Alteration of the Fecal but Not Salivary Microbiome in Patients with Behçet’s Disease According to Disease Activity Shift
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Fecal and Salivary Sampling
2.3. DNA Extraction, Polymerase Chain Reaction Amplification, and Sequencing
2.4. Sequence Analysis
2.5. Statistical Analysis
2.6. Ethics Approval
3. Results
3.1. Subject Characteristics
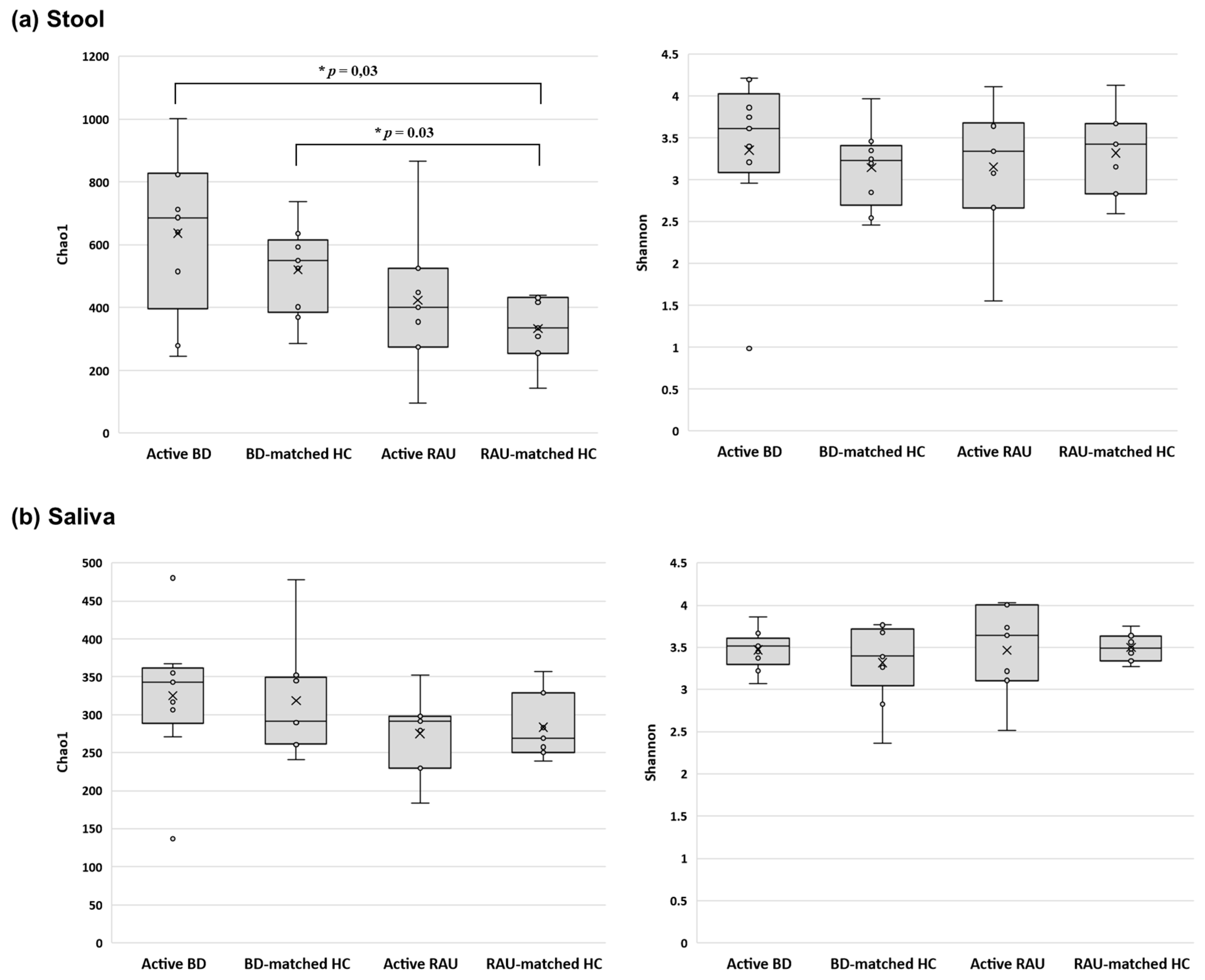
3.2. Alpha Diversity of the Fecal Microbiome in Patients with Active BD Was Significantly Different from That of Unmatched HCs but Not Matched HCs or Patients with Active RAUs
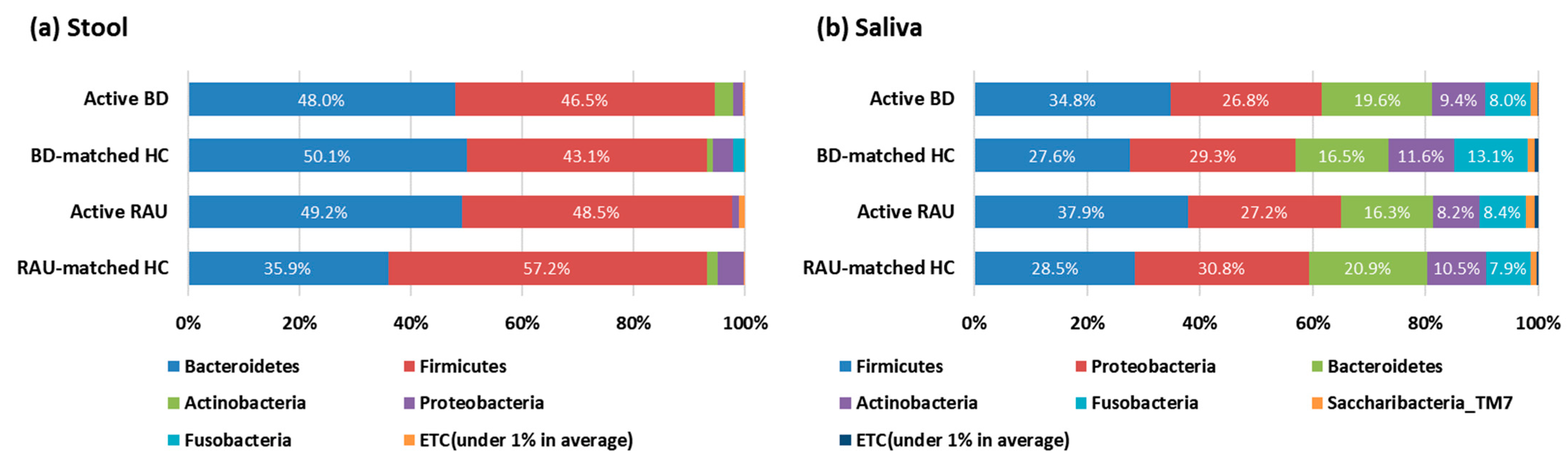
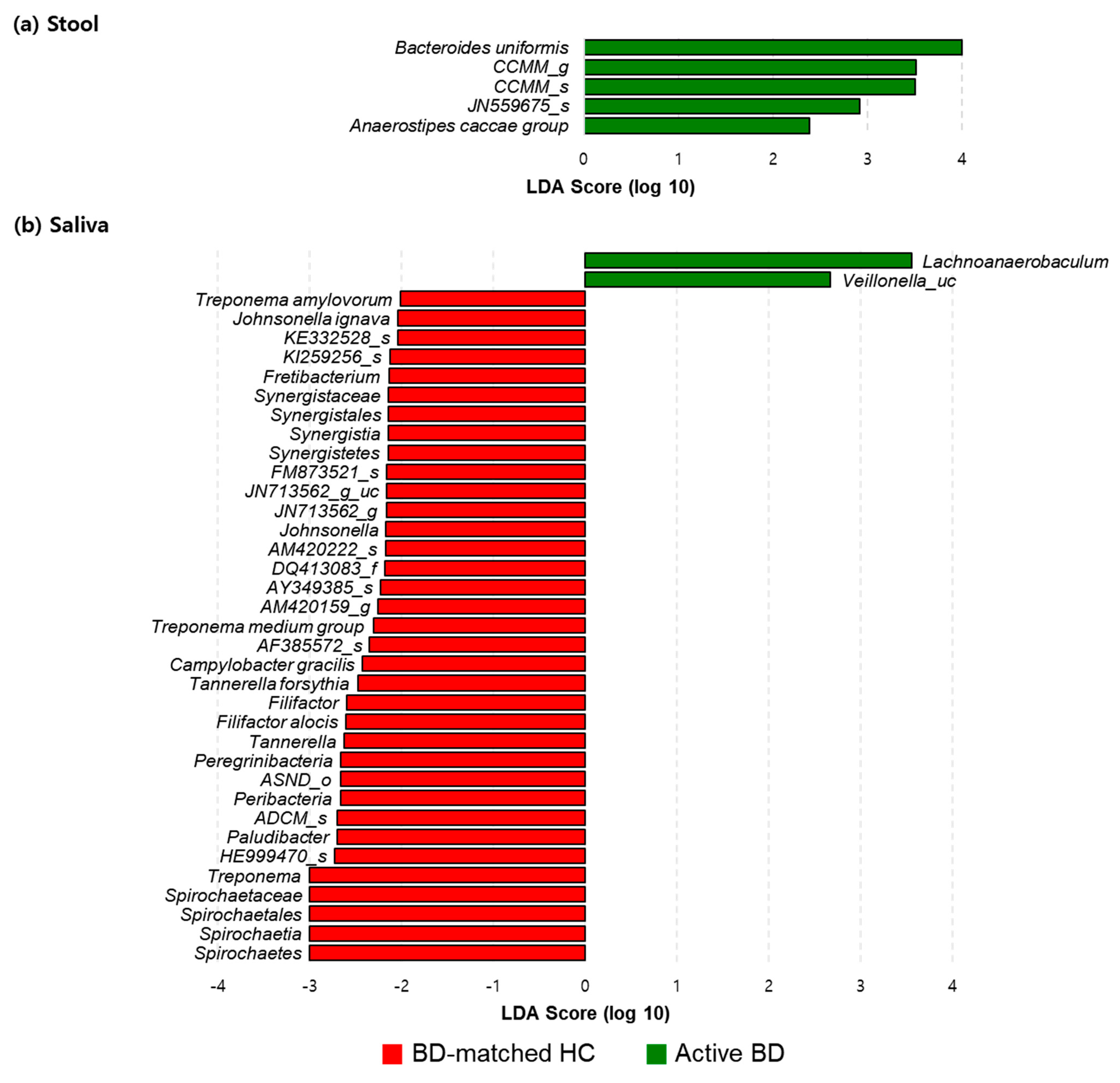
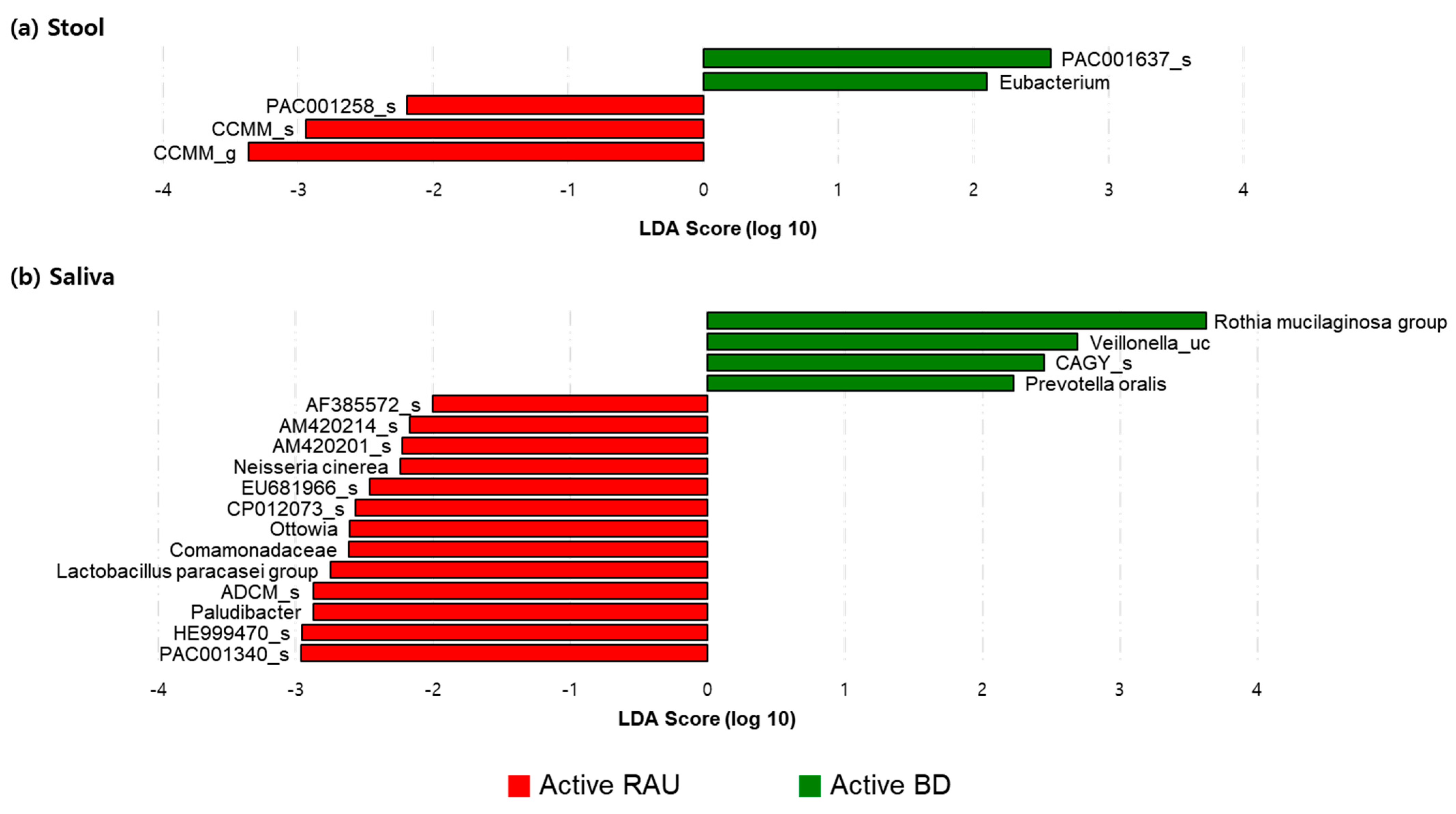
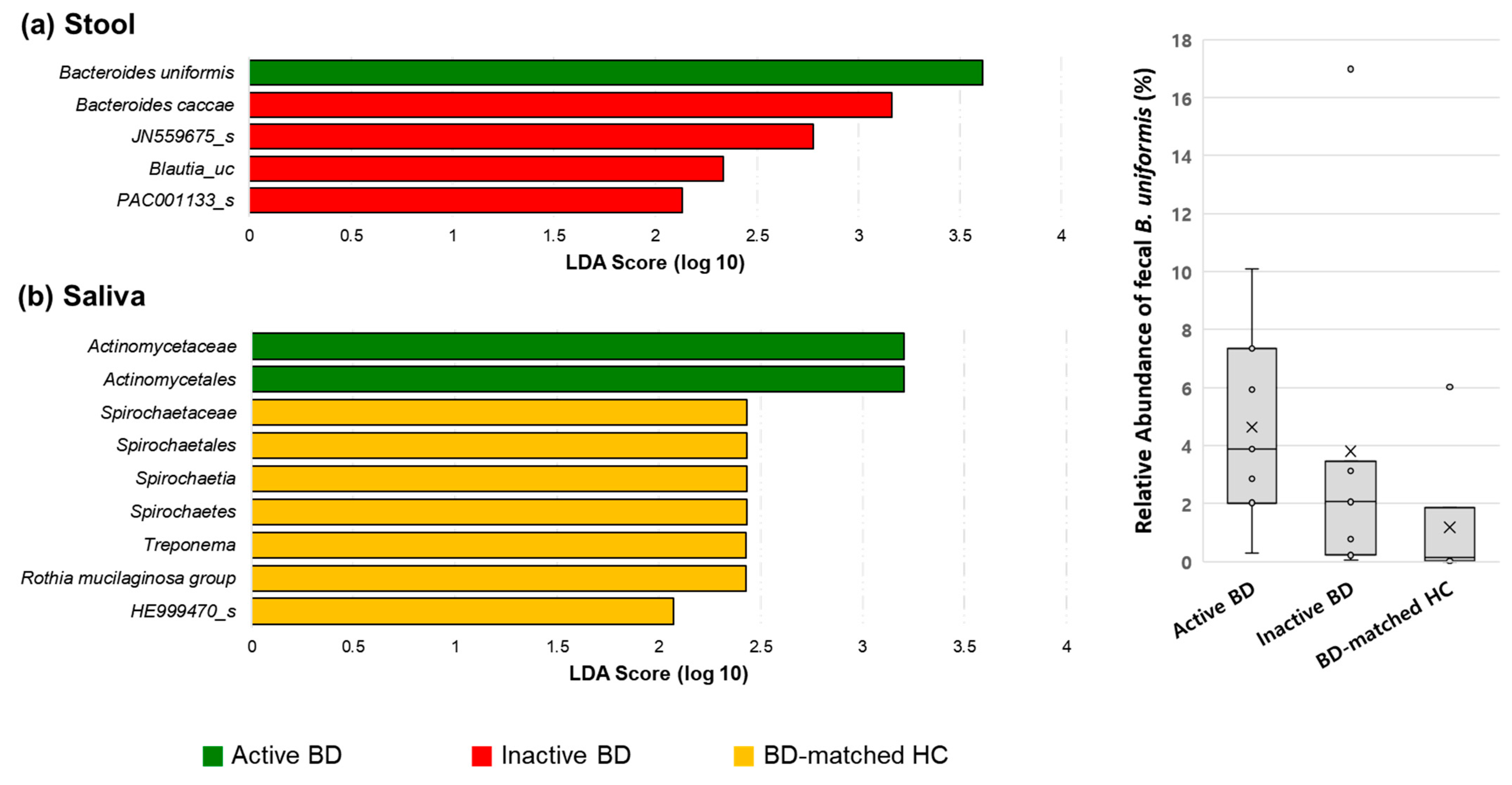
3.3. Fecal Bacteroides Uniformis and Salivary Rothia Mucilaginosa Levels Were Higher in Patients with BD Than in BD-Matched HCs and Patients with RAUs, Respectively
3.4. Alpha Diversity of the Fecal Microbiome Decreased in Patients with BD When the Disease Changed from the Active to Inactive Phase
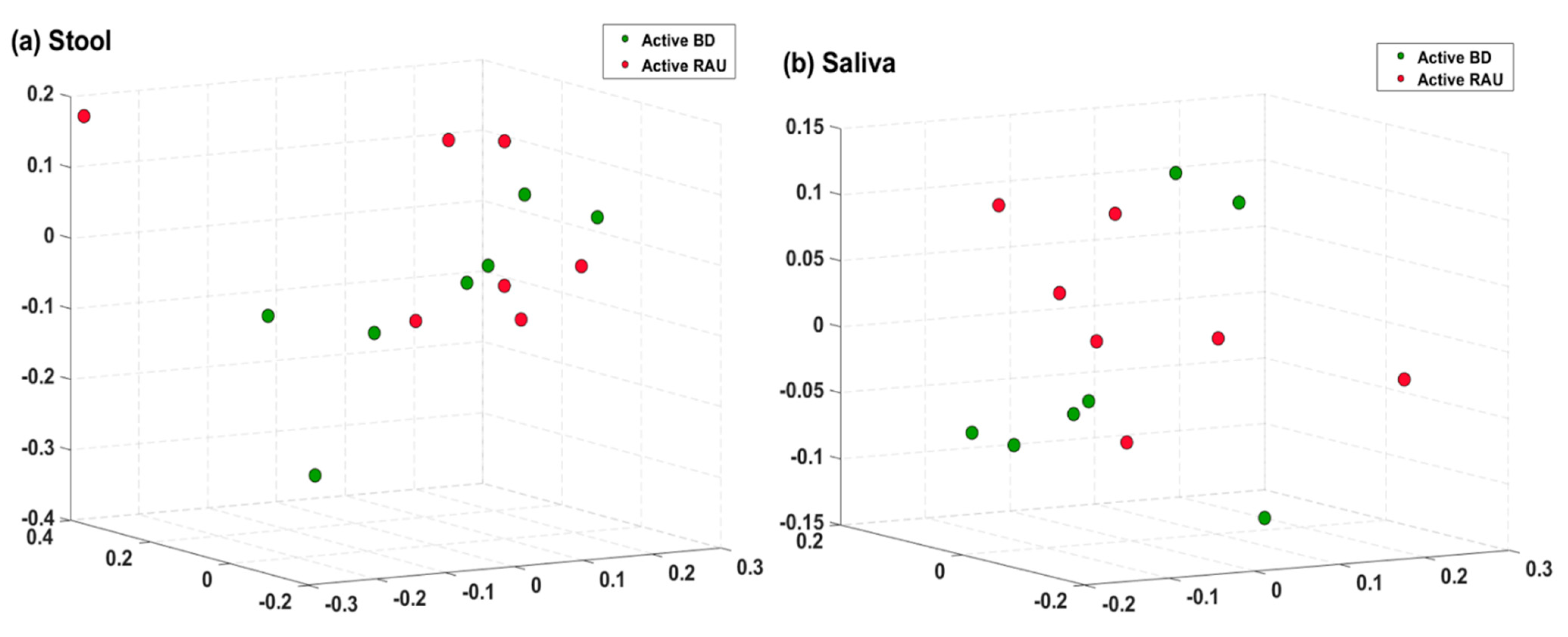
3.5. The Relative Compositions in the Fecal and Salivary Microbiomes Were Different in Patients with BD According to Their Having Uveitis or Not
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Segre, J.A. The human microbiome: Our second genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssen, A.W.; Kersten, S. The role of the gut microbiota in metabolic health. FASEB J. 2015, 29, 3111–3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, F.; Backhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Akdeniz, N.; Elmas, O.F.; Karadag, A.S. Behcet syndrome: A great imitator. Clin. Dermatol. 2019, 37, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, K.; Alrawi, Z.; Persson, A.; Jonsson, G.; Marsal, J. Intestinal dysbiosis is common in systemic sclerosis and associated with gastrointestinal and extraintestinal features of disease. Arthritis Res. Ther. 2016, 18, 278. [Google Scholar] [CrossRef] [Green Version]
- Arron, S.T.; Dimon, M.T.; Li, Z.; Johnson, M.E.; Wood, T.A.; Feeney, L.; Angeles, J.G.; Lafyatis, R.; Whitfield, M.L. High Rhodotorula sequences in skin transcriptome of patients with diffuse systemic sclerosis. J. Investig. Dermatol. 2014, 134, 2138–2145. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, A.S.; Manzo, V.E.; Pedamallu, C.S.; Duke, F.; Cai, D.; Bienfang, D.C.; Padera, R.F.; Meyerson, M.; Docken, W.P. In search of a candidate pathogen for giant cell arteritis: Sequencing-based characterization of the giant cell arteritis microbiome. Arthritis Rheumatol. 2014, 66, 1939–1944. [Google Scholar] [CrossRef] [Green Version]
- Coit, P.; Mumcu, G.; Ture-Ozdemir, F.; Unal, A.U.; Alpar, U.; Bostanci, N.; Ergun, T.; Direskeneli, H.; Sawalha, A.H. Sequencing of 16S rRNA reveals a distinct salivary microbiome signature in Behcet’s disease. Clin. Immunol. 2016, 169, 28–35. [Google Scholar] [CrossRef]
- Consolandi, C.; Turroni, S.; Emmi, G.; Severgnini, M.; Fiori, J.; Peano, C.; Biagi, E.; Grassi, A.; Rampelli, S.; Silvestri, E.; et al. Behcet’s syndrome patients exhibit specific microbiome signature. Autoimmun. Rev. 2015, 14, 269–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correa, J.D.; Calderaro, D.C.; Ferreira, G.A.; Mendonca, S.M.; Fernandes, G.R.; Xiao, E.; Teixeira, A.L.; Leys, E.J.; Graves, D.T.; Silva, T.A. Subgingival microbiota dysbiosis in systemic lupus erythematosus: Association with periodontal status. Microbiome 2017, 5, 34. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Shao, T.; Li, H.; Xie, Z.; Wen, C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016, 8, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hevia, A.; Milani, C.; Lopez, P.; Cuervo, A.; Arboleya, S.; Duranti, S.; Turroni, F.; Gonzalez, S.; Suarez, A.; Gueimonde, M.; et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 2014, 5, e01548-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinumaki, A.; Sekizuka, T.; Hamada, H.; Kato, K.; Yamashita, A.; Kuroda, M. Characterization of the gut microbiota of Kawasaki disease patients by metagenomic analysis. Front. Microbiol. 2015, 6, 824. [Google Scholar] [CrossRef] [Green Version]
- Seoudi, N.; Bergmeier, L.A.; Drobniewski, F.; Paster, B.; Fortune, F. The oral mucosal and salivary microbial community of Behcet’s syndrome and recurrent aphthous stomatitis. J. Oral Microbiol. 2015, 7, 27150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, J.; Kubota, T.; Takada, E.; Takai, K.; Fujiwara, N.; Arimitsu, N.; Ueda, Y.; Wakisaka, S.; Suzuki, T.; Suzuki, N. Bifidobacteria abundance-featured gut microbiota compositional change in patients with Behcet’s disease. PLoS ONE 2016, 11, e0153746. [Google Scholar] [CrossRef] [Green Version]
- Volkmann, E.R.; Chang, Y.L.; Barroso, N.; Furst, D.E.; Clements, P.J.; Gorn, A.H.; Roth, B.E.; Conklin, J.L.; Getzug, T.; Borneman, J.; et al. Association of systemic sclerosis with a unique colonic microbial consortium. Arthritis Rheumatol. 2016, 68, 1483–1492. [Google Scholar] [CrossRef] [Green Version]
- Clausen, M.L.; Agner, T.; Lilje, B.; Edslev, S.M.; Johannesen, T.B.; Andersen, P.S. Association of disease severity with skin microbiome and filaggrin gene mutations in adult atopic dermatitis. JAMA Dermatol. 2018, 154, 293–300. [Google Scholar] [CrossRef]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polkowska-Pruszynska, B.; Gerkowicz, A.; Krasowska, D. The gut microbiome alterations in allergic and inflammatory skin diseases—An update. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 455–464. [Google Scholar] [CrossRef]
- Thompson, K.G.; Rainer, B.M.; Antonescu, C.; Florea, L.; Mongodin, E.F.; Kang, S.; Chien, A.L. Minocycline and Its Impact on Microbial Dysbiosis in the Skin and Gastrointestinal Tract of Acne Patients. Ann. Dermatol. 2020, 32, 21–30. [Google Scholar] [CrossRef]
- Davatchi, F.; Chams-Davatchi, C.; Shams, H.; Shahram, F.; Nadji, A.; Akhlaghi, M.; Faezi, T.; Ghodsi, Z.; Abdollahi, B.S.; Ashofteh, F.; et al. Behcet’s disease: Epidemiology, clinical manifestations, and diagnosis. Expert Rev. Clin. Immunol. 2017, 13, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Bai, R. Clinical characteristics of conduction disturbance in patients with Behcet’s disease. Clin. Rheumatol. 2018, 37, 1921–1925. [Google Scholar] [CrossRef]
- de Menthon, M.; Lavalley, M.P.; Maldini, C.; Guillevin, L.; Mahr, A. HLA-B51/B5 and the risk of Behcet’s disease: A systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 2009, 61, 1287–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durrani, K.; Papaliodis, G.N. The genetics of Adamantiades-Behcet’s disease. Semin. Ophthalmol. 2008, 23, 73–79. [Google Scholar] [CrossRef]
- Fietta, P. Behcet’s disease: Familial clustering and immunogenetics. Clin. Exp. Rheumatol. 2005, 23, S96–S105. [Google Scholar]
- Park, K.S.; Baek, J.A.; Do, J.E.; Bang, D.; Lee, E.S. CTLA4 gene polymorphisms and soluble CTLA4 protein in Behcet’s disease. Tissue Antigens 2009, 74, 222–227. [Google Scholar] [CrossRef]
- Park, K.S.; Nam, J.H.; Lee, E.S.; Choi, J.S.; Bang, D.; Lee, S. Increased risk of human leukocyte antigen-G gene variants in Behcet’s disease. Clin. Exp. Rheumatol. 2006, 24, S126–S127. [Google Scholar] [PubMed]
- Remmers, E.F.; Cosan, F.; Kirino, Y.; Ombrello, M.J.; Abaci, N.; Satorius, C.; Le, J.M.; Yang, B.; Korman, B.D.; Cakiris, A.; et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat. Genet. 2010, 42, 698–702. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, F.; Togashi, A.; Saito, S.; Sakuma, H.; Oyama, N.; Nakamura, K.; Yokota, K.; Oguma, K. Behcet’s disease (Adamantiades-Behcet’s disease). Clin. Dev. Immunol. 2011, 2011, 681956. [Google Scholar] [CrossRef]
- Lee, S.; Bang, D.; Cho, Y.H.; Lee, E.S.; Sohn, S. Polymerase chain reaction reveals herpes simplex virus DNA in saliva of patients with Behcet’s disease. Arch. Dermatol. Res. 1996, 288, 179–183. [Google Scholar] [CrossRef]
- Chi, W.; Zhu, X.; Yang, P.; Liu, X.; Lin, X.; Zhou, H.; Huang, X.; Kijlstra, A. Upregulated IL-23 and IL-17 in Behcet patients with active uveitis. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3058–3064. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cho, S.B.; Bang, D.; Oh, S.H.; Ahn, K.J.; Kim, J.; Park, Y.B.; Lee, S.K.; Lee, K.H. Human anti-alpha-enolase antibody in sera from patients with Behcet’s disease and rheumatologic disorders. Clin. Exp. Rheumatol. 2009, 27, S63–S66. [Google Scholar] [PubMed]
- Turan, B.; Gallati, H.; Erdi, H.; Gurler, A.; Michel, B.A.; Villiger, P.M. Systemic levels of the T cell regulatory cytokines IL-10 and IL-12 in Behcet’s disease; soluble TNFR-75 as a biological marker of disease activity. J. Rheumatol. 1997, 24, 128–132. [Google Scholar] [PubMed]
- Alipour, S.; Nouri, M.; Sakhinia, E.; Samadi, N.; Roshanravan, N.; Ghavami, A.; Khabbazi, A. Epigenetic alterations in chronic disease focusing on Behcet’s disease: Review. Biomed. Pharmacother. 2017, 91, 526–533. [Google Scholar] [CrossRef]
- ISG. Criteria for diagnosis of Behcet’s disease. International Study Group for Behcet’s Disease. Lancet 1990, 335, 1078–1080. [Google Scholar]
- Kurokawa, M.S.; Suzuki, N. Behcet’s disease. Clin. Exp. Med. 2004, 4, 10–20. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Ye, Z.; Zhang, N.; Wu, C.; Zhang, X.; Wang, Q.; Huang, X.; Du, L.; Cao, Q.; Tang, J.; Zhou, C.; et al. A metagenomic study of the gut microbiome in Behcet’s disease. Microbiome 2018, 6, 135. [Google Scholar] [CrossRef] [PubMed]
- van der Houwen, T.B.; van Laar, J.A.M.; Kappen, J.H.; van Hagen, P.M.; de Zoete, M.R.; van Muijlwijk, G.H.; Berbers, R.M.; Fluit, A.C.; Rogers, M.; Groot, J.; et al. Behcet’s Disease under Microbiotic Surveillance? A Combined Analysis of Two Cohorts of Behcet’s Disease Patients. Front. Immunol. 2020, 11, 1192. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S.; Gallini, C.A.; Yatsunenko, T.; Michaud, M.; DuBois, A.; Delaney, M.L.; Punit, S.; Karlsson, M.; Bry, L.; Glickman, J.N.; et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 2010, 8, 292–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.J.; Kim, K.; Lee, E.-S.; Park, S. The Suppressive Effect of Butyrate and Bromopyruvate on Inflammatory Cytokine Production and Short Chain Fatty Acid Receptor Expression by Blood Mononuclear Cells in Patients with Behçet’s Disease. Ann. Dermatol. 2018, 30, 566–574. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Chen, Y.; Chen, D. The Role of Gut Microbiome in Autoimmune Uveitis. Ophthalmic Res. 2021, 64, 168–177. [Google Scholar] [CrossRef]
- Kodati, S.; Sen, H.N. Uveitis and the gut microbiota. Best Pract. Res. Clin. Rheumatol. 2019, 33, 101500. [Google Scholar] [CrossRef]
- Chakravarthy, S.K.; Jayasudha, R.; Prashanthi, G.S.; Ali, M.H.; Sharma, S.; Tyagi, M.; Shivaji, S. Dysbiosis in the Gut Bacterial Microbiome of Patients with Uveitis, an Inflammatory Disease of the Eye. Indian J. Microbiol. 2018, 58, 457–469. [Google Scholar] [CrossRef]



| Fusion Primers | |
|---|---|
| 341F | 5′-AATGATACGGCGACCACCGAGATCTACAC-XXXXXXX-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3′ |
| 805R | 5′- CAAGCAGAAGACGGCATACGAGAT-XXXXXXXX-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGACAG-GACTACHVGGGTATCTAATCC-3′ |
| Active BD (n = 9) | BD-Matched HC (n = 9) | Active RAU (n = 7) | RAU-Matched HC (n = 7) | |
|---|---|---|---|---|
| Age, median (IQR), years | 33 (28.5–45.5) | 53 (33–56.5) | 47 (40–66) | 44 (36–67) |
| Sex | ||||
| Male, n (%) | 1 (11.1) | 4 (44.4) | 2 (28.6) | 3 (42.9) |
| Female, n (%) | 8 (88.9) | 5 (55.6) | 5 (71.4) | 4 (57.1) |
| Alcohol history, n (%) | 2 (22.2) | 5 (55.6) | 1 (14.3) | 4 (57.1) |
| Smoking | ||||
| Smokers, n (%) | 2 (22.2) | 0 (0) | 0 (0) | 0 (0) |
| Ex-smokers, n (%) | 1 (11.1) | 1 (11.1) | 2 (28.6) | 2 (28.6) |
| Nonsmokers, n (%) | 6 (66.7) | 8 (88.9) | 5 (71.4) | 0 (0) |
| Family history of each disease, n (%) | 0 (0) | N/A | 0 (0) | N/A |
| Disease duration, median (IQR), years | 5.0 (1.5–9.0) | N/A | 1.0 (0.5–2.0) | N/A |
| Disease characteristics | ||||
| Oral aphthous ulcers, n (%) | 9(100) | 0 (0) | 7 (100) | 0 (0) |
| Genital ulcers, n (%) | 9 (100) | 0 (0) | 0 (0) | 0 (0) |
| Skin lesions, n (%) | 9 (100) | 0 (0) | 0 (0) | 0 (0) |
| Uveitis, n (%) | 2 (22.2) | 0 (0) | 0 (0) | 0 (0) |
| Arthritis, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| GI lesions, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| CNS lesions, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| ESR, median (IQR), mm/h | 22.0 (13.5–26.0) | N/A | 9.0 (2.0–37.0) | N/A |
| CRP, median (IQR), mg/dL | 0.27 (0.07–1.15) | N/A | 0.12 (0.03–0.30) | N/A |
| HLA-B51 | ||||
| Positive, n (%) | 9 (100) | N/A | 1 (14.3) | N/A |
| Negative, n (%) | 0 (100) | N/A | 6 (85.7) | N/A |
| Previous treatment | ||||
| Systemic steroids, n (%) | 9 (100) | N/A | 7 (100) | N/A |
| Colchicine, n (%) | 8 (88.9) | N/A | 2 (28.6) | N/A |
| Azathioprine, n (%) | 1 (14.3) | N/A | 0 (0) | N/A |
| Cyclosporine, n (%) | 1 (14.3) | N/A | 0 (0) | N/A |
| Antibiotic, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Previous probiotic use, n (%) | 2 (22.2) | 2 (22.2) | 2 (28.6) | 2 (28.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.C.; Park, M.J.; Park, S.; Lee, E.-S. Alteration of the Fecal but Not Salivary Microbiome in Patients with Behçet’s Disease According to Disease Activity Shift. Microorganisms 2021, 9, 1449. https://doi.org/10.3390/microorganisms9071449
Kim JC, Park MJ, Park S, Lee E-S. Alteration of the Fecal but Not Salivary Microbiome in Patients with Behçet’s Disease According to Disease Activity Shift. Microorganisms. 2021; 9(7):1449. https://doi.org/10.3390/microorganisms9071449
Chicago/Turabian StyleKim, Jin Cheol, Mi Jin Park, Sun Park, and Eun-So Lee. 2021. "Alteration of the Fecal but Not Salivary Microbiome in Patients with Behçet’s Disease According to Disease Activity Shift" Microorganisms 9, no. 7: 1449. https://doi.org/10.3390/microorganisms9071449
APA StyleKim, J. C., Park, M. J., Park, S., & Lee, E.-S. (2021). Alteration of the Fecal but Not Salivary Microbiome in Patients with Behçet’s Disease According to Disease Activity Shift. Microorganisms, 9(7), 1449. https://doi.org/10.3390/microorganisms9071449






