Eggshell and Feed Microbiota Do Not Represent Major Sources of Gut Anaerobes for Chickens in Commercial Production
Abstract
:1. Introduction
2. Materials and Methods
2.1. Comparison of Eggshell Microbiota with Caecal Microbiota of 7-Day-Old Chicks
2.2. Housing Conditions
2.3. DNA Purification from Eggshells
2.4. Feed as a Source of Gut Microbiota
2.5. Definition of Optimal Age for Administration of Bacterial Strains Intended for Gut Colonisation
2.6. DNA Purification
2.7. Quantitative Real-Time PCR
2.8. Microbiota Characterisation by Sequencing of V3/V4 Variable Region of 16S rRNA Genes
2.9. Statistics and Data Availability
3. Results
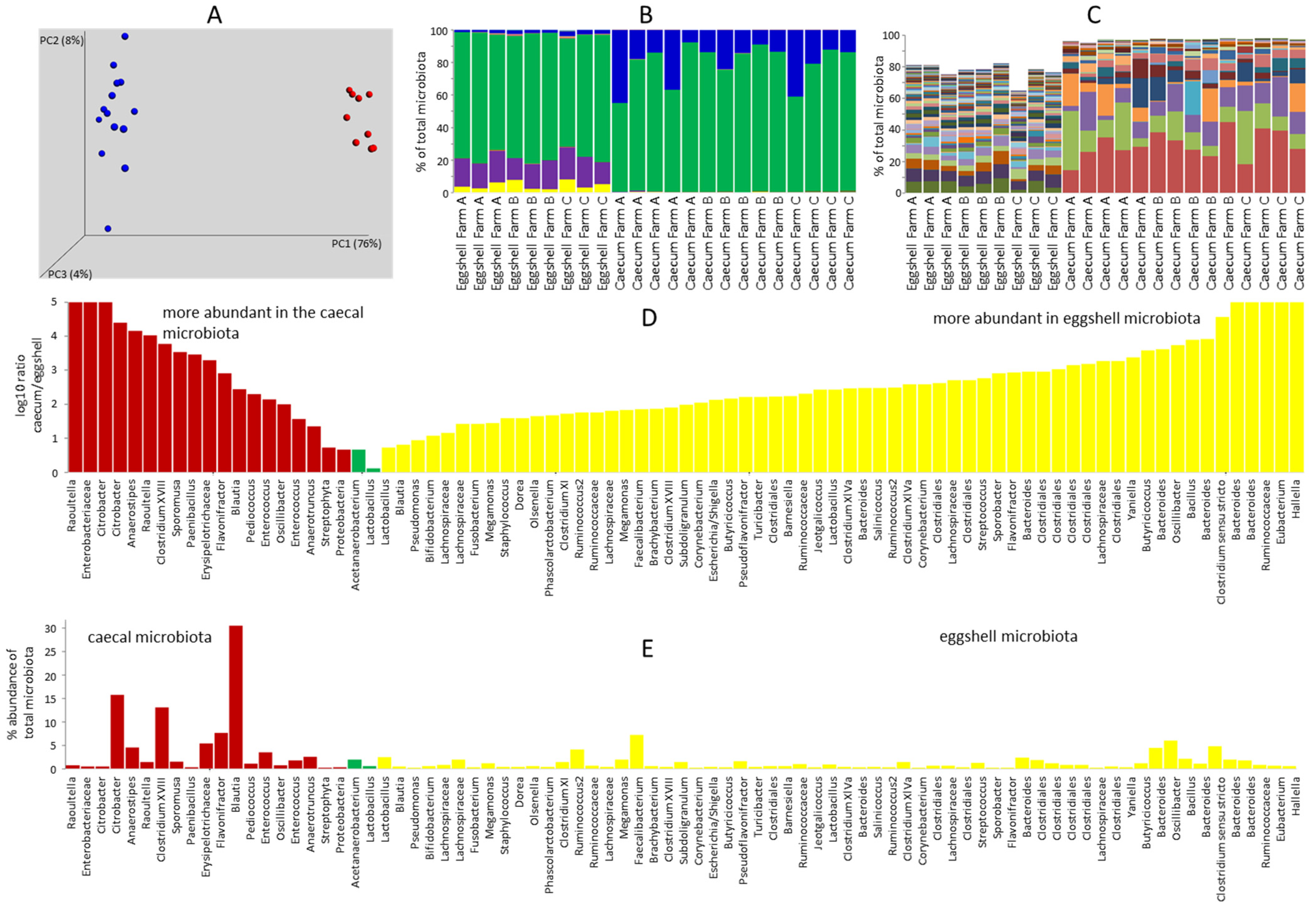
3.1. Comparison of Eggshell Microbiota and Caecal Microbiota of 7-Day-Old Chicks
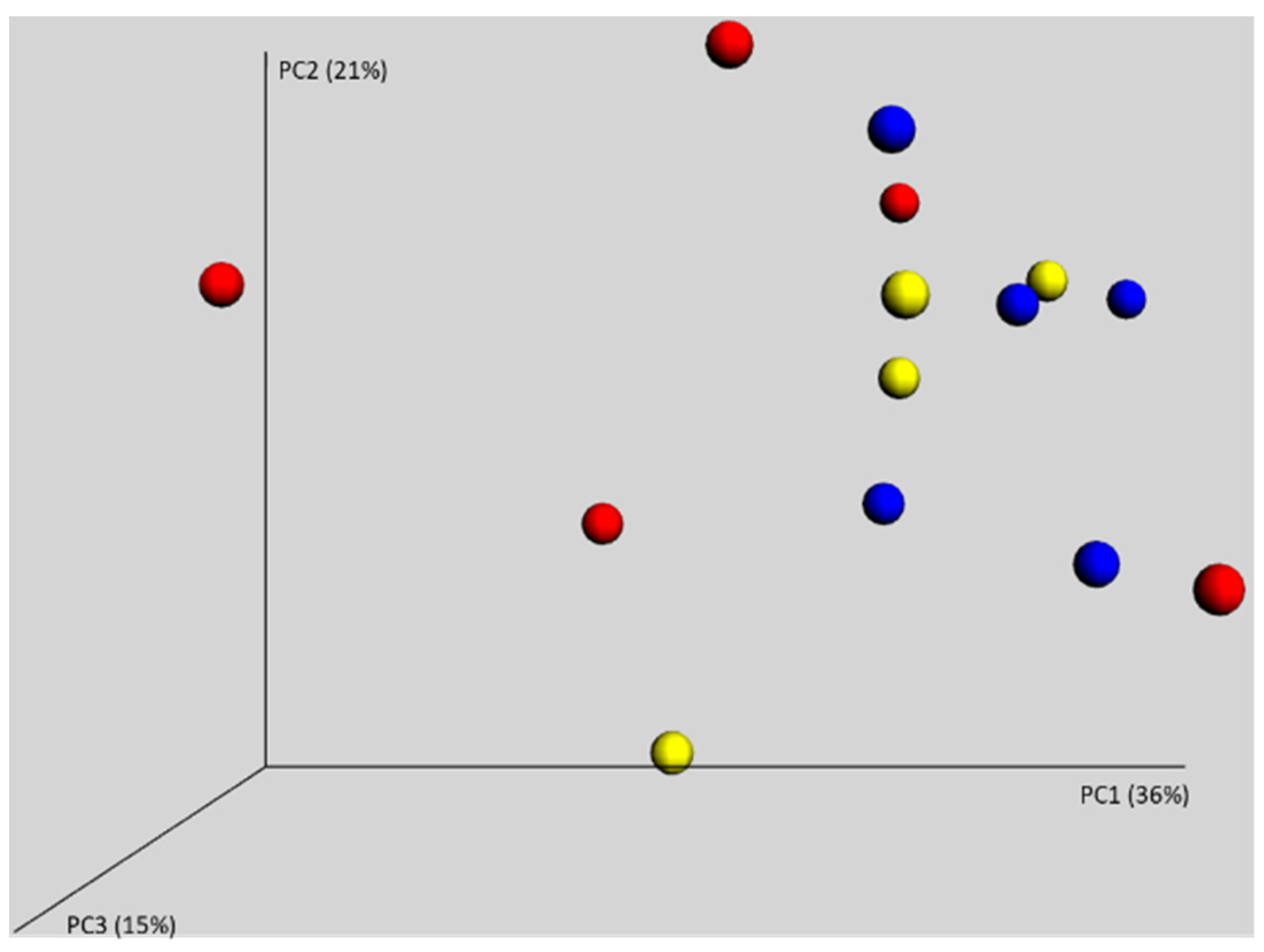
3.2. Gut Microbiota of 7-Day-Old Chickens Originating from Different Parent Flocks
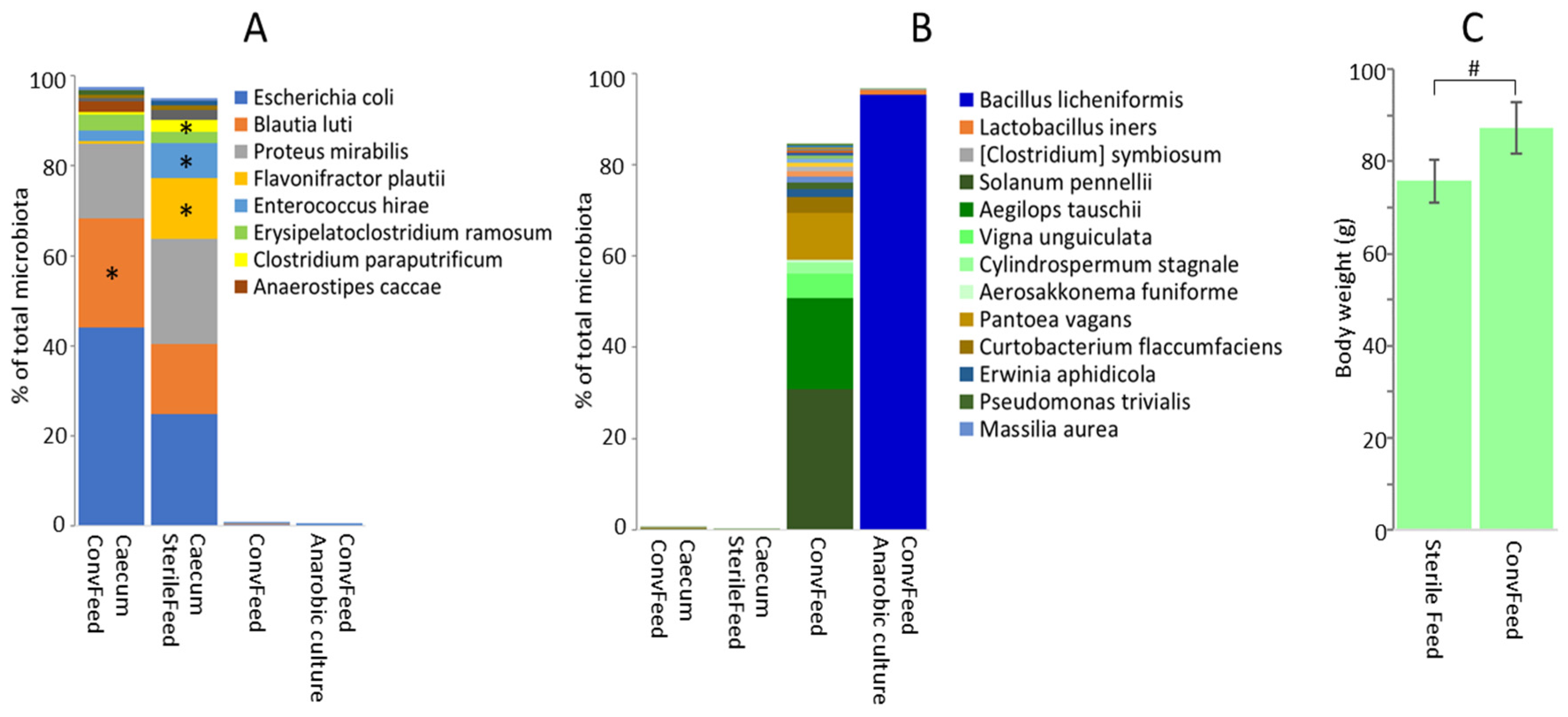
3.3. Feed as a Source of Gut Microbiota
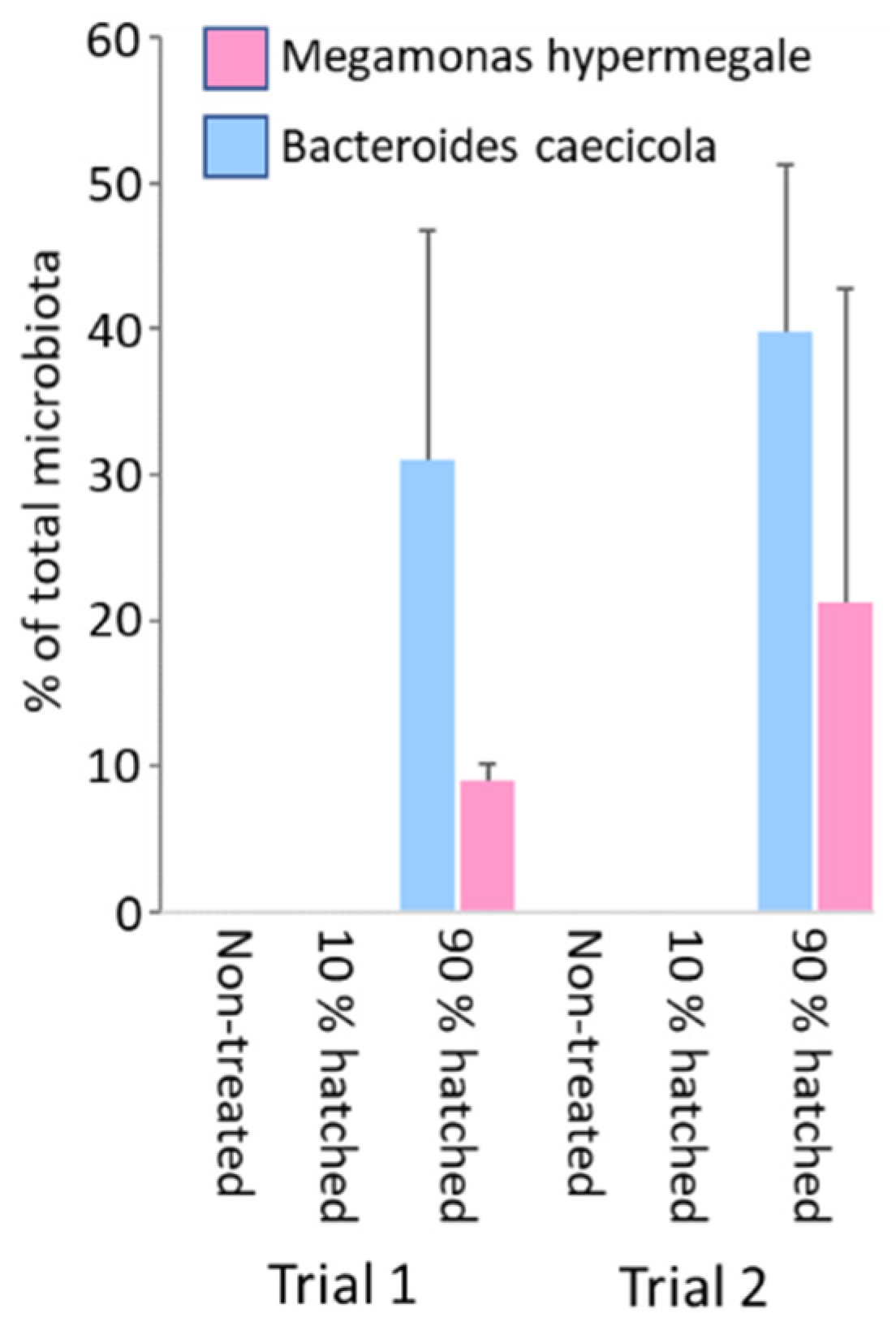
3.4. Definition of an Optimal Time Point for Administration of Bacterial Strains during Hatching
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Videnska, P.; Sedlar, K.; Lukac, M.; Faldynova, M.; Gerzova, L.; Cejkova, D.; Sisak, F.; Rychlik, I. Succession and Replacement of Bacterial Populations in the Caecum of Egg Laying Hens over Their Whole Life. PLoS ONE 2014, 9, e115142. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Ma, C.; Sun, Z.; Wang, L.; Huang, S.; Su, X.; Xu, J.; Zhang, H. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome 2017, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Shuling, N.; Kunyuan, T.; Qiuyang, Z.; Hewen, D.; ChenCheng, G.; Tianhe, Y.; Liancheng, L.; Xin, F. Characteristics of the intestinal flora of specific pathogen free chickens with age. Microb. Pathog. 2019, 132, 325–334. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Hughes, R.J.; Denman, S.; Moore, R.J. Highly Variable Microbiota Development in the Chicken Gastrointestinal Tract. PLoS ONE 2013, 8, e84290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjitkar, S.; Lawley, B.; Tannock, G.; Engberg, R.M. Bacterial Succession in the Broiler Gastrointestinal Tract. Appl. Environ. Microbiol. 2016, 82, 2399–2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubasova, T.; Kollarcikova, M.; Crhanova, M.; Karasova, D.; Cejkova, D.; Sebkova, A.; Matiasovicova, J.; Faldynova, M.; Pokorna, A.; Cizek, A.; et al. Contact with adult hen affects development of caecal microbiota in newly hatched chicks. PLoS ONE 2019, 14, e0212446. [Google Scholar] [CrossRef] [PubMed]
- Varmuzova, K.; Kubasova, T.; Davidova-Gerzova, L.; Sisak, F.; Havlickova, H.; Sebkova, A.; Faldynova, M.; Rychlik, I. Composition of Gut Microbiota Influences Resistance of Newly Hatched Chickens to Salmonella Enteritidis Infection. Front. Microbiol. 2016, 7, 957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rantala, M.; Nurmi, E. Prevention of the growth ofSalmonella infantisin chicks by the flora of the alimentary tract of chickens. Br. Poult. Sci. 1973, 14, 627–630. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the Gastrointestinal Tract Microbiota Correlated with Improved Growth and Feed Conversion: Challenges Presented for the Identification of Performance Enhancing Probiotic Bacteria. Front. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef] [Green Version]
- Rikimaru, K.; Takahashi, H. A simple and efficient method for extraction of PCR-amplifiable DNA from chicken eggshells. Anim. Sci. J. 2009, 80, 220–223. [Google Scholar] [CrossRef]
- Medvecky, M.; Cejkova, D.; Polansky, O.; Karasova, D.; Kubasova, T.; Cizek, A.; Rychlik, I. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genom. 2018, 19, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Kubasova, T.; Kollarcikova, M.; Crhanova, M.; Karasova, D.; Cejkova, D.; Sebkova, A.; Matiasovicova, J.; Faldynova, M.; Sisak, F.; Babak, V.; et al. Gut Anaerobes Capable of Chicken Caecum Colonisation. Microorganisms 2019, 7, 597. [Google Scholar] [CrossRef] [Green Version]
- Kollarcikova, M.; Faldynova, M.; Matiasovicova, J.; Jahodarova, E.; Kubasova, T.; Seidlerova, Z.; Babak, V.; Videnska, P.; Cizek, A.; Rychlik, I. Different Bacteroides Species Colonise Human and Chicken Intestinal Tract. Microorganisms 2020, 8, 1483. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, T.J.; Youmans, B.P.; Noll, S.; Cardona, C.; Evans, N.P.; Karnezos, T.P.; Ngunjiri, J.M.; Abundo, M.C.; Lee, C.-W. A Consistent and Predictable Commercial Broiler Chicken Bacterial Microbiota in Antibiotic-Free Production Displays Strong Correlations with Performance. Appl. Environ. Microbiol. 2018, 84, e00362-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucher, M.G.; Zwirzitz, B.; Oladeinde, A.; Cook, K.; Plymel, C.; Zock, G.; Lakin, S.; Aggrey, S.E.; Ritz, C.; Looft, T.; et al. Reused poultry litter microbiome with competitive exclusion potential against Salmonella Heidelberg. J. Environ. Qual. 2020, 49, 869–881. [Google Scholar] [CrossRef] [Green Version]
- De Cesare, A.; Caselli, E.; Lucchi, A.; Sala, C.; Parisi, A.; Manfreda, G.; Mazzacane, S. Impact of a probiotic-based cleaning product on the microbiological profile of broiler litters and chicken caeca microbiota. Poult. Sci. 2019, 98, 3602–3610. [Google Scholar] [CrossRef] [PubMed]
- Martín, E.; Fallschissel, K.; Kampfer, P.; Jäckel, U. Detection of Jeotgalicoccus spp. in poultry house air. Syst. Appl. Microbiol. 2010, 33, 188–192. [Google Scholar] [CrossRef]
- Cressman, M.D.; Yu, Z.; Nelson, M.C.; Moeller, S.J.; Lilburn, M.S.; Zerby, H.N. Interrelations between the Microbiotas in the Litter and in the Intestines of Commercial Broiler Chickens. Appl. Environ. Microbiol. 2010, 76, 6572–6582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adkins, P.; Dufour, S.; Spain, J.; Calcutt, M.; Reilly, T.; Stewart, G.; Middleton, J. Cross-sectional study to identify staphylococcal species isolated from teat and inguinal skin of different-aged dairy heifers. J. Dairy Sci. 2018, 101, 3213–3225. [Google Scholar] [CrossRef]
- Callaway, T.R.; Dowd, S.; Wolcott, R.D.; Sun, Y.; McReynolds, J.L.; Edrington, T.S.; Byrd, J.A.; Anderson, R.C.; Krueger, N.; Nisbet, D.J. Evaluation of the bacterial diversity in cecal contents of laying hens fed various molting diets by using bacterial tag-encoded FLX amplicon pyrosequencing. Poult. Sci. 2009, 88, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Nordentoft, S.; Mølbak, L.; Bjerrum, L.; De Vylder, J.; Van Immerseel, F.; Pedersen, K. The influence of the cage system and colonisation of Salmonella Enteritidis on the microbial gut flora of laying hens studied by T-RFLP and 454 pyrosequencing. BMC Microbiol. 2011, 11, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Videnska, P.; Rahman, M.; Faldynova, M.; Babak, V.; Matulova, M.E.; Prukner-Radovcic, E.; Krizek, I.; Smole-Mozina, S.; Kovac, J.; Szmolka, A.; et al. Characterization of Egg Laying Hen and Broiler Fecal Microbiota in Poultry Farms in Croatia, Czech Republic, Hungary and Slovenia. PLoS ONE 2014, 9, e110076. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lilburn, M.; Yu, Z. Intestinal Microbiota of Broiler Chickens As Affected by Litter Management Regimens. Front. Microbiol. 2016, 7, 593. [Google Scholar] [CrossRef] [Green Version]
- Oppliger, A.; Charrière, N.; Droz, P.-O.; Rinsoz, T. Exposure to Bioaerosols in Poultry Houses at Different Stages of Fattening; Use of Real-time PCR for Airborne Bacterial Quantification. Ann. Occup. Hyg. 2008, 52, 405–412. [Google Scholar] [CrossRef]
- Olsen, R.; Kudirkiene, E.; Thøfner, I.; Pors, S.; Karlskov-Mortensen, P.; Li, L.; Papasolomontos, S.; Angastiniotou, C.; Christensen, J. Impact of egg disinfection of hatching eggs on the eggshell microbiome and bacterial load. Poult. Sci. 2017, 96, 3901–3911. [Google Scholar] [CrossRef] [PubMed]
- Trudeau, S.; Thibodeau, A.; Côté, J.-C.; Gaucher, M.-L.; Fravalo, P. Contribution of the Broiler Breeders’ Fecal Microbiota to the Establishment of the Eggshell Microbiota. Front. Microbiol. 2020, 11, 666. [Google Scholar] [CrossRef] [Green Version]
- Maki, J.J.; Bobeck, E.A.; Sylte, M.J.; Looft, T. Eggshell and environmental bacteria contribute to the intestinal microbiota of growing chickens. J. Anim. Sci. Biotechnol. 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Wilson, K.M.; Rodrigues, D.; Briggs, W.N.; Duff, A.F.; Chasser, K.M.; Bielke, L.R. Evaluation of the impact of in ovo administered bacteria on microbiome of chicks through 10 days of age. Poult. Sci. 2019, 98, 5949–5960. [Google Scholar] [CrossRef]
- de Oliveira, J.E.; van der Hoeven-Hangoor, E.; van de Linde, I.B.; Montijn, R.C.; van der Vossen, J.M.B.M. In ovo inoculation of chicken embryos with probiotic bacteria and its effect on posthatch Salmonella susceptibility. Poult. Sci. 2014, 93, 818–829. [Google Scholar] [CrossRef]

| Target Species | Forward Primer | Reverse Primer |
|---|---|---|
| Pseudoflavonifractor capillosus | GCCAGGAATTTCCCTTCTTC | GGCAGTCTCATGGAGGTAGC |
| Anaerotruncus colihominis | GCAAATGGGCGTATCCTCAA | CTGTTCAATTTCCCGGCAC |
| Butyricicoccus pullicaecorum | CGAGCAGGCAAACGACAA | CCAGGTCTTGGTACCGTCC |
| Blautia producta | GAGAAAAAGGGGCAACAACA | TCAGCATCTTTTCCCCAATC |
| Clostridium lactatifermentans | GGTATCCCCCACGCTTATTT | CCTGCGGTCAATTCTTTGAT |
| Clostridium glycyrrhizinilyticum | CTGCAGATAACGCACAGGAA | GTACCGGCAGGCATATCTGT |
| Oscillibacter valericigenes | TGTGAGTCCCAGCTCTACGA | GCAGGGCCTCTCTCCATTTG |
| Megamonas hypermegale | ATTCGCCTTCACGACAATTC | TTACGATATCGGCGAGAACC |
| Bacteroides caecicola | TATACCATGCACGATGAACC | AAATACCTTCTTCCCTCACG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volf, J.; Crhanova, M.; Karasova, D.; Faldynova, M.; Kubasova, T.; Seidlerova, Z.; Sebkova, A.; Zeman, M.; Juricova, H.; Matiasovicova, J.; et al. Eggshell and Feed Microbiota Do Not Represent Major Sources of Gut Anaerobes for Chickens in Commercial Production. Microorganisms 2021, 9, 1480. https://doi.org/10.3390/microorganisms9071480
Volf J, Crhanova M, Karasova D, Faldynova M, Kubasova T, Seidlerova Z, Sebkova A, Zeman M, Juricova H, Matiasovicova J, et al. Eggshell and Feed Microbiota Do Not Represent Major Sources of Gut Anaerobes for Chickens in Commercial Production. Microorganisms. 2021; 9(7):1480. https://doi.org/10.3390/microorganisms9071480
Chicago/Turabian StyleVolf, Jiri, Magdalena Crhanova, Daniela Karasova, Marcela Faldynova, Tereza Kubasova, Zuzana Seidlerova, Alena Sebkova, Michal Zeman, Helena Juricova, Jitka Matiasovicova, and et al. 2021. "Eggshell and Feed Microbiota Do Not Represent Major Sources of Gut Anaerobes for Chickens in Commercial Production" Microorganisms 9, no. 7: 1480. https://doi.org/10.3390/microorganisms9071480
APA StyleVolf, J., Crhanova, M., Karasova, D., Faldynova, M., Kubasova, T., Seidlerova, Z., Sebkova, A., Zeman, M., Juricova, H., Matiasovicova, J., Foltyn, M., Tvrdon, Z., & Rychlik, I. (2021). Eggshell and Feed Microbiota Do Not Represent Major Sources of Gut Anaerobes for Chickens in Commercial Production. Microorganisms, 9(7), 1480. https://doi.org/10.3390/microorganisms9071480






