Acid Stable Yeast Cell-Associated Tannase with High Capability in Gallated Catechin Biotransformation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
2.2. Microorganisms and Culture Conditions
2.3. Effect of Temperature on Growth of Tannin-Tolerant Yeasts
2.4. Production of CAT from Tannin-Tolerant Yeasts
2.5. CAT Preparation
2.6. Assay of CAT Activity and Determination of Gallic Acid
2.7. Effect of pH on CAT Activity and Stability
2.8. Effect of Temperature on CAT Activity Stability
2.9. Effect of Cations on CAT Activity
2.10. Effect of Organic Solvents on CAT Stability
2.11. Substrate Specificity
2.12. Feasible Evaluation for the Biotransformation of Catechin Derivatives
2.13. Operational Stability Determination
2.14. Statistical Analysis
3. Results
3.1. Effect of Temperature on the Growth of Tannin-Tolerant Yeasts
3.2. Profile of CAT Production by Tannin-Tolerant Yeasts
3.3. Temperature Optima and Stabilities, and pH Optima and Stabilities of CATs
3.4. Effects of Cations on CAT Activity
3.5. Effects of Organic Solvents on Enzyme Stability
3.6. Substrate Specificity and Biotransformation of Catechin Derivatives
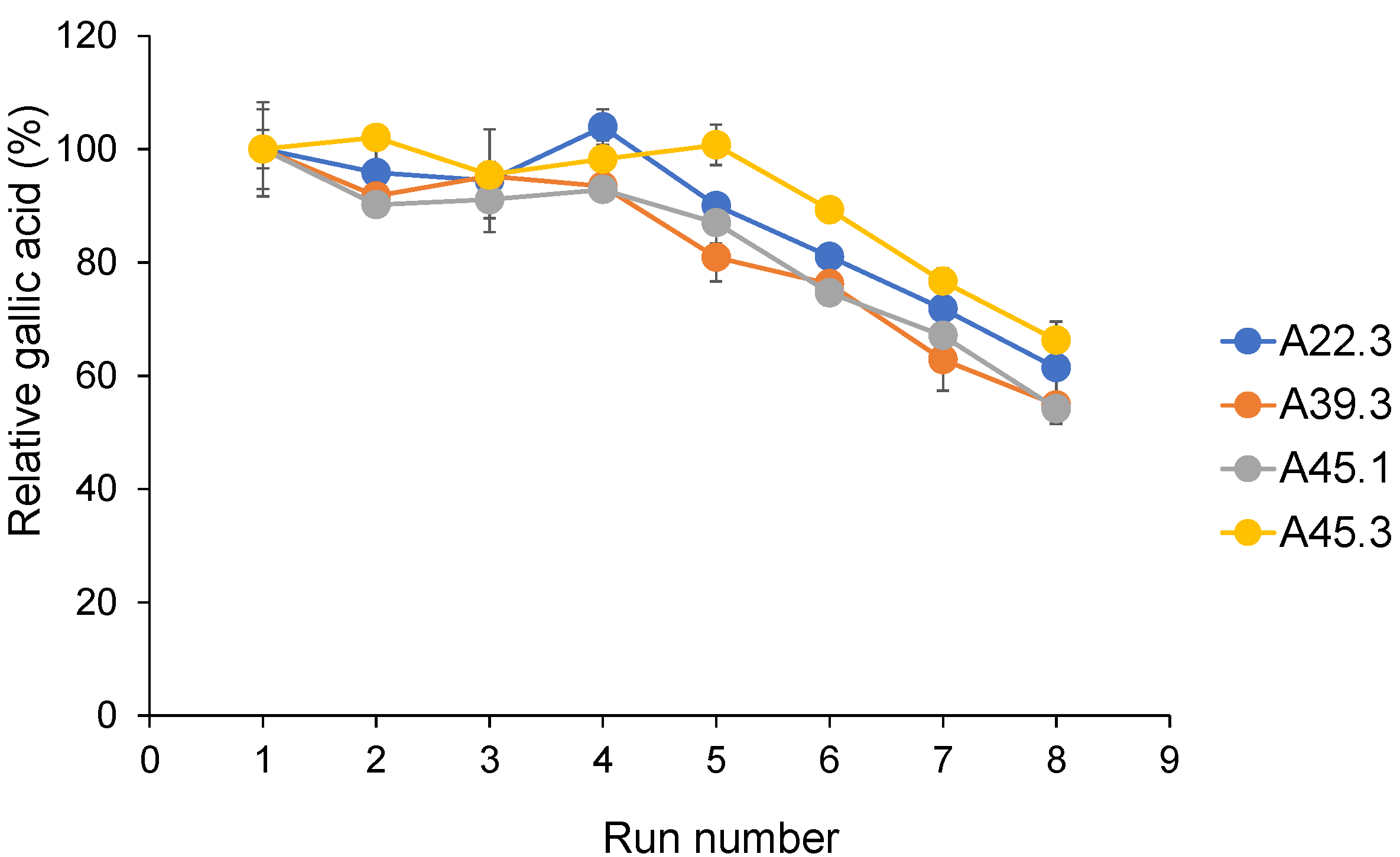
3.7. Repeatability of CATs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Govindarajan, R.K.; Mathivanan, K.; Khanongnuch, C.; Srinivasan, R.; Unban, K.; Charli Deepak, A.; Al Farraj, D.A.; Mohammed Alarjani, K.; Al Qahtany, F.S. Tannin acyl-hydrolase production by Bacillus subtilis KMS2-2: Purification, characterization, and cytotoxicity studies. J. King Saud Univ. Sci. 2021, 33, 101359. [Google Scholar] [CrossRef]
- Dhiman, S.; Mukherjee, G.; Kumar, A.; Mukherjee, P.; Verekar, S.A.; Deshmukh, S.K. Fungal tannase: Recent advances and industrial applications. In Developments in Fungal Biology and Applied Mycology; Satyanarayana, T., Deshmukh, S.K., Johri, B.N., Eds.; Springer Nature: Singapore, 2017; pp. 295–313. [Google Scholar]
- Bhat, T.K.; Singh, B.; Sharma, O.P. Microbial degradation of tannins—A current perspective. Biodegradation 1998, 9, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Yin, J.-F.; Chen, J.-X.; Wang, F.; Du, Q.-Z.; Jiang, Y.-W.; Xu, Y.-Q. Improving the sweet aftertaste of green tea infusion with tannase. Food Chem. 2016, 192, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Guo, G.S.; Ren, G.H.; Liu, Y.H. Production, characterization and applications of tannase. J. Mol. Catal. B Enzym. 2014, 101, 137–147. [Google Scholar] [CrossRef]
- Aoki, K.; Shinke, R.; Nishira, H. Purification and some properties of yeast tannase. Agric. Biol. Chem. 1976, 40, 79–85. [Google Scholar] [CrossRef]
- Zhang, L.L.; Li, J.; Wang, Y.L.; Liu, S.; Wang, Z.P.; Yu, X.J. Integrated approaches to reveal genes crucial for tannin degradation in Aureobasidium melanogenum T9. Biomolecules 2019, 9, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böer, E.; Bode, R.; Mock, H.-P.; Piontek, M.; Kunze, G. Atan1p-an extracellular tannase from the dimorphic yeast Arxula adeninivorans: Molecular cloning of the ATAN1 gene and characterization of the recombinant enzyme. Yeast 2009, 26, 323–337. [Google Scholar] [CrossRef]
- Mahmoud, A.E.; Fathy, S.A.; Rashad, M.M.; Ezz, M.K.; Mohammed, A.T. Purification and characterization of a novel tannase produced by Kluyveromyces marxianus using olive pomace as solid support, and its promising role in gallic acid production. Int. J. Biol. Macromol. 2018, 107, 2342–2350. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Khanongnuch, C.; Lumyong, S.; Haltrich, D.; Nguyen, T.-H.; Kittibunchakul, S. Co-production of gallic acid and a novel cell-associated tannase by a pigment-producing yeast, Sporidiobolus ruineniae A45.2. Microb. Cell Factories 2020, 19, 95. [Google Scholar] [CrossRef]
- Pan, J.; Wang, N.-N.; Yin, X.-J.; Liang, X.-L.; Wang, Z.-P. Characterization of a robust and pH-stable tannase from mangrove-derived yeast Rhodosporidium diobovatum Q95. Mar. Drugs 2020, 18, 546. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.; Serban, S. Industrial applications of immobilized enzymes-A review. Mol. Catal. 2019, 479, 110607. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Chui-Chai, N.; Chaikaew, S.; Khanongnuch, C. Distribution of tannin-tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int. J. Food. Microbiol. 2016, 238, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Khanongnuch, C.; Unban, K.; Kanpiengjai, A.; Saenjum, C. Recent research advances and ethno-botanical history of miang, a traditional fermented tea (Camellia sinensis var. assamica) of northern Thailand. J. Ethn. Foods 2017, 4, 135–144. [Google Scholar] [CrossRef]
- Unban, K.; Khatthongngam, N.; Shetty, K.; Khanongnuch, C. Nutritional biotransformation in traditional fermented tea (Miang) from north Thailand and its impact on antioxidant and antimicrobial activities. J. Food Sci. Technol. 2019, 56, 2687–2699. [Google Scholar] [CrossRef]
- Unban, K.; Khatthongngam, N.; Pattananandecha, T.; Saenjum, C.; Shetty, K.; Khanongnuch, C. Microbial community dynamics during the non-filamentous fungi growth-based fermentation process of miang, a traditional fermented tea of north thailand and their product characterizations. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Kodchasee, P.; Nain, K.; Abdullahi, A.D.; Unban, K.; Saenjum, C.; Shetty, K.; Khanongnuch, C. Microbial dynamics-linked properties and functional metabolites during Miang fermentation using the filamentous fungi growth-based process. Food Biosci. 2021, 41, 100998. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Khanongnuch, C.; Lumyong, S.; Kummasook, A.; Kittibunchakul, S. Characterization of Sporidiobolus ruineniae A45.2 Cultivated in Tannin Substrate for Use as a Potential Multifunctional Probiotic Yeast in Aquaculture. J. Fungi 2020, 6, 378. [Google Scholar] [CrossRef]
- Rigoldi, F.; Donini, S.; Redaelli, A.; Parisini, E.; Gautieri, A. Review: Engineering of thermostable enzymes for industrial applications. APL Bioeng. 2018, 2, 011501. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Kar, R. Characterization and application of tannase produced by Aspergillus niger ITCC 6514.07 on pomegranate rind. Braz. J. Microbiol. 2009, 40, 782–789. [Google Scholar] [CrossRef] [Green Version]
- Aguilar-Zárate, P.; Cruz, M.A.; Montañez, J.; Rodríguez-Herrera, R.; Wong-Paz, J.E.; Belmares, R.E.; Aguilar, C.N. Gallic acid production under anaerobicsubmerged fermentation by two bacilli strains. Microb. Cell Factories 2015, 14, 209. [Google Scholar] [CrossRef] [Green Version]
- Meier, A.K.; Worch, S.; Böer, E.; Hartmann, A.; Mascher, M.; Marzec, M.; Scholz, U.; Riechen, J.; Baronian, K.; Schauer, F.; et al. Agdc1p—A gallic acid decarboxylase involved in the degradation of tannic acid in the yeast Blastobotrys (Arxula) adeninivorans. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Dhiman, S.; Mukherjee, G.; Singh, A.K. Recent trends and advancements in microbial tannase-catalyzed biotransformation of tannins: A review. Int. Microbiol. 2018, 21, 175–195. [Google Scholar] [CrossRef] [PubMed]
- Ueda, S.; Nomoto, R.; Yoshida, K.-i.; Osawa, R. Comparison of three tannases cloned from closely related lactobacillus species: L. plantarum, L. paraplantarum, and L. pentosus. BMC Microbiol. 2014, 14, 87. [Google Scholar] [CrossRef] [Green Version]
- Kanpiengjai, A.; Unban, K.; Nguyen, T.-H.; Haltrich, D.; Khanongnuch, C. Expression and biochemical characterization of a new alkaline tannase from Lactobacillus pentosus. Protein Expr. Purif. 2019. [Google Scholar] [CrossRef] [PubMed]
- Jana, A.; Maity, C.; Halder, S.K.; Das, A.; Pati, B.R.; Mondal, K.C.; Das Mohapatra, P.K. Structural characterization of thermostable, solvent tolerant, cytosafe tannase from Bacillus subtilis PAB2. Biochem. Eng. J. 2013, 77, 161–170. [Google Scholar] [CrossRef]
- Jana, A.; Halder, S.K.; Banerjee, A.; Paul, T.; Pati, B.R.; Mondal, K.C.; Das Mohapatra, P.K. Biosynthesis, structural architecture and biotechnological potential of bacterial tannase: A molecular advancement. Bioresour. Technol. 2014, 157, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Wu, M.; Wang, Q.; Peng, X.; Wen, H.; McKinstry, W.J.; Chen, Q. Crystal structure of tannase from Lactobacillus plantarum. J. Mol. Biol. 2013, 425, 2737–2751. [Google Scholar] [CrossRef]
- Xu, J.Z.; Yeung, S.Y.; Chang, Q.; Huang, Y.; Chen, Z.Y. Comparison of antioxidant activity and bioavailability of tea epicatechins with their epimers. Br. J. Nutr. 2004, 91, 873–881. [Google Scholar] [CrossRef]
- Fang, X.; Du, M.; Liu, T.; Fang, Q.a.; Liao, Z.; Zhong, Q.; Chen, J.; Meng, X.; Zhou, S.; Wang, J. Changes in the biotransformation of green tea catechins induced by different carbon and nitrogen sources in Aspergillus niger RAF106. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Baik, J.H.; Shin, K.-S.; Park, Y.; Yu, K.-W.; Suh, H.J.; Choi, H.-S. Biotransformation of catechin and extraction of active polysaccharide from green tea leaves via simultaneous treatment with tannase and pectinase. J. Sci. Food Agric. 2015, 95, 2337–2344. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, Y.-H.; Zhang, F.; Yang, Q.-M.; Weng, H.-F.; Xiao, Q.; Xiao, A.-F. Thermostable tannase from Aspergillus niger and its application in the enzymatic extraction of green tea. Molecules 2020, 25, 952. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Cations (5 mM) | Relative Activity (%) | |||
|---|---|---|---|---|
| Cy. rhodanensis A22.3 | Candida sp. A39.3 | D. hansenii A45.1 | Cy. rhodanensis A45.3 | |
| Na+ | 91.3 ± 4.3 d | 97.9 ± 3.6 cd | 105.4 ± 6.9 cd | 98.6 ± 6.8 d |
| K+ | 97.6 ± 4.5 cd | 102.1 ± 3.2 cd | 96.2 ± 2.3 de | 88.9 ± 2.8 de |
| Ba2+ | 90.5 ± 0.4 d | 89.7 ± 4.8 d | 84.1 ± 7.7 e | 87.8 ± 2.7 de |
| Ca2+ | 99.5 ± 0.9 cd | 131.9 ± 2.5 b | 100.6 ± 0.1 cde | 98.0 ± 5.6 d |
| Mg2+ | 102.1 ± 2.5 cd | 111.3 ± 6.0 c | 86.9 ± 1.7 de | 80.7 ± 1.4 e |
| Cu2+ | 70.7 ± 6.0 e | 63.4 ± 2.2 e | 88.2 ± 1.0 de | 60.3 ± 2.4 f |
| Mn2+ | 118.9 ± 0.8 b | 154.1 ± 5.9 a | 139.9 ± 8.6 b | 171.8 ± 5.3 a |
| Co2+ | 119.5 ± 1.1 b | 137.9 ± 3.1 b | 117.7 ± 2.2 c | 144.1 ± 0.0 b |
| Ni2+ | 119.0 ± 0.8 b | 111.9 ± 1.1 c | 100.5 ± 6.0 cde | 128.7 ± 7.2 c |
| Zn2+ | 107.2 ± 1.5 bc | 90.5 ± 1.0 d | 94.8 ± 4.7 de | 102.0 ± 2.2 d |
| Al3+ | 53.5 ± 0.2 f | 36.8 ± 0.9 f | 27.8 ± 4.0 f | 18.2 ± 2.0 g |
| Fe3+ | 146.3 ± 4.7 a | 142.5 ± 4.8 ab | 170.7 ± 3.2 a | 141.6 ± 0.0 bc |
| Control | 100.0 ± 6.8 cd | 100.0 ± 0.3 cd | 100.0 ± 3.1 cde | 100.0 ± 2.1 d |
| Solvents | Residual Activity (%) | |||
|---|---|---|---|---|
| Cy. rhodanensis A22.3 | Candida sp. A39.3 | D. hansenii A45.1 | Cy. rhodanensis A45.3 | |
| Methanol | 64.4 ± 1.3 b | 64.9 ± 5.6 bc | 69.4 ± 2.3 b | 90.5 ± 3.3 b |
| Ethanol | 65.4 ± 3.9 b | 55.7 ± 1.9 bc | 55.4 ± 3.0 c | 79.2 ± 1.7 c |
| Propanol | 49.3 ± 2.0 c | 34.4 ± 2.5 d | 13.4 ± 1.9 e | 52.1 ± 1.8 d |
| Butanol | 62.8 ± 1.0 b | 47.8 ± 4.4 cd | 25.2 ± 3.2 d | 74.4 ± 2.3 c |
| Acetone | 47.0 ± 4.0 c | 17.4 ± 1.9 d | 10.9 ± 1.4 e | 32.8 ± 1.5 e |
| Control | 100.0 ± 1.1 a | 100.0 ± 1.5 a | 100.0 ± 0.7 a | 100.0 ± 1.3 a |
| Strains | Relative Activity (%) | |||
|---|---|---|---|---|
| Methyl Gallate | Propyl Gallate | EGCG | ECG | |
| Cy. rhodanensis A22.3 | 100.0 ± 3.1 b | 95.5 ± 4.3 b | 139.2 ± 3.1 a | 141.0 ± 2.8 a |
| Candida sp. A39.3 | 100.0 ± 5.2 c | 75.7 ± 5.5 d | 160.3 ± 2.3 b | 185.6 ± 8.4 a |
| D. hansenii A45.1 | 100.0 ± 5.5 c | 73.6 ± 7.8 d | 149.6 ± 1.0 b | 290.3 ± 0.3 a |
| Cy. rhodanensis A45.3 | 100.0 ± 4.1 c | 75.4 ± 7.5 d | 153.7 ± 5.5 b | 201.6 ± 2.8 a |
| Microorganisms | Optimal pH | Optimal Temperature | pH Stability | (t1/2) Thermostability | References |
|---|---|---|---|---|---|
| Candida sp. | 6.0 | 50 °C | 4.0–8.0 at 13 °C, 16 h | 60 °C, 10 min | [6] |
| B. adeninivorans | 7.0 | 40 °C | 5.0–7.0, 10 min | Stable at 30–40 °C, 10 min | [8] |
| K. marxianus | 4–4.5 | 35 °C | 4–5.5, at 30 °C, 30 min | 70 °C, 1 h | [9] |
| S. ruineniae A45.2 | 7.0 | 40 °C | 5.0–9.0 at 37 °C, 6 h | 75 °C, 1 h | [10] |
| R. diobovatum | 4.5 | 40 °C | 4.5–5.5, at 40 °C, 12 h | 55–60 °C, 12 h | [11] |
| Cy. rhodanensis A22.3 | 2.0 | 45 °C | 2.0–4.0, at 37 °C, 6 h | 70 °C, 1 h | This study |
| Cy. rhodanensis A45.3 | 6.0 | 20–40 °C | 4.0–9.0, at 37 °C, 6 h | 60 °C, 1 h | This study |
| Candida sp. A39.3 | 7.0 | 20–30 °C | 5.0–9.0, at 37 °C, 6 h | 50 °C, 1 h | This study |
| D. hansenii A45.1 | 5.0 | 20–40 °C | 4.0–9.0, at 37 °C, 6 h | 50 °C, 1 h | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leangnim, N.; Aisara, J.; Unban, K.; Khanongnuch, C.; Kanpiengjai, A. Acid Stable Yeast Cell-Associated Tannase with High Capability in Gallated Catechin Biotransformation. Microorganisms 2021, 9, 1418. https://doi.org/10.3390/microorganisms9071418
Leangnim N, Aisara J, Unban K, Khanongnuch C, Kanpiengjai A. Acid Stable Yeast Cell-Associated Tannase with High Capability in Gallated Catechin Biotransformation. Microorganisms. 2021; 9(7):1418. https://doi.org/10.3390/microorganisms9071418
Chicago/Turabian StyleLeangnim, Nalapat, Jakkrit Aisara, Kridsada Unban, Chartchai Khanongnuch, and Apinun Kanpiengjai. 2021. "Acid Stable Yeast Cell-Associated Tannase with High Capability in Gallated Catechin Biotransformation" Microorganisms 9, no. 7: 1418. https://doi.org/10.3390/microorganisms9071418
APA StyleLeangnim, N., Aisara, J., Unban, K., Khanongnuch, C., & Kanpiengjai, A. (2021). Acid Stable Yeast Cell-Associated Tannase with High Capability in Gallated Catechin Biotransformation. Microorganisms, 9(7), 1418. https://doi.org/10.3390/microorganisms9071418






