Bacterial Communities in Alkaline Saline Soils Amended with Young Maize Plants or Its (Hemi)Cellulose Fraction
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of the Maize and Neutral Detergent Extraction
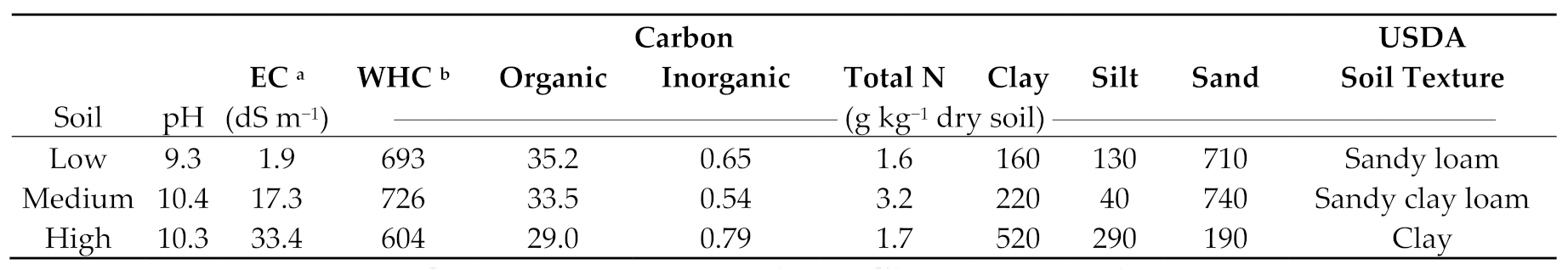
2.2. Soil Sampling
2.3. Experimental Design and Microcosms Setup
2.4. DNA Extraction and PCR Amplification of Bacterial 16S rRNA Genes
2.5. Analysis of Pyrosequencing Data
2.6. Statistical Analysis
3. Results
3.1. Carbon Mineralization
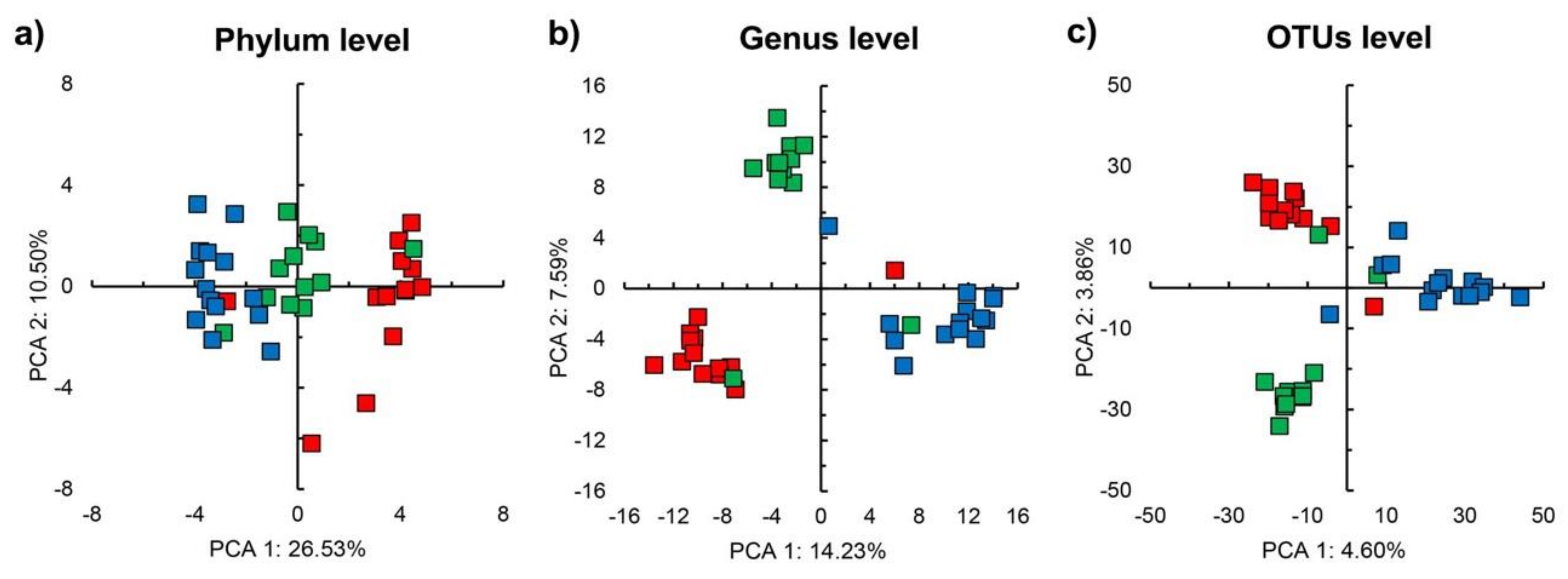
3.2. Sequencing Results and Bacterial Diversity
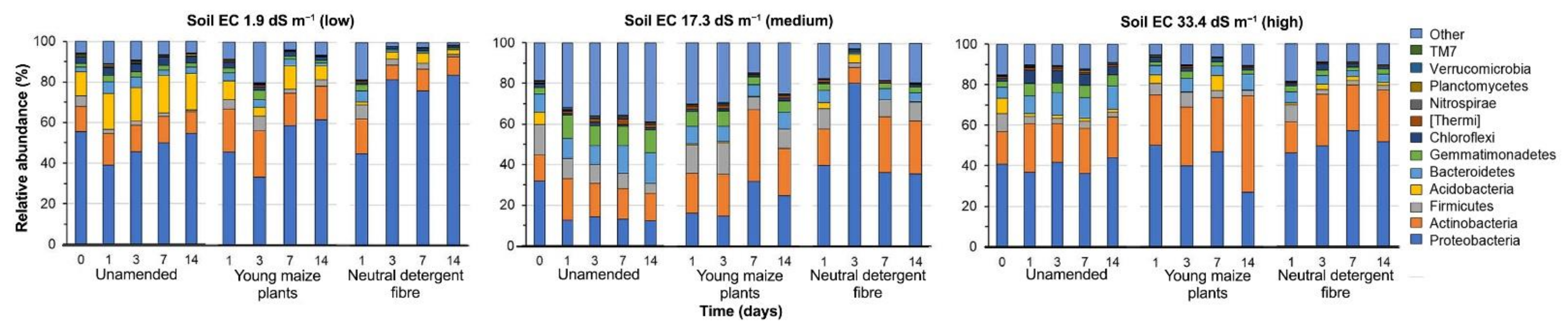
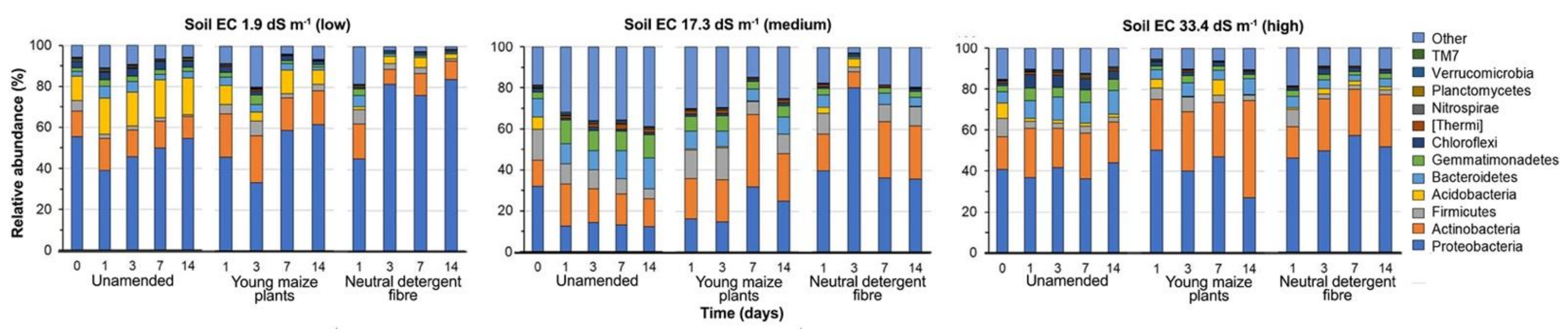
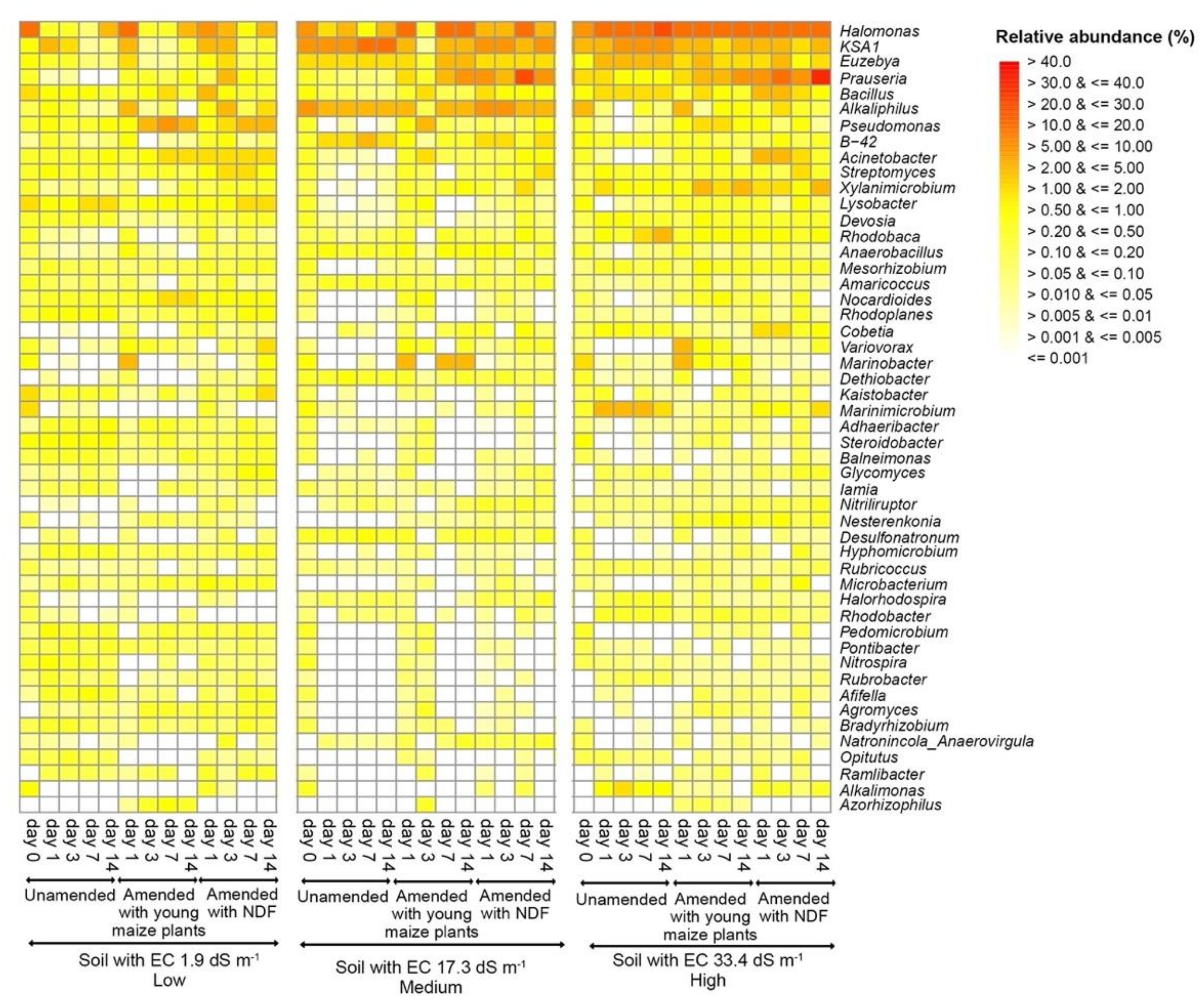
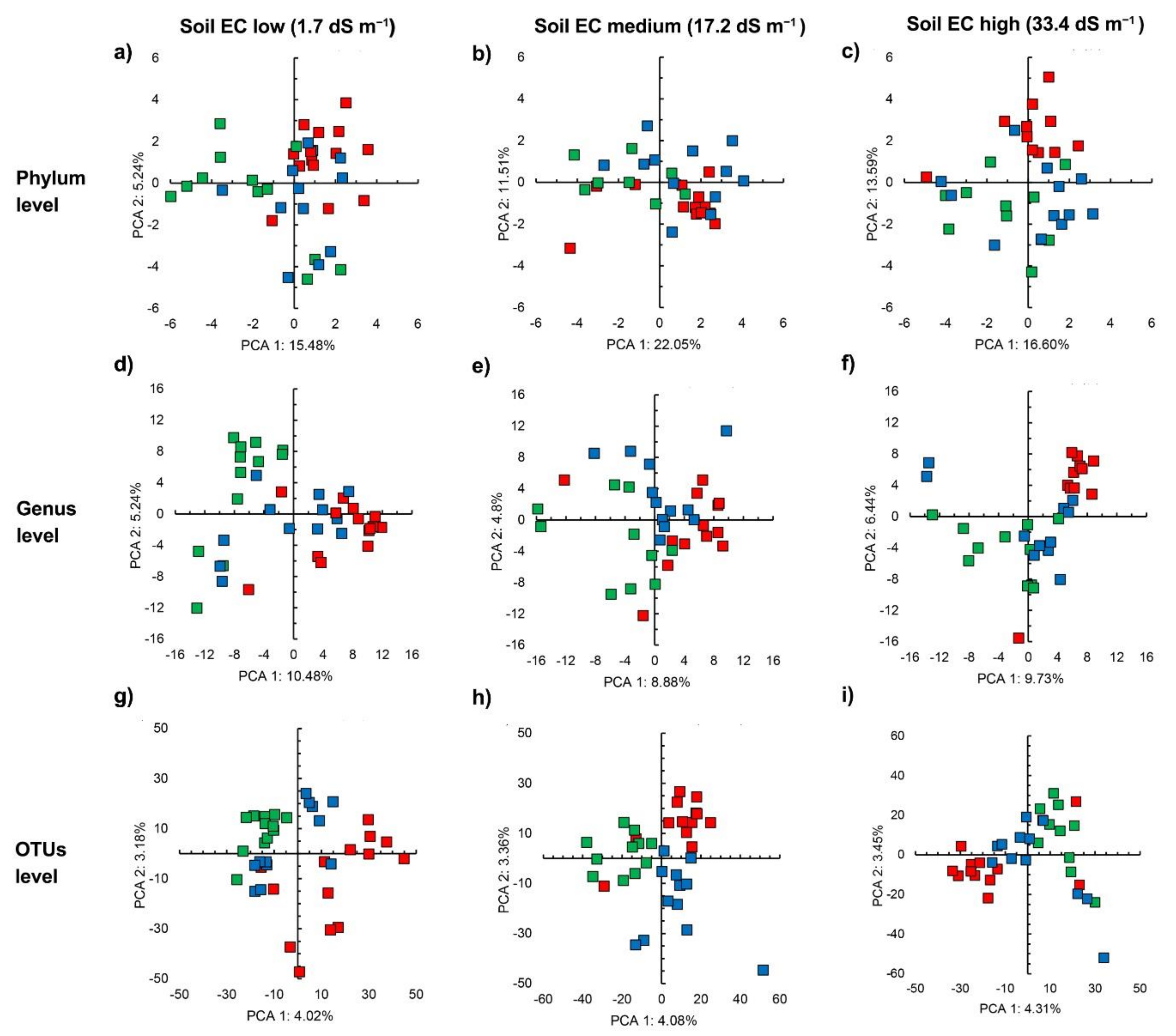
3.3. Bacterial Community Structure in the Unamended Soil
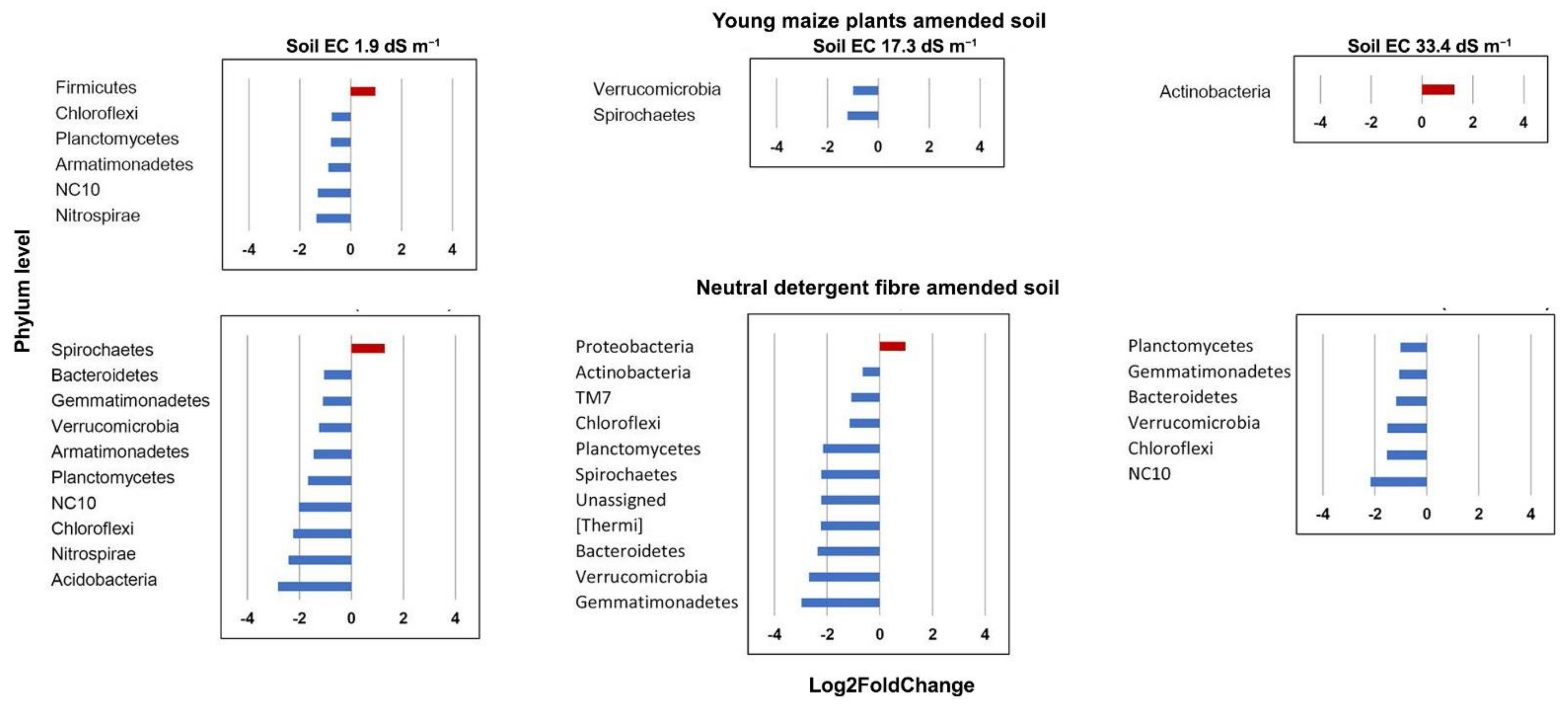
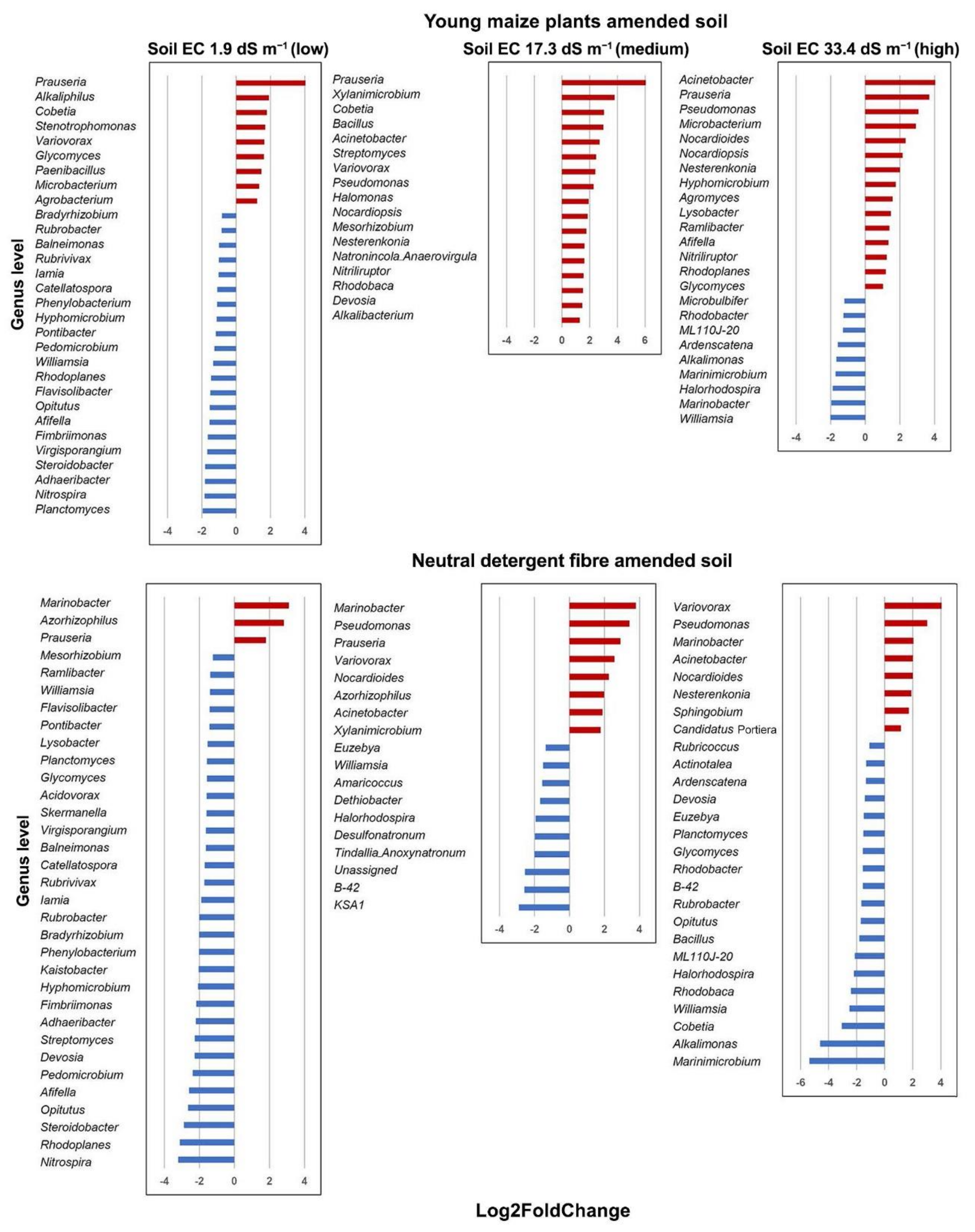
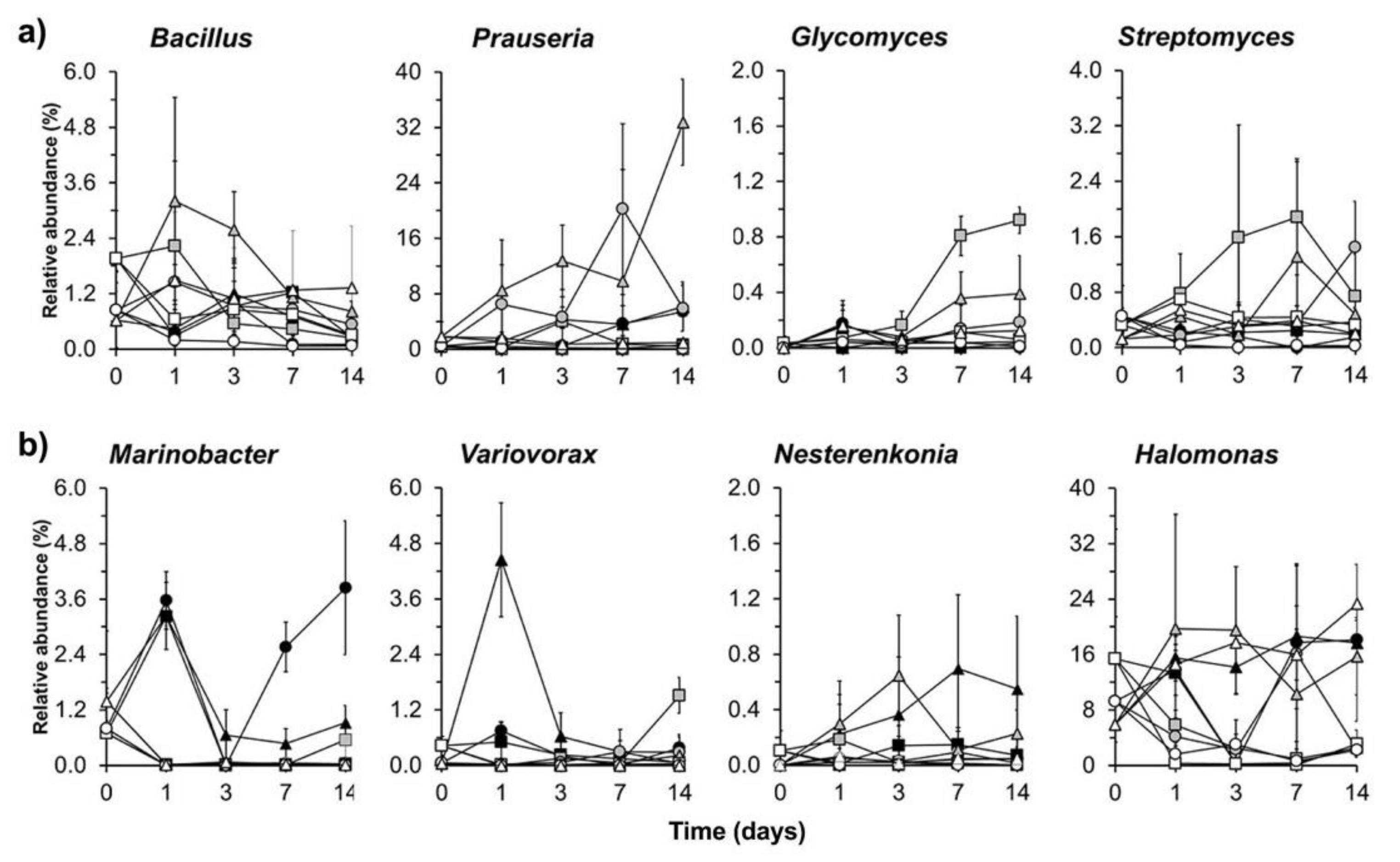
3.4. Bacterial Community Structure in the Organic Material Amended Soil
3.5. Bacterial Functional Prediction Analysis
4. Discussion
4.1. Bacterial Community in Unamended Soil with Varying pH and EC
4.2. Bacterial Community Structure in Organic Material Amended Soil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahid, S.A.; Zaman, M.; Heng, L. Soil salinity: Historical perspectives and a world overview of the problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Zaman, M., Shahid, S.A., Heng, L., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 43–53. ISBN 978-3-319-96190-3. [Google Scholar]
- Edelstein, M.; Plaut, Z.; Ben-Hur, M. Water salinity and sodicity effects on soil structure and hydraulic properties. Adv. Hortic. Sci. 2010, 24, 154–160. [Google Scholar]
- Bidalia, A.; Vikram, K.; Yamal, G.; Rao, K.S. Effect of salinity on soil nutrients and plant health. In Salt Stress, Microbes, and Plant Interactions: Causes and Solution; Akhtar, M.S., Ed.; Springer: Singapore, 2019; pp. 273–297. ISBN 978-981-13-8801-9. [Google Scholar]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and bacterial diversity: To what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef]
- Bui, E.N. Soil salinity: A neglected factor in plant ecology and biogeography. J. Arid Environ. 2013, 92, 14–25. [Google Scholar] [CrossRef]
- Dendooven, L.; Alcántara-Hernández, R.J.; Valenzuela-Encinas, C.; Luna-Guido, M.; Perez-Guevara, F.; Marsch, R. Dynamics of carbon and nitrogen in an extreme alkaline saline soil: A review. Soil Biol. Biochem. 2010, 42, 865–877. [Google Scholar] [CrossRef]
- Luna-Guido, M.L.; Vega-Estrada, J.; Ponce-Mendoza, A.; Hernandez-Hernandez, H.; Montes-Horcasitas, M.-C.; Vaca-Mier, M.; Dendooven, L. Mineralization of 14C-labelled maize in alkaline saline soils. Plant Soil 2003, 250, 29–38. [Google Scholar] [CrossRef]
- Ruiz-Romero, E.; Alcántara-Hernández, R.; Cruz-Mondragon, C.; Marsch, R.; Luna-Guido, M.L.; Dendooven, L. Denitrification in extreme alkaline saline soils of the former lake Texcoco. Plant Soil 2009, 319, 247–257. [Google Scholar] [CrossRef]
- Martínez-Olivas, M.A.; Jiménez-Bueno, N.G.; Hernández-García, J.A.; Fusaro, C.; Luna-Guido, M.; Navarro-Noya, Y.E.; Dendooven, L. Bacterial and archaeal spatial distribution and its environmental drivers in an extremely haloalkaline soil at the landscape scale. PeerJ 2019, 7, e6127. [Google Scholar] [CrossRef]
- Lopez-Ramirez, M.P.; Sanchez-Lopez, K.B.; Sarria-Guzman, Y.; Bello-Lopez, J.M.; Cano-Garcia, V.L.; Ruiz-Valdiviezo, V.M.; Dendooven, L. Haloalkalophilic cellulose-degrading bacteria isolated from an alkaline saline soil. J. Pure Appl. Microbiol. 2015, 9, 2879–2886. [Google Scholar]
- De León-Lorenzana, A.S.; Delgado-Balbuena, L.; Domínguez-Mendoza, C.A.; Navarro-Noya, Y.E.; Luna-Guido, M.; Dendooven, L. Soil salinity controls relative abundance of specific bacterial groups involved in the decomposition of maize plant residues. Front. Ecol. Evol. 2018, 6, 51. [Google Scholar] [CrossRef]
- Ramirez-Villanueva, D.A.; Bello-López, J.M.; Navarro-Noya, Y.E.; Luna-Guido, M.; Verhulst, N.; Govaerts, B.; Dendooven, L. Bacterial community structure in maize residue amended soil with contrasting management practices. Appl. Soil Ecol. 2015, 90, 49–59. [Google Scholar] [CrossRef]
- Steiner, A.A. A universal method for preparing nutrient solutions of a certain desired composition. Plant Soil 1961, 15, 134–154. [Google Scholar] [CrossRef]
- Van Soest, P.J. Use of detergents in the analysis of fibrous feeds. II. A rapid method for the determination of fiber and lignin. J. Assoc. Off. Agric. Chem. 1963, 46, 829–835. [Google Scholar]
- Van Soest, P.J.; Wine, R.H. Use of detergents in the analysis of fibrous feeds. IV. Determination of plant cell-wall constituents. J. AOAC Int. 1967, 50, 50–55. [Google Scholar] [CrossRef]
- Heffner, R.A.; Butler, M.J.; Reilly, C.K. Pseudoreplication revisited. Ecology 1996, 77, 2558–2562. [Google Scholar] [CrossRef]
- Gee, G.W.; Bauder, J.W. Particle-size Analysis. In Methods of Soil Analysis: Part 1—Physical and Mineralogical Methods; Klute, A., Ed.; SSSA Book Series SV—5.1; Soil Science Society of America: Madison, WI, USA; American Society of Agronomy: Madison, WI, USA, 1972; pp. 383–411. ISBN 978-0-89118-864-3. [Google Scholar]
- Bundy, L.G.; Bremner, J.M. A simple titrimetric method for determination of inorganic carbon in soils. Soil Sci. Soc. Am. J. 1972, 36, 273–275. [Google Scholar] [CrossRef]
- Bremner, J.M. Nitrogen-Total. In Methods of Soil Analysis Part 3—Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; SSSA Book Series SV—5.3; Soil Science Society of America: Madison, WI, USA; American Society of Agronomy: Madison, WI, USA, 1996; pp. 1085–1121. ISBN 978-0-89118-866-7. [Google Scholar]
- Rhoades, J.D. Salinity: Electrical Conductivity and Total Dissolved Solids. In Methods of Soil Analysis Part 3—Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; SSSA Book Series SV—5.3; Soil Science Society of America: Madison, WI, USA; American Society of Agronomy: Madison, WI, USA, 1996; pp. 417–435. ISBN 978-0-89118-866-7. [Google Scholar]
- Thomas, G.W. Soil pH and Soil Acidity. In Methods of Soil Analysis Part 3—Chemical Methods; Sparks, D.L., Page, A.L., Helmke, P.A., Loeppert, R.H., Soltanpour, P.N., Tabatabai, M.A., Johnston, C.T., Sumner, M.E., Eds.; SSSA Book Series SV—5.3; Soil Science Society of America: Madison, WI, USA; American Society of Agronomy: Madison, WI, USA, 1996; pp. 475–490. ISBN 978-0-89118-866-7. [Google Scholar]
- Inubushi, K.; Brookes, P.C.; Jenkinson, D.S. Soil microbial biomass C, N and ninhydrin-N in aerobic and anaerobic soils measured by the fumigation-extraction method. Soil Biol. Biochem. 1991, 23, 737–741. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Rivera, F.N.; Patiño-Zúñiga, L.; Govaerts, B.; Marsch, R.; Vila-Sanjurjo, A.; Dendooven, L. Molecular characterization of soil bacterial communities in contrasting zero tillage systems. Plant Soil 2010, 329, 127–137. [Google Scholar] [CrossRef]
- Valenzuela-Encinas, C.; Neria-González, I.; Alcántara-Hernández, R.J.; Enríquez-Aragón, J.A.; Estrada-Alvarado, I.; Hernández-Rodríguez, C.; Dendooven, L.; Marsch, R. Phylogenetic analysis of the archaeal community in an alkaline-saline soil of the former lake Texcoco (Mexico). Extremophiles 2008, 12, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. Plasmid and their usefulness in molecular cloning. In Molecular Cloning, a Laboratory Manual; Cold Spring Harbor Laboratory: New York, NY, USA, 2001; Volume 1, pp. 1–31. ISBN 978-0879695774. [Google Scholar]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformaion of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Navarro-Noya, Y.E.; Suárez-Arriaga, M.C.; Rojas-Valdes, A.; Montoya-Ciriaco, N.M.; Gómez-Acata, S.; Fernández-Luqueño, F.; Dendooven, L. Pyrosequencing analysis of the bacterial community in drinking water wells. Microb. Ecol. 2013, 66, 19–29. [Google Scholar] [CrossRef]
- Acinas, S.G.; Klepac-Ceraj, V.; Hunt, D.E.; Pharino, C.; Ceraj, I.; Distel, D.L.; Polz, M.F. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 2004, 430, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, S.J.; Yu, C.; Claypool, J.T.; Singer, S.W.; Simmons, B.A.; Thelen, M.P.; Simmons, C.W.; VanderGheynst, J.S. Development and characterization of a thermophilic, lignin degrading microbiota. Process Biochem. 2017, 63, 193–203. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Ma, Z.; Li, L. Measuring metagenome diversity and similarity with Hill numbers. Mol. Ecol. Resour. 2018, 18, 1339–1355. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing (Versión 4.0.2); R Foundation for Statistical Computing: Vienna, Austria, 2000; Available online: https//www.R-project.org (accessed on 30 July 2020).
- Kolde, R. Pheatmap: Pretty Heatmaps. 2018. Available online: https//CRAN.R-project.org/package=pheatmap (accessed on 30 July 2020).
- Gloor, G.; Fernandes, A.; Macklaim, J.; Albert, A.; Links, M.; Quinn, T.; Wu, J.R.; Wong, R.G.; Lieng, B. ALDEx2: Analysis of Differential Abundance Taking Sample Variation into Account; Bioconductor: Seattle, WA, USA, 2020. [Google Scholar]
- Husson, F.; Josse, J.; Le, S.; Mazet, J.; Husson, M.F. Package ‘FactoMineR’. 2020. Available online: https://CRAN.R-project.org/package=FactoMineR (accessed on 30 July 2020).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘Vegan.’ Community Ecol. Package. Version 2.5; 2020; pp. 1–295. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yan, N.; Marschner, P.; Cao, W.; Zuo, C.; Qin, W. Influence of salinity and water content on soil microorganisms. Int. Soil Water Conserv. Res. 2015, 3, 316–323. [Google Scholar] [CrossRef]
- Rengasamy, P. Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: An overview. Aust. J. Exp. Agric. 2002, 42, 351–361. [Google Scholar] [CrossRef]
- Sparks, D.L. (Ed.) The chemistry of saline and sodic soils. In Environmental Soil Chemistry, 2nd ed.; Academic Press: Burlington, VT, USA, 2003; pp. 285–300. ISBN 978-0-12-656446-4. [Google Scholar]
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Fageria, N.K. Role of soil organic matter in maintaining sustainability of cropping systems. Commun. Soil Sci. Plant Anal. 2012, 43, 2063–2113. [Google Scholar] [CrossRef]
- Leogrande, R.; Vitti, C. Use of organic amendments to reclaim saline and sodic soils: A review. Arid Land Res. Manag. 2019, 33, 1–21. [Google Scholar] [CrossRef]
- Wang, L.; Sun, X.; Li, S.; Zhang, T.; Zhang, W.; Zhai, P. Application of organic amendments to a coastal saline soil in North China: Effects on soil physical and chemical properties and tree growth. PLoS ONE 2014, 9, e89185. [Google Scholar]
- Lakhdar, A.; Rabhi, M.; Ghnaya, T.; Montemurro, F.; Jedidi, N.; Abdelly, C. Effectiveness of compost use in salt-affected soil. J. Hazard. Mater. 2009, 171, 29–37. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, T.; Liu, K.S.; Wang, L.; Wang, K.; Zhou, Y. Effects of different amendments for the reclamation of coastal saline soil on soil nutrient dynamics and electrical conductivity responses. Agric. Water Manag. 2015, 159, 115–122. [Google Scholar] [CrossRef]
- Keshri, J.; Mishra, A.; Jha, B. Microbial population index and community structure in saline–alkaline soil using gene targeted metagenomics. Microbiol. Res. 2013, 168, 165–173. [Google Scholar] [CrossRef]
- Mhete, M.; Eze, P.N.; Rahube, T.O.; Akinyemi, F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Sci. Afr. 2020, 7, e00246. [Google Scholar] [CrossRef]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2019, 13, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, J.-J.; Banerjee, S.; Zhou, N.; Zhao, Z.-Y.; Zhang, K.; Tian, C.-Y. Soil pH is equally important as salinity in shaping bacterial communities in saline soils under halophytic vegetation. Sci. Rep. 2018, 8, 4550. [Google Scholar] [CrossRef] [PubMed]
- Begmatov, S.A.; Selitskaya, O.V.; Vasileva, L.V.; Berestovskaja, Y.Y.; Manucharova, N.A.; Drenova, N. V Morphophysiological features of some cultivable bacteria from saline soils of the aral sea region. Eurasian Soil Sci. 2020, 53, 90–96. [Google Scholar] [CrossRef]
- Kearl, J.; McNary, C.; Lowman, J.S.; Mei, C.; Aanderud, Z.T.; Smith, S.T.; West, J.; Colton, E.; Hamson, M.; Nielsen, B.L. Salt-tolerant halophyte rhizosphere bacteria stimulate growth of alfalfa in salty soil. Front. Microbiol. 2019, 10, 1849. [Google Scholar] [CrossRef]
- Oueriaghli, N.; González-Domenech, C.M.; Martínez-Checa, F.; Muyzer, G.; Ventosa, A.; Quesada, E.; Béjar, V. Diversity and distribution of Halomonas in Rambla Salada, a hypersaline environment in the southeast of Spain. FEMS Microbiol. Ecol. 2014, 87, 460–474. [Google Scholar] [CrossRef]
- Bull, A.T. Extremophiles Handbook; Horikoshi, K., Ed.; Springer: Tokyo, Japan, 2011; pp. 1203–1240. ISBN 978-4-431-53898-1. [Google Scholar]
- Gohel, S.D.; Singh, S.P. Molecular Phylogeny and diversity of the salt-tolerant alkaliphilic Actinobacteria inhabiting coastal Gujarat, India. Geomicrobiol. J. 2018, 35, 775–789. [Google Scholar] [CrossRef]
- Ibarra-Sánchez, C.L.; Prince, L.; Aguirre-Noyola, J.L.; Sánchez-Cerda, K.E.; Navaro-Noya, Y.E.; Luna-Guido, M.; Conde-Barajas, E.; Dendooven, L.; Gomez-Acata, E.S. The microbial community in an alkaline saline sediment of a former maar lake bed. J. Soils Sediments 2020, 20, 542–555. [Google Scholar] [CrossRef]
- Osman, K.T. Physical Properties of Soil. In Soils Principles, Properties and Management; Osman, K.T., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 49–65. ISBN 978-94-007-5663-2. [Google Scholar]
- Wolf, A.B.; Vos, M.; de Boer, W.; Kowalchuk, G.A. Impact of matric potential and pore size distribution on growth dynamics of filamentous and non-filamentous soil Bacteria. PLoS ONE 2014, 8, e83661. [Google Scholar] [CrossRef]
- Osman, K.T. Soil Organic Matter. In Soils Principles, Properties and Management; Osman, K.T., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 89–96. ISBN 978-94-007-5663-2. [Google Scholar]
- Schostag, M.; Priemé, A.; Jacquiod, S.; Russel, J.; Ekelund, F.; Jacobsen, C.S. Bacterial and protozoan dynamics upon thawing and freezing of an active layer permafrost soil. ISME J. 2019, 13, 1345–1359. [Google Scholar] [CrossRef]
- Vanlauwe, B.; Dendooven, L.; Merckx, R. Residue fractionation and decomposition: The significance of the active fraction. Plant Soil 1994, 158, 263–274. [Google Scholar] [CrossRef]
- Silva, M.S.; Sales, A.N.; Magalhães-Guedes, K.T.; Dias, D.R.; Schwan, R.F. Brazilian Cerrado soil Actinobacteria ecology. Biomed Res. Int. 2013, 2013, 503805. [Google Scholar] [CrossRef]
- Drzyzga, O. The strengths and weaknesses of Gordonia: A review of an emerging genus with increasing biotechnological potential. Crit. Rev. Microbiol. 2012, 38, 300–316. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, G.; Bhalla, A.; Adhikari, A.; Bischoff, K.M.; Hughes, S.R.; Christopher, L.P.; Sani, R.K. Characterization of thermostable cellulases produced by Bacillus and Geobacillus strains. Bioresour. Technol. 2010, 101, 8798–8806. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef]
- Guan, T.-W.; Lin, Y.-J.; Ou, M.-Y.; Chen, K.-B. Isolation and diversity of sediment bacteria in the hypersaline aiding lake, China. PLoS ONE 2020, 15, e0236006. [Google Scholar]
- Wongwilaiwalin, S.; Rattanachomsri, U.; Laothanachareon, T.; Eurwilaichitr, L.; Igarashi, Y.; Champreda, V. Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzyme Microb. Technol. 2010, 47, 283–290. [Google Scholar] [CrossRef]
- Albdaiwi, R.N.; Khyami-Horani, H.; Ayad, J.Y.; Alananbeh, K.M.; Al-Sayaydeh, R. Isolation and characterization of halotolerant plant growth promoting rhizobacteria from durum wheat (Triticum turgidum subsp. durum) cultivated in saline areas of the Dead Sea region. Front. Microbiol. 2019, 10, 1639. [Google Scholar] [CrossRef]
- Han, J.-I.; Choi, H.-K.; Lee, S.-W.; Orwin, P.M.; Kim, J.; LaRoe, S.L.; Kim, T.-G.; O’Neil, J.; Leadbetter, J.R.; Hur, C.-G.; et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J. Bacteriol. 2011, 193, 1183–1190. [Google Scholar] [CrossRef]
- Orhan, F.; Demirci, A. salt stress mitigating potential of halotolerant/halophilic plant growth promoting. Geomicrobiol. J. 2020, 37, 663–669. [Google Scholar] [CrossRef]
- Cortes-Tolalpa, L.; Norder, J.; van Elsas, J.D.; Falcao Salles, J. Halotolerant microbial consortia able to degrade highly recalcitrant plant biomass substrate. Appl. Microbiol. Biotechnol. 2018, 102, 2913–2927. [Google Scholar] [CrossRef]
- Shivanand, P.; Mugeraya, G.; Kumar, A. Utilization of renewable agricultural residues for the production of extracellular halostable cellulase from newly isolated Halomonas sp. strain PS47. Ann. Microbiol. 2013, 63, 1257–1263. [Google Scholar] [CrossRef]
- De León-Lorenzana, A.S.; Delgado-Balbuena, L.; Domínguez-Mendoza, C.; Navarro-Noya, Y.E.; Luna-Guido, M.; Dendooven, L. Reducing salinity by flooding an extremely alkaline and saline soil changes the Bacterial community but its effect on the Archaeal community is limited. Front. Microbiol. 2017, 8, 466. [Google Scholar] [CrossRef] [PubMed]
- Soto-Padilla, M.Y.; Valenzuela-Encinas, C.; Dendooven, L.; Marsch, R.; Gortáres-Moroyoqui, P.; Estrada-Alvarado, M.I. Isolation and phylogenic identification of soil haloalkaliphilic strains in the former Texcoco Lake. Int. J. Environ. Health Res. 2014, 24, 82–90. [Google Scholar] [CrossRef]
- Green, P.N. Methylobacterium. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 257–265. ISBN 978-0-387-30745-9. [Google Scholar]
- Wang, L.; Nie, Y.; Tang, Y.-Q.; Song, X.-M.; Cao, K.; Sun, L.-Z.; Wang, Z.-J.; Wu, X.-L. Diverse bacteria with lignin degrading potentials isolated from two ranks of coal. Front. Microbiol. 2016, 7, 1428. [Google Scholar] [CrossRef]
- Zhang, X.; Yao, Q.; Cai, Z.; Xie, X.; Zhu, H. Isolation and identification of myxobacteria from saline-alkaline soils in Xinjiang, China. PLoS ONE 2013, 8, e70466. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.N.; Porto, A.L.M. Chapter eight—Recent advances in marine enzymes for biotechnological processes. In Marine Enzymes Biotechnology: Production and Industrial Applications, Part I—Production of Enzymes; Kim, S.-K., Toldrá, F.B.T.-A., Eds.; Academic Press: Cambridge, MA, USA, 2016; Volume 78, pp. 153–192. ISBN 1043-4526. [Google Scholar]
- Puentes-Téllez, P.E.; Salles, J.F. Dynamics of abundant and rare bacteria during degradation of lignocellulose from sugarcane biomass. Microb. Ecol. 2020, 79, 312–325. [Google Scholar] [CrossRef]
- Valenzuela-Encinas, C.; Neria-González, I.; Alcántara-Hernández, R.J.; Estrada-Alvarado, I.; Zavala-Díaz de la Serna, F.J.; Dendooven, L.; Marsch, R. Changes in the bacterial populations of the highly alkaline saline soil of the former lake Texcoco (Mexico) following flooding. Extremophiles 2009, 13, 609–621. [Google Scholar] [CrossRef]
- Ma, B.; Gong, J. A meta-analysis of the publicly available bacterial and archaeal sequence diversity in saline soils. World J. Microbiol. Biotechnol. 2013, 29, 2325–2334. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
 |
| Comparison | F Value | p Value a |
|---|---|---|
| Phylum | ||
| Soil | 4.071 | <0.001 |
| Time | 1.503 | 0.080 |
| Interaction | 1.143 | 0.273 |
| All taxonomic groups assigned to the taxonomic level of genus | ||
| Soil | 3.865 | <0.001 |
| Time | 0.996 | 0.396 |
| Interaction | 1.059 | 0.284 |
| All operational taxonomic units (OTUs) | ||
| Soil | 1.577 | <0.001 |
| Time | 1.003 | 0.364 |
| Interaction | 0.992 | 0.579 |
| Soil EC 1.9 dS m−1 | Soil EC 17.2 dS m−1 | Soil EC 33.4 dS m−1 | ||||
|---|---|---|---|---|---|---|
| Comparison | F Value | p Value a | F Value | p Value | F Value | p Value |
| Phyla | ||||||
| Treatment | 2.070 | <0.001 | 1.551 | <0.019 | 1.848 | <0.001 |
| Time | 1.003 | 0.434 | 1.261 | 0.197 | 1.242 | 0.174 |
| Interaction | 1.028 | 0.412 | 1.446 | 0.065 | 1.062 | 0.341 |
| All Taxonomic Groups Assigned up to the Level of Genus | ||||||
| Treatment | 1.829 | <0.001 | 1.560 | <0.001 | 1.556 | <0.001 |
| Time | 1.148 | <0.018 | 1.185 | <0.004 | 1.191 | <0.004 |
| Interaction | 1.101 | <0.031 | 1.135 | <0.004 | 0.968 | 0.723 |
| All Operational Taxonomic Units (OTUs) | ||||||
| Treatment | 1.179 | <0.001 | 1.184 | <0.001 | 1.196 | <0.001 |
| Time | 1.021 | 0.171 | 1.025 | 0.146 | 0.989 | 0.621 |
| Interaction | 0.995 | 0.568 | 1.023 | 0.102 | 1.015 | 0.219 |
| Soil EC 1.9 dS m−1 | Soil EC 17.3 dS m−1 | Soil EC 33.4 dS m−1 | |||
|---|---|---|---|---|---|
| Phyla | |||||
| Taxonomic group | p Value a | Taxonomic group | p Value | Taxonomic group | p Value |
| Acidobacteria | 0.0076 | Verrucomicrobia | 0.0062 | Actinobacteria | 0.0365 |
| Chloroflexi | 0.0105 | Gemmatimonadetes | 0.0039 | ||
| Nitrospirae | 0.0140 | Proteobacteria | 0.0073 | ||
| Proteobacteria | 0.0199 | Bacteroidetes | 0.0089 | ||
| NC10 | 0.0424 | Planctomycetes | 0.0190 | ||
| Genera | |||||
| Pseudomonas | 0.0025 | Prauseria | 0.0006 | Marinobacter | 0.0133 |
| Azorhizophilus | 0.0026 | KSA1 | 0.0059 | Acinetobacter | 0.0154 |
| Rhodoplanes | 0.0037 | Streptomyces | 0.0200 | Marinimicrobium | 0.0195 |
| Nitrospira | 0.0063 | Bacillus | 0.0279 | Bacillus | 0.0303 |
| Opitutus | 0.0099 | B-42 | 0.0299 | Alkalimonas | 0.0317 |
| Steroidobacter | 0.0102 | Cobetia | 0.0351 | ||
| Acinetobacter | 0.0125 | Variovorax | 0.0405 | ||
| Glycomyces | 0.0132 | ||||
| Streptomyces | 0.0136 | ||||
| Prauseria | 0.0196 | ||||
| Devosia | 0.0367 | ||||
| Nocardioides | 0.0400 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Hernández, V.; Hernández-Guzmán, M.; Luna-Guido, M.; Navarro-Noya, Y.E.; Romero-Tepal, E.M.; Dendooven, L. Bacterial Communities in Alkaline Saline Soils Amended with Young Maize Plants or Its (Hemi)Cellulose Fraction. Microorganisms 2021, 9, 1297. https://doi.org/10.3390/microorganisms9061297
Pérez-Hernández V, Hernández-Guzmán M, Luna-Guido M, Navarro-Noya YE, Romero-Tepal EM, Dendooven L. Bacterial Communities in Alkaline Saline Soils Amended with Young Maize Plants or Its (Hemi)Cellulose Fraction. Microorganisms. 2021; 9(6):1297. https://doi.org/10.3390/microorganisms9061297
Chicago/Turabian StylePérez-Hernández, Valentín, Mario Hernández-Guzmán, Marco Luna-Guido, Yendi E. Navarro-Noya, Elda M. Romero-Tepal, and Luc Dendooven. 2021. "Bacterial Communities in Alkaline Saline Soils Amended with Young Maize Plants or Its (Hemi)Cellulose Fraction" Microorganisms 9, no. 6: 1297. https://doi.org/10.3390/microorganisms9061297
APA StylePérez-Hernández, V., Hernández-Guzmán, M., Luna-Guido, M., Navarro-Noya, Y. E., Romero-Tepal, E. M., & Dendooven, L. (2021). Bacterial Communities in Alkaline Saline Soils Amended with Young Maize Plants or Its (Hemi)Cellulose Fraction. Microorganisms, 9(6), 1297. https://doi.org/10.3390/microorganisms9061297






