An Internal Promoter Drives the Expression of a Truncated Form of CCC1 Capable of Protecting Yeast from Iron Toxicity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Growth Conditions
2.2. Plasmids
2.3. Immunoblotting
2.4. Protein Stability Assays
2.5. β-Galactosidase Assays
2.6. Fluorescence Microscopy
2.7. Synchrotron X-ray Fluorescence Imaging (SXRF)
2.8. Northern Blot
2.9. Real-Time RT-PCR Analyses (qRT-PCR)
3. Results
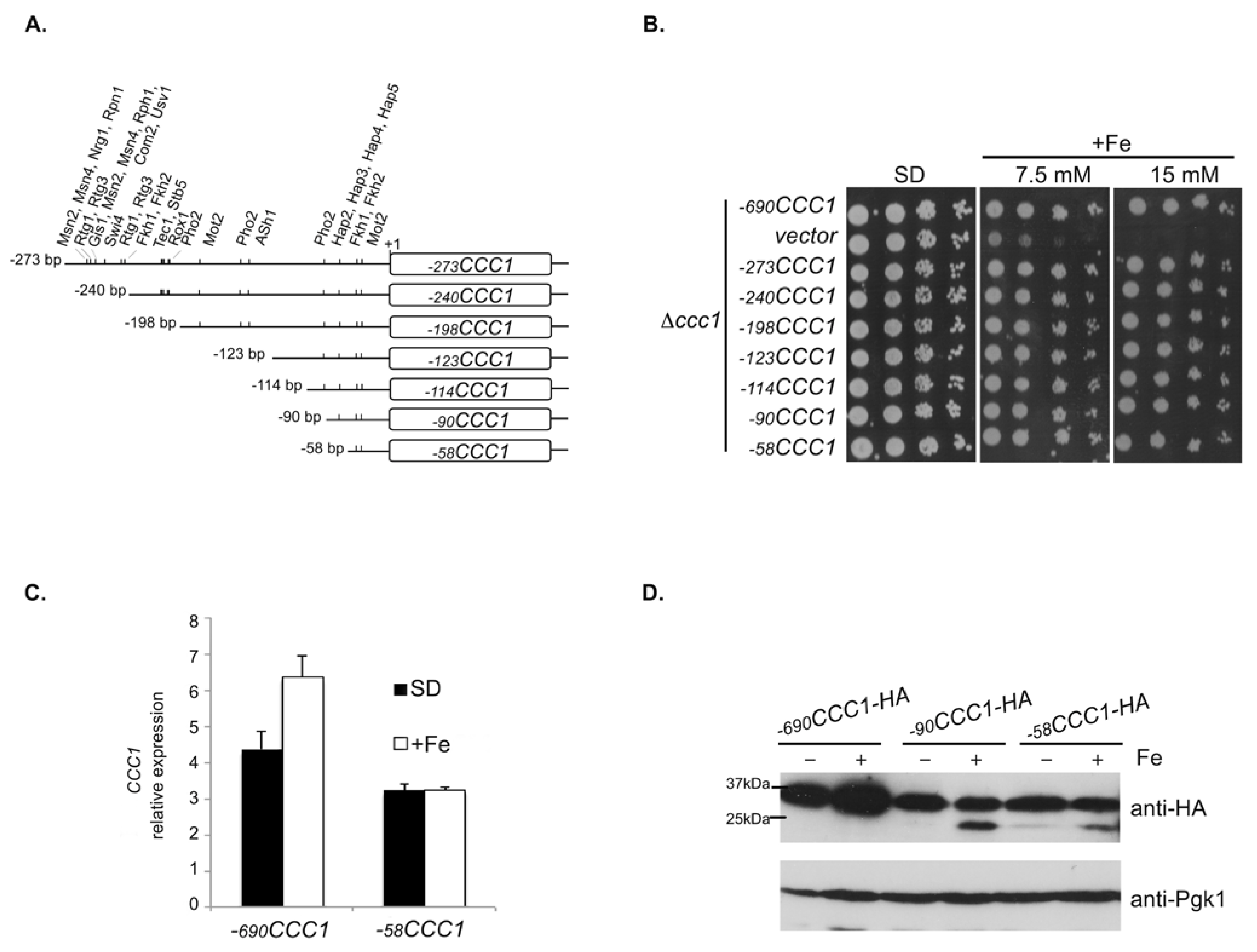
3.1. A Promoter Region of 58 bp Maintains the Expression of CCC1 at Levels That Allow Cells to Overcome Iron Toxicity
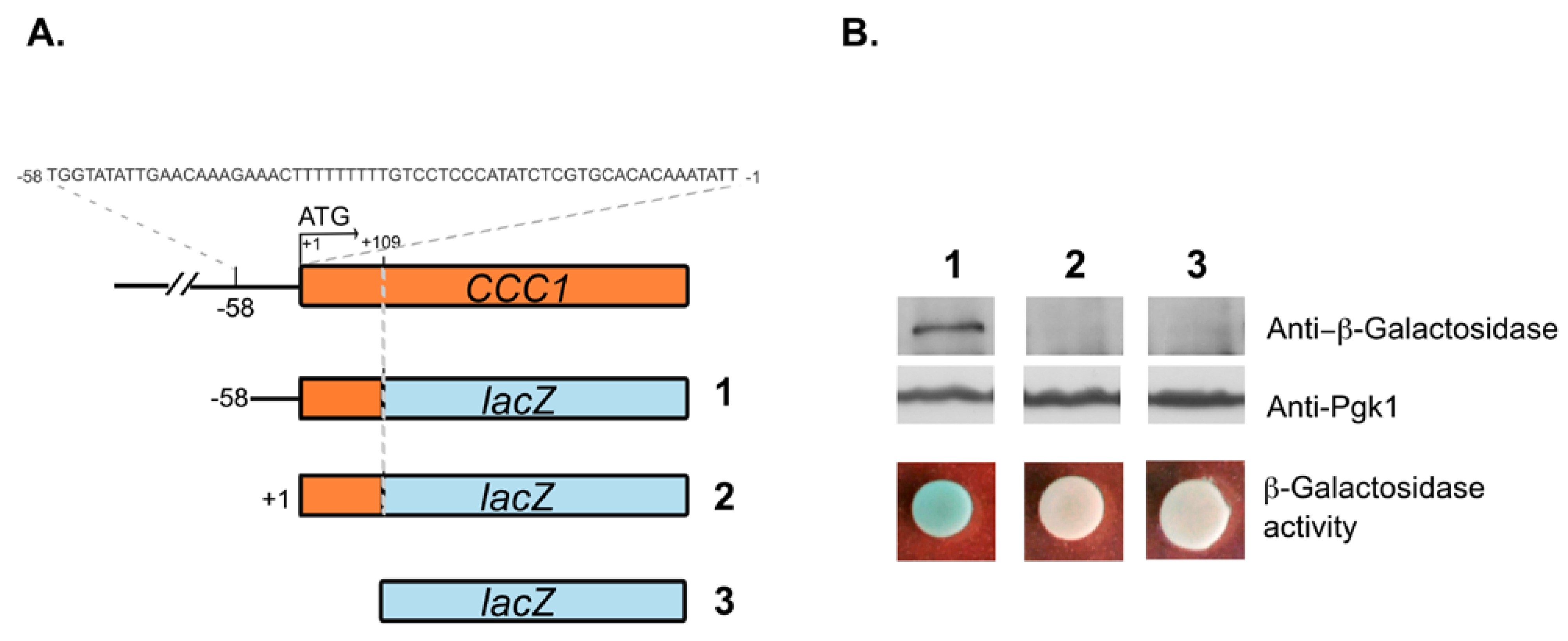
3.2. Deletions Extending into the CCC1 Coding Sequence Lead to the Expression of a Functional In-Frame N-Terminal Truncated Form of Ccc1
3.3. The Genomic Context Controls s-CCC1 Expression
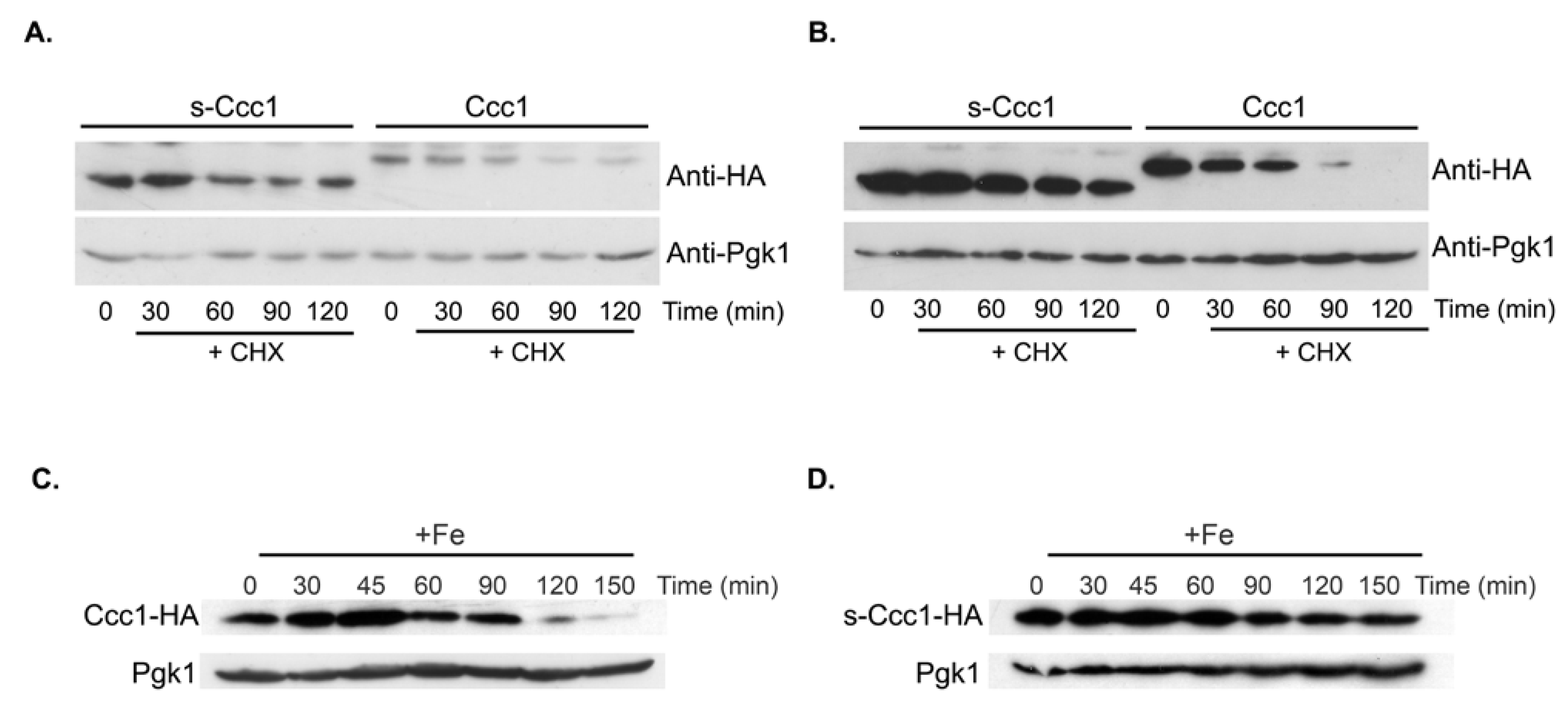
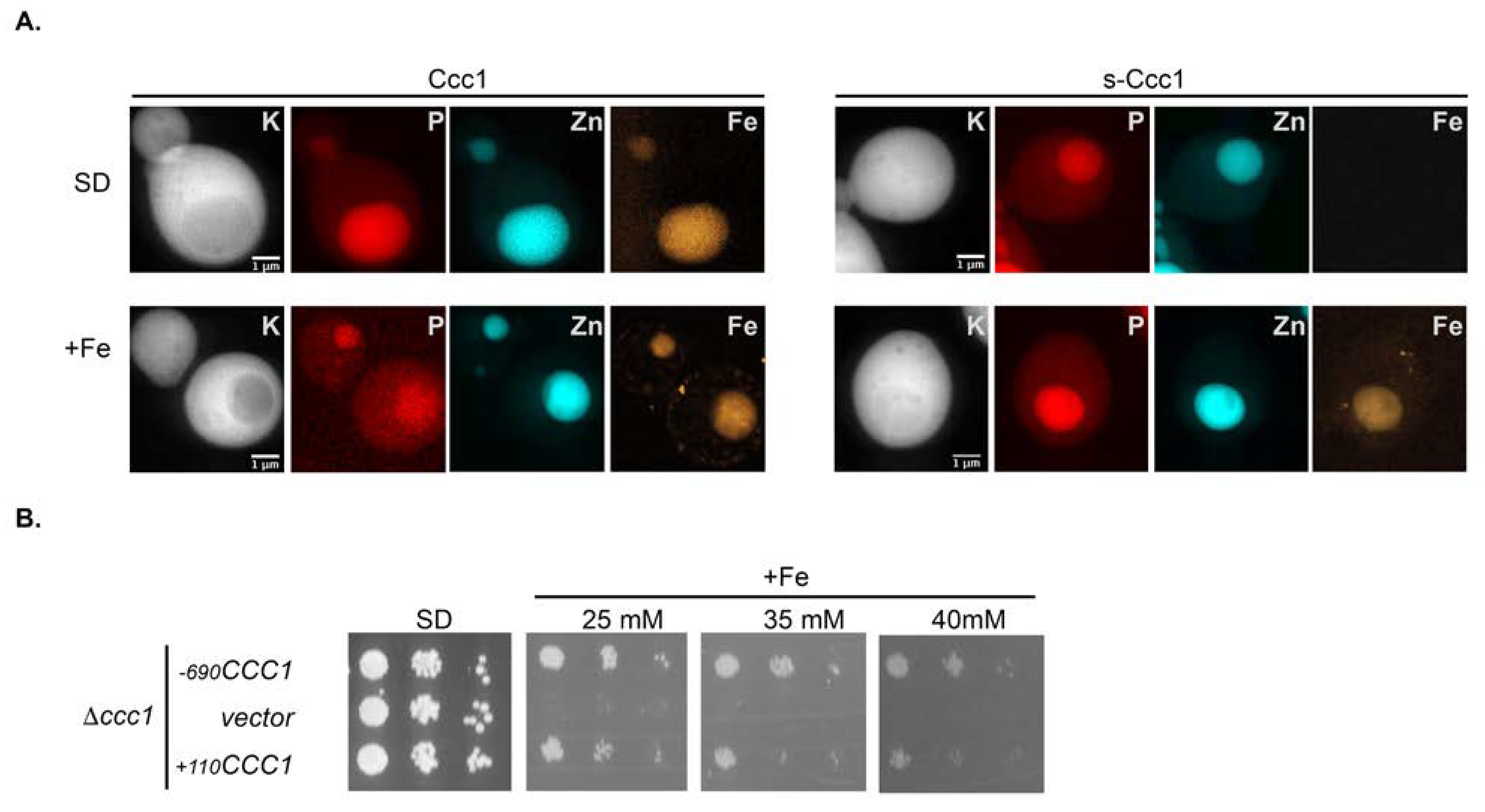
3.4. s-Ccc1 Locates in the Endoplasmic Reticulum and Vacuole and Is More Stable but Less Efficient Than Ccc1 in Iron Detoxification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Meneghini, R. Iron Homeostasis, Oxidative Stress, and DNA Damage. Free. Radic. Biol. Med. 1997, 23, 783–792. [Google Scholar] [CrossRef]
- Kaplan, C.D.; Kaplan, J. Iron Acquisition and Transcriptional Regulation. Chem. Rev. 2009, 109, 4536–4552. [Google Scholar] [CrossRef]
- Martínez-Pastor, M.T.; Perea-García, A.; Puig, S. Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2017, 33, 75. [Google Scholar] [CrossRef] [Green Version]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to Tango: Regulation of Mammalian Iron Metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dlouhy, A.C.; Outten, C.E. The Iron Metallome in Eukaryotic Organisms. Met. Ions Life Sci. 2013, 12, 241–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueta, R.; Fujiwara, N.; Iwai, K.; Yamaguchi-Iwai, Y. Mechanism Underlying the Iron-dependent Nuclear Export of the Iron-responsive Transcription Factor Aft1p in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2980–2990. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi-Iwai, Y.; Ueta, R.; Fukunaka, A.; Sasaki, R. Subcellular Localization of Aft1 Transcription Factor Responds to Iron Status in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 18914–18918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig, S.; Vergara, S.V.; Thiele, D.J. Cooperation of Two mRNA-Binding Proteins Drives Metabolic Adaptation to Iron Deficiency. Cell Metab. 2008, 7, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic Response to Iron Deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef] [Green Version]
- Shakoury-Elizeh, M.; Tiedeman, J.; Rashford, J.; Ferea, T.; Demeter, J.; Garcia, E.; Rolfes, R.; Brown, A.J.; Botstein, D.; Philpott, C.C. Transcriptional Remodeling in Response to Iron Deprivation inSaccharomyces cerevisiae. Mol. Biol. Cell 2004, 15, 1233–1243. [Google Scholar] [CrossRef] [Green Version]
- Felice, M.R.; De Domenico, I.; Li, L.; Ward, D.M.; Bartok, B.; Musci, G.; Kaplan, J. Post-transcriptional Regulation of the Yeast High Affinity Iron Transport System. J. Biol. Chem. 2005, 280, 22181–22190. [Google Scholar] [CrossRef] [Green Version]
- Holmes-Hampton, G.P.; Jhurry, N.D.; McCormick, S.P.; Lindahl, P.A. Iron Content ofSaccharomyces cerevisiaeCells Grown under Iron-Deficient and Iron-Overload Conditions. Biochemistry 2013, 52, 105–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.; Henras, A.K.; Chanfreau, G. Multiple RNA Surveillance Pathways Limit Aberrant Expression of Iron Uptake mRNAs and Prevent Iron Toxicity in S. cerevisiae. Mol. Cell 2005, 19, 39–51. [Google Scholar] [CrossRef]
- Yun, C.-W.; Ferea, T.; Rashford, J.; Ardon, O.; Brown, P.O.; Botstein, D.; Kaplan, J.; Philpott, C.C. Desferrioxamine-mediated Iron Uptake in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 10709–10715. [Google Scholar] [CrossRef] [Green Version]
- Bode, H.P.; Dumschat, M.; Garotti, S.; Fuhrmann, G.F. Iron sequestration by the yeast vacuole. A study with vacuolar mutants of Saccharomyces cerevisiae. Eur. J. Biochem. 1995, 228, 337–342. [Google Scholar] [CrossRef]
- Raguzzi, F.; Lesuisse, E.; Crichton, R.R. Iron storage in Saccharomyces cerevisiae. Febs Lett. 1988, 231, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Chen, O.S.; Ward, D.M.; Kaplan, J. CCC1 Is a Transporter That Mediates Vacuolar Iron Storage in Yeast. J. Biol. Chem. 2001, 276, 29515–29519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gsaller, F.; Eisendle, M.; Lechner, B.E.; Schrettl, M.; Lindner, H.; Müller, D.; Geley, S.; Haas, H. The interplay between vacuolar and siderophore-mediated iron storage in Aspergillus fumigatus. Metallom. Integr. Biomet. Sci. 2012, 4, 1262–1270. [Google Scholar] [CrossRef] [Green Version]
- Blaby-Haas, C.E.; Merchant, S.S. Lysosome-related Organelles as Mediators of Metal Homeostasis. J. Biol. Chem. 2014, 289, 28129–28136. [Google Scholar] [CrossRef] [Green Version]
- Connorton, J.M.; Jones, E.R.; Rodríguez-Ramiro, I.; Fairweather-Tait, S.; Uauy, C.; Balk, J. Wheat Vacuolar Iron Transporter TaVIT2 Transports Fe and Mn and Is Effective for Biofortification. Plant. Physiol. 2017, 174, 2434–2444. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.A.; Punshon, T.; Lanzirotti, A.; Li, L.; Alonso, J.M.; Ecker, J.R.; Kaplan, J.; Guerinot, M.L. Localization of Iron in Arabidopsis Seed Requires the Vacuolar Membrane Transporter VIT1. Science 2006, 314, 1295–1298. [Google Scholar] [CrossRef]
- Kato, T.; Kumazaki, K.; Wada, M.; Taniguchi, R.; Nakane, T.; Yamashita, K.; Hirata, K.; Ishitani, R.; Ito, K.; Nishizawa, T.; et al. Crystal structure of plant vacuolar iron transporter Vitnat. Plants 2019, 5, 308–315. [Google Scholar] [CrossRef]
- Sankari, S.; O’Brian, M.R. A Bacterial Iron Exporter for Maintenance of Iron Homeostasis. J. Biol. Chem. 2014, 289, 16498–16507. [Google Scholar] [CrossRef] [Green Version]
- Bhubhanil, S.; Chamsing, J.; Sittipo, P.; Chaoprasid, P.; Sukchawalit, R.; Mongkolsuk, S. Roles of Agrobacterium tumefaciens membrane-bound ferritin (MbfA) in iron transport and resistance to iron under acidic conditions. Microbiology 2014, 160, 863–871. [Google Scholar] [CrossRef]
- Huang, G.; Ulrich, P.N.; Storey, M.; Johnson, D.; Tischer, J.; Tovar, J.A.; Moreno, S.N.J.; Orlando, R.; Docampo, R. Proteomic Analysis of the Acidocalcisome, an Organelle Conserved from Bacteria to Human Cells. Plos Pathog. 2014, 10, e1004555. [Google Scholar] [CrossRef] [Green Version]
- Slavic, K.; Krishna, S.; Lahree, A.; Bouyer, G.; Hanson, K.K.; Vera, I.; Pittman, J.K.; Staines, H.M.; Mota, M.M. A vacuolar iron-transporter homologue acts as a detoxifier in Plasmodium. Nat. Commun. 2016, 7, 10403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Xu, Y.-H.; Yi, H.-Y.; Gong, J.-M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Rietzschel, N.; Pierik, A.J.; Bill, E.; Lill, R.; Mühlenhoff, U. The Basic Leucine Zipper Stress Response Regulator Yap5 Senses High-Iron Conditions by Coordination of [2Fe-2S] Clusters. Mol. Cell. Biol. 2015, 35, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Bagley, D.; Ward, D.M.; Kaplan, J. Yap5 Is an Iron-Responsive Transcriptional Activator That Regulates Vacuolar Iron Storage in Yeast. Mol. Cell. Biol. 2008, 28, 1326–1337. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Miao, R.; Bertram, S.; Jia, X.; Ward, D.M.; Kaplan, J. A Role for Iron-Sulfur Clusters in the Regulation of Transcription Factor Yap5-dependent High Iron Transcriptional Responses in Yeast. J. Biol. Chem. 2012, 287, 35709–35721. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, C.; Vicente, C.; Menezes, R.; Caetano, S.; Carreto, L.; Rodrigues-Pousada, C. The Role of the Yap5 Transcription Factor in Remodeling Gene Expression in Response to Fe Bioavailability. PLoS ONE 2012, 7, e37434. [Google Scholar] [CrossRef]
- Li, L.; Kaplan, J.; Ward, D.M. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J. Biol. Chem. 2017, 292, 15577–15586. [Google Scholar] [CrossRef] [Green Version]
- Guldener, U.; Heck, S.; Fielder, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucl. Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [Green Version]
- Menezes, R.; Pimentel, C.; Silva, A.R.C.; Amaral, C.; Merhej, J.; Devaux, F.; Rodrigues-Pousada, C. Mediator, SWI/SNF and SAGA complexes regulate Yap8-dependent transcriptional activation of ACR2 in response to arsenate. Biochim. Biophys. Acta Bioenerg. 2017, 1860, 472–481. [Google Scholar] [CrossRef]
- Morawe, C.B.R.; Cloetens, P.; Lantelme, B.; Peffen, J.-C.; Vivo, A. Graded multilayers for figured Kirkpatrick-Baez mirrors on the new ESRF end station ID16A. In Proceedings of the Advances in X-ray/Euv Optics and Components, San Diego, CA, USA, 26 August 2015. [Google Scholar] [CrossRef]
- Solé, V.; Papillon, E.; Cotte, M.; Walter, P.; Susini, J. A multiplatform code for the analysis of energy-dispersive X-ray fluorescence spectra. Spectrochim. Acta Part B Spectrosc. 2007, 62, 63–68. [Google Scholar] [CrossRef]
- Teixeira, M.; Monteiro, P.T.; Palma, M.; Costa, C.; Godinho, C.P.; Pais, P.; Cavalheiro, M.; Antunes, M.; Lemos, A.; Pedreira, T.; et al. YEASTRACT: An upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, D348–D353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Mizianty, M.J.; Filipow, P.L.; Uversky, V.N.; Kurgan, L. RAPID: Fast and accurate sequence-based prediction of intrinsic disorder content on proteomic scale. Biochim. Biophys. Acta Proteins Proteom. 2013, 1834, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Roudeau, S.; Carmona, A.; Perrin, L.; Ortega, R. Correlative organelle fluorescence microscopy and synchrotron X-ray chemical element imaging in single cells. Anal. Bioanal. Chem. 2014, 406, 6979–6991. [Google Scholar] [CrossRef]
- Simm, C.; Lahner, B.; Salt, D.; LeFurgey, A.; Ingram, P.; Yandell, B.; Eide, D.J. Saccharomyces cerevisiae Vacuole in Zinc Storage and Intracellular Zinc Distribution. Eukaryot. Cell 2007, 6, 1166–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redden, H.; Alper, H.S. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015, 6, 7810. [Google Scholar] [CrossRef] [Green Version]
- Fournier, C.T.; Cherny, J.J.; Truncali, K.; Robbins-Pianka, A.; Lin, M.S.; Krizanc, D.; Weir, M.P. Amino Termini of Many Yeast Proteins Map to Downstream Start Codons. J. Proteome Res. 2012, 11, 5712–5719. [Google Scholar] [CrossRef] [Green Version]
- Lycette, B.E.; Glickman, J.W.; Roth, S.J.; Cram, A.E.; Kim, T.H.; Krizanc, D.; Weir, M.P. N-Terminal Peptide Detection with Optimized Peptide-Spectrum Matching and Streamlined Sequence Libraries. J. Proteome Res. 2016, 15, 2891–2899. [Google Scholar] [CrossRef]
- Wei, W.; Hennig, B.P.; Wang, J.; Zhang, Y.; Piazza, I.; Sanchez, Y.P.; Chabbert, C.D.; Adjalley, S.H.; Steinmetz, L.M.; Pelechano, V. Chromatin-sensitive cryptic promoters putatively drive expression of alternative protein isoforms in yeast. Genome Res. 2019, 29, 1974–1984. [Google Scholar] [CrossRef]
- Arribere, J.A.; Gilbert, W.V. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013, 23, 977–987. [Google Scholar] [CrossRef] [Green Version]
- Beltzer, J.P.; Chang, L.F.; E Hinkkanen, A.; Kohlhaw, G.B. Structure of yeast LEUThe 5’ flanking region contains features that predict two modes of control and two productive translation starts. J. Biol. Chem. 1986, 261, 5160–5167. [Google Scholar] [CrossRef]
- Carlson, M.; Botstein, D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell 1982, 28, 145–154. [Google Scholar] [CrossRef]
- Chatton, B.; Walter, P.; Ebel, J.P.; Lacroute, F.; Fasiolo, F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J. Biol. Chem. 1988, 263, 52–57. [Google Scholar] [CrossRef]
- Chiu, M.I.; Mason, T.L.; Fink, G.R. Hts1 Encodes Both the Cytoplasmic and Mitochondrial Histidyl-Trna Synthetase of Saccharomyces Cerevisiae: Mutations Alter the Specificity of Compartmentation. Genetics 1992, 132, 987–1001. [Google Scholar] [CrossRef]
- Ellis, S.R.; Hopper, A.K.; Martin, N.C. Amino-terminal extension generated from an upstream AUG codon is not required for mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferase. Proc. Natl. Acad. Sci. USA 1987, 84, 5172–5176. [Google Scholar] [CrossRef] [Green Version]
- Mikros, E.; Diallinas, G. Tales of tails in transporters. Open Biol. 2019, 9, 190083. [Google Scholar] [CrossRef] [Green Version]
- Gammie, A.E.; Stewart, B.G.; Scott, C.F.; Rose, M.D. The Two Forms of Karyogamy Transcription Factor Kar4p Are Regulated by Differential Initiation of Transcription, Translation, and Protein Turnover. Mol. Cell. Biol. 1999, 19, 817–825. [Google Scholar] [CrossRef] [Green Version]
- Gawron, D.; Ndah, E.; Gevaert, K.; Van Damme, P. Positional proteomics reveals differences in N-terminal proteoform stability. Mol. Syst. Biol. 2016, 12, 858. [Google Scholar] [CrossRef]
- Sorribes-Dauden, R.; Peris, D.; Martínez-Pastor, M.T.; Puig, S. Structure and function of the vacuolar Ccc1/VIT1 family of iron transporters and its regulation in fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3712–3722. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amaral, C.; Vicente, C.T.; Caetano, S.M.; Gaspar-Cordeiro, A.; Yang, Y.; Cloetens, P.; Romão, C.V.; Rodrigues-Pousada, C.; Pimentel, C. An Internal Promoter Drives the Expression of a Truncated Form of CCC1 Capable of Protecting Yeast from Iron Toxicity. Microorganisms 2021, 9, 1337. https://doi.org/10.3390/microorganisms9061337
Amaral C, Vicente CT, Caetano SM, Gaspar-Cordeiro A, Yang Y, Cloetens P, Romão CV, Rodrigues-Pousada C, Pimentel C. An Internal Promoter Drives the Expression of a Truncated Form of CCC1 Capable of Protecting Yeast from Iron Toxicity. Microorganisms. 2021; 9(6):1337. https://doi.org/10.3390/microorganisms9061337
Chicago/Turabian StyleAmaral, Catarina, Cristina Teixeira Vicente, Soraia Marques Caetano, Ana Gaspar-Cordeiro, Yang Yang, Peter Cloetens, Célia V. Romão, Claudina Rodrigues-Pousada, and Catarina Pimentel. 2021. "An Internal Promoter Drives the Expression of a Truncated Form of CCC1 Capable of Protecting Yeast from Iron Toxicity" Microorganisms 9, no. 6: 1337. https://doi.org/10.3390/microorganisms9061337
APA StyleAmaral, C., Vicente, C. T., Caetano, S. M., Gaspar-Cordeiro, A., Yang, Y., Cloetens, P., Romão, C. V., Rodrigues-Pousada, C., & Pimentel, C. (2021). An Internal Promoter Drives the Expression of a Truncated Form of CCC1 Capable of Protecting Yeast from Iron Toxicity. Microorganisms, 9(6), 1337. https://doi.org/10.3390/microorganisms9061337





