996, 78, 51–58. [Google Scholar] [CrossRef]
- Louca, S.; Jacques, S.M.S.; Pires, A.P.F.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol. 2017, 1, 15. [Google Scholar] [CrossRef]
- Xu, Z.; Malmer, D.; Langille, M.G.I.; Way, S.F.; Knight, R. Which is more important for classifying microbial communities: Who’s there or what they can do? ISME J. 2014, 8, 2357–2359. [Google Scholar] [CrossRef] [Green Version]
- Koo, H.; Hakim, J.A.; Morrow, C.D.; Eipers, P.G.; Davila, A.; Andersen, D.T.; Bej, A.K. Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee, Antarctica. J. Microbiol. Methods 2017, 140, 15–22. [Google Scholar] [CrossRef]
- Jankowiak, J.G.; Gobler, C.J. The composition and function of microbiomes within Microcystis colonies are significantly different than native bacterial assemblages in two North American lakes. Front. Microbiol. 2020, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Zhang, Y.; Cheng, G.; Li, X.; Qin, Y.; He, D. Dredging mitigates cyanobacterial bloom in eutrophic Lake Nanhu: Shifts in associations between the bacterioplankton community and sediment biogeochemistry. Environ. Res. 2020, 188, 109799. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Sun, Y.; Yu, M.; Shi, Z.; Zhang, H.; Peng, R.; Li, Z.; Cui, J.; Luo, X. Linking shifts in bacterial community composition and function with changes in the dissolved organic matter pool in ice-covered Baiyangdian Lake, northern China. Microorganisms 2020, 8, 883. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Imlay, J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 2012, 525, 145–160. [Google Scholar] [CrossRef] [Green Version]
- Andrade, A.C.; Fróes, A.; Lopes, F.Á.C.; Thompson, F.L.; Krüger, R.H.; Dinsdale, E.; Bruce, T. Diversity of microbial carbohydrate-active enzymes (Cazymes) associated with freshwater and soil samples from Caatinga biome. Microb. Ecol. 2017, 74, 89–105. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piel, T.; Sandrini, G.; Muyzer, G.; Brussaard, C.P.D.; Slot, P.C.; van Herk, M.J.; Huisman, J.; Visser, P.M. Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms. Microorganisms 2021, 9, 1495. https://doi.org/10.3390/microorganisms9071495
Piel T, Sandrini G, Muyzer G, Brussaard CPD, Slot PC, van Herk MJ, Huisman J, Visser PM. Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms. Microorganisms. 2021; 9(7):1495. https://doi.org/10.3390/microorganisms9071495
Chicago/Turabian StylePiel, Tim, Giovanni Sandrini, Gerard Muyzer, Corina P. D. Brussaard, Pieter C. Slot, Maria J. van Herk, Jef Huisman, and Petra M. Visser. 2021. "Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms" Microorganisms 9, no. 7: 1495. https://doi.org/10.3390/microorganisms9071495
APA StylePiel, T., Sandrini, G., Muyzer, G., Brussaard, C. P. D., Slot, P. C., van Herk, M. J., Huisman, J., & Visser, P. M. (2021). Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms. Microorganisms, 9(7), 1495. https://doi.org/10.3390/microorganisms9071495





_Brussaard.png)

