Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors
Abstract
1. Introduction
- Why are a high percentage of pathogen moonlighting proteins involved in virulence?
- Why do most of the canonical functions of these moonlighting proteins belong to primary metabolism? Moreover, why are they common in many pathogen species?
- How are these different protein sequences and structures able to bind the same set of host ECM protein targets, mainly plasminogen (PLG), and colonize host tissues?
2. Materials and Methods
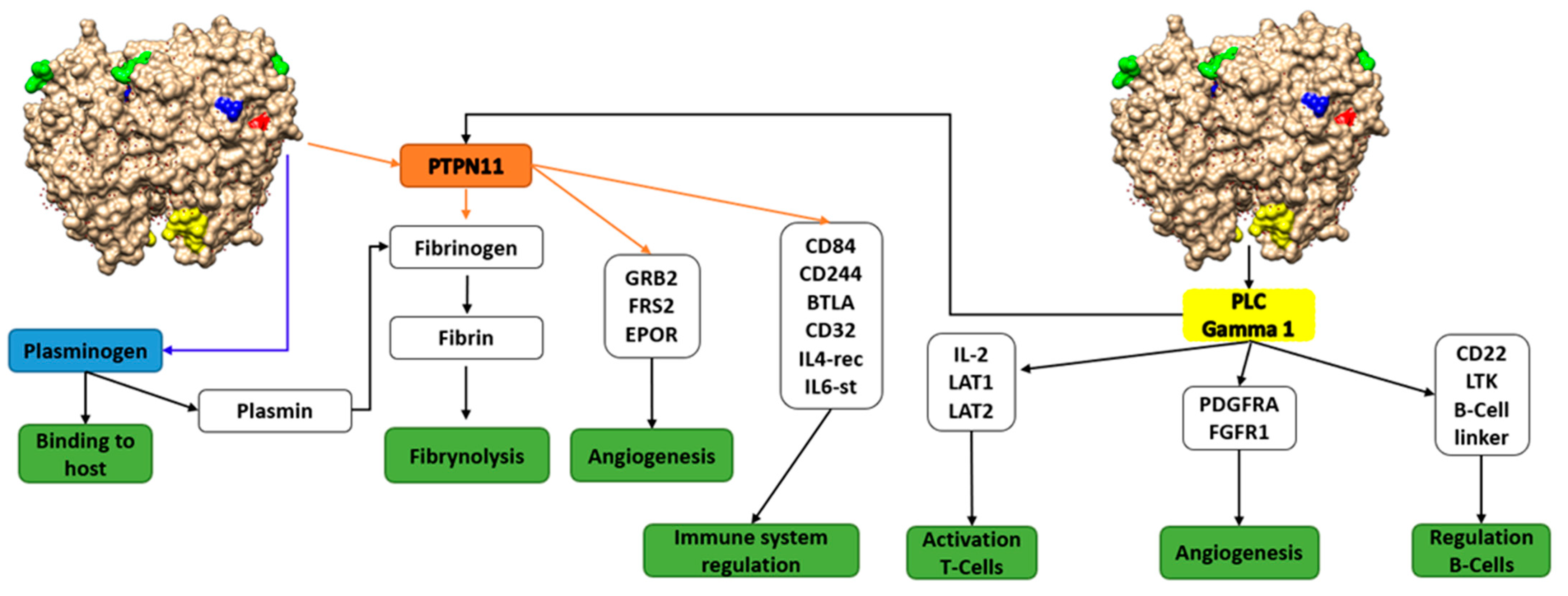
3. Results and Discussion
3.1. Moonlighting Proteins and Virulence: Questions 1 and 2
3.2. Putative Involvement of Moonlighting Proteins in the Interaction with ECM Proteins, Tissue Colonization, and Metastasis: Question 3
3.3. Tissue Remodeling Strongly Suggests a Parallelism between Microbial Tissue Colonization and Cancer Metastasis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PLG | Plasminogen |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| ECM | Extracellular matrix |
| GO | Gene ontology |
| PLC Gamma1 | Phospholipase C-gamma 1 |
| PTPN11 | Protein tyrosine phosphatase non-receptor type 11 |
References
- Huberts, D.H.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta 2010, 1803, 520–525. [Google Scholar] [CrossRef]
- Copley, S.D. Moonlighting is mainstream: Paradigm adjustment required. Bioessays 2012, 34, 578–588. [Google Scholar] [CrossRef]
- Jeffery, C.J. An introduction to protein moonlighting. Biochem. Soc. Trans. 2014, 42, 1679–1683. [Google Scholar] [CrossRef]
- Flores, C.L.; Gancedo, C. Unraveling moonlighting functions with yeasts. IUBMB Life 2011, 63, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Espinosa-Cantu, A.; Cruz-Bonilla, E.; Noda-Garcia, L.; DeLuna, A. Multiple Forms of Multifunctional Proteins in Health and Disease. Front. Cell Dev. Biol. 2020, 8, 451. [Google Scholar] [CrossRef] [PubMed]
- Franco-Serrano, L.; Herández, S.; Calvo, A.; Severi, M.A.; Ferragut, G.; Pérez-Pons, J.A.; Piñol, J.; Pich, O.; Mozo-Villarias, A.; Amela, I.; et al. MultitaskProtDB-II: An update of a database of multitasking/moonlighting proteins. Nucleic Acids Res. 2018, 46, D645–D648. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zabad, S.; Liu, H.; Wang, W.; Jeffery, C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 2018, 46, D640–D644. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Martin, A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011, 79, 3476–3491. [Google Scholar] [CrossRef]
- Amblee, V.; Jeffery, C.J. Physical Features of Intracellular Proteins that Moonlight on the Cell Surface. PLoS ONE 2015, 10, e0130575. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.J.; Philippe, B.; Lipke, P.N. Enzymatic Analysis of Yeast Cell Wall-Resident GAPDH and Its Secretion. mSphere 2020, 5. [Google Scholar] [CrossRef]
- Moreau, C.; Terrasse, R.; Thielens, N.M.; Vernet, T.; Gaboriaud, C.; Di Guilmi, A.M. Deciphering Key Residues Involved in the Virulence-Promoting Interactions between Streptococcus pneumoniae and Human Plasminogen. J. Biol. Chem. 2017, 292, 2217–2225. [Google Scholar] [CrossRef]
- Amela, I.; Cedano, J.; Querol, E. Pathogen proteins eliciting antibodies do not share epitopes with host proteins: A bioinformatics approach. PLoS ONE 2007, 2, e512. [Google Scholar] [CrossRef] [PubMed]
- Franco-Serrano, L.; Cedano, J.; Perez-Pons, J.A.; Mozo-Villarias, A.; Piñol, J.; Amela, I.; Querol, E. A hypothesis explaining why so many pathogen virulence proteins are moonlighting proteins. Pathog. Dis. 2018, 76. [Google Scholar] [CrossRef] [PubMed]
- Benoist, C.; Mathis, D. Autoimmunity provoked by infection: How good is the case for T cell epitope mimicry? Nat. Immunol. 2001, 2, 797–801. [Google Scholar] [CrossRef]
- Finney, J.; Watanabe, A.; Kelsoe, G.; Kuraoka, M. Minding the gap: The impact of B-cell tolerance on the microbial antibody repertoire. Immunol. Rev. 2019, 292, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Collado, R.; Prenafeta, A.; González-González, L.; Pérez-Pons, J.A.; Sitjà, M. Probing vaccine antigens against bovine mastitis caused by Streptococcus uberis. Vaccine 2016, 34, 3848–3854. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Li, W.; Cowley, A.; Uludag, M.; Gur, T.; McWilliam, H.; Squizzato, S.; Park, Y.M.; Buso, N.; Lopez, R. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015, 43, W580–W584. [Google Scholar] [CrossRef] [PubMed]
- Mi, T.; Merlin, J.C.; Deversaetty, S.; Gryk, M.R.; Bill, T.J.; Brooks, A.W.; Lee, L.Y.; Rathnayake, V.; Ross, C.A.; Sargeant, D.P.; et al. Minimotif Miner 3.0: Database expansion and significantly improved reduction of false-positive predictions from consensus sequences. Nucleic Acids Res. 2012, 40, D252–D260. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Larsen, J.E.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef][Green Version]
- He, Y.; He, Y.; Racz, R.; Sayers, S.; Lin, Y.; Todd, T.; Hur, J.; Li, X.; Patel, M.; Zhao, B.; et al. Updates on the web-based VIOLIN vaccine database and analysis system. Nucleic Acids Res. 2014, 42, D1124–D1132. [Google Scholar] [CrossRef] [PubMed]
- Hemmadi, V.; Biswas, M. An overview of moonlighting proteins in Staphylococcus aureus infection. Arch. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Tunio, S.A.; Oldfield, N.J.; Berry, A.; Ala’Aldeen, D.A.; Wooldridge, K.G.; Turner, D.P. The moonlighting protein fructose-1, 6-bisphosphate aldolase of Neisseria meningitidis: Surface localization and role in host cell adhesion. Mol. Microbiol. 2010, 76, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Agarwal, S.; Agarwal, S.; Pancholi, V. Surface export of GAPDH/SDH, a glycolytic enzyme, is essential for Streptococcus pyogenes virulence. mBio 2011, 2, e00068-11. [Google Scholar] [CrossRef]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-Negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef]
- Chauhan, A.S.; Kumar, M.; Chaudhary, S.; Patidar, A.; Dhiman, A.; Sheokand, N.; Malhotra, H.; Raje, C.I.; Raje, M. Moonlighting glycolytic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH): An evolutionarily conserved plasminogen receptor on mammalian cells. FASEB J. 2017, 31, 2638–2648. [Google Scholar] [CrossRef]
- Gomez, A.; Domedel, N.; Cedano, J.; Pinol, J.; Querol, E. Do current sequence analysis algorithms disclose multifunctional (moonlighting) proteins? Bioinformatics 2003, 19, 895–896. [Google Scholar] [CrossRef]
- Hernandez, S.; Gomez, A.; Cedano, J.; Querol, E. Bioinformatics annotation of the hypothetical proteins found by omics techniques can help to disclose additional virulence factors. Curr. Microbiol. 2009, 59, 451–456. [Google Scholar] [CrossRef]
- Hernandez, S.; Calvo, A.; Ferragut, G.; Franco, L.; Hermoso, A.; Amela, I.; Gomez, A.; Querol, E.; Cedano, J. Can bioinformatics help in the identification of moonlighting proteins? Biochem. Soc. Trans. 2014, 42, 1692–1697. [Google Scholar] [CrossRef]
- Hernandez, S.; Franco, L.; Calvo, A.; Ferragut, G.; Hermoso, A.; Amela, I.; Gomez, A.; Querol, E.; Cedano, J. Bioinformatics and Moonlighting Proteins. Front. Bioeng. Biotechnol. 2015, 3, 90. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Gehring, C.; Irving, H.R. Conserved Functional Motifs and Homology Modeling to Predict Hidden Moonlighting Functional Sites. Front. Bioeng. Biotechnol. 2015, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- Zanzoni, A.; Ribeiro, D.M.; Brun, C. Understanding protein multifunctionality: From short linear motifs to cellular functions. Cell Mol. Life Sci. 2019, 76, 4407–4412. [Google Scholar] [CrossRef] [PubMed]
- Hulo, N.; Bairoch, A.; Bulliard, V.; Cerutti, L.; De Castro, E. The PROSITE database. Nucleic Acids Res. 2006, 34, D227–D230. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Grimmer, J.; Dumke, R. Organization of multi-binding to host proteins: The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Mycoplasma pneumoniae. Microbiol. Res. 2019, 218, 22–31. [Google Scholar] [CrossRef]
- Satala, D.; Satala, G.; Karkowska-Kuleta, J.; Bukowski, M.; Kluza, A.; Rapala-Kozik, M.; Kozik, A. Structural Insights into the Interactions of Candidal Enolase with Human Vitronectin, Fibronectin and Plasminogen. Int. J. Mol. Sci. 2020, 21, 7843. [Google Scholar] [CrossRef]
- Derbise, A.; Song, Y.P.; Parikh, S.; Fischetti, V.A.; Pancholi, V. Role of the C-Terminal lysine residues of streptococcal surface enolase in Glu- and Lys-plasminogen-binding activities of group A streptococci. Infect. Immun. 2004, 72, 94–105. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R.; et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef]
- Alonso-Lopez, D.; Campos-Laborie, F.J.; Gutierrez, M.A.; Lambourne, L.; Calderwood, M.A.; Vidal, M.; De Las Rivas, J. APID database: Redefining protein-protein interaction experimental evidences and binary interactomes. Database 2019, 2019. [Google Scholar] [CrossRef]
- Lahteenmaki, K.; Kuusela, P.; Korhonen, T.K. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 2001, 25, 531–552. [Google Scholar] [CrossRef]
- Degen, J.L.; Bugge, T.H.; Goguen, J.D. Fibrin and fibrinolysis in infection and host defense. J. Thromb Haemost. 2007, 5 (Suppl. 1), 24–31. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Ploplis, V.A.; Castellino, F.J. Bacterial plasminogen receptors utilize host plasminogen system for effective invasion and dissemination. J. Biomed. Biotechnol. 2012, 2012, 482096. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Steinert, M.; Bergmann, S. Interaction of streptococcal plasminogen binding proteins with the host fibrinolytic system. Front. Cell Infect. Microbiol. 2013, 3, 85. [Google Scholar] [CrossRef]
- Peetermans, M.; Vanassche, T.; Liesenborghs, L.; Lijnen, R.H.; Verhamme, P. Bacterial pathogens activate plasminogen to breach tissue barriers and escape from innate immunity. Crit. Rev. Microbiol. 2016, 42, 866–882. [Google Scholar] [CrossRef]
- Ayon-Nunez, D.A.; Fragoso, G.; Bobes, R.J.; Laclette, J.P. Plasminogen-Binding proteins as an evasion mechanism of the host’s innate immunity in infectious diseases. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Foo, R.S.; Nam, Y.J.; Ostreicher, M.J.; Metzl, M.D.; Whelan, R.S.; Peng, C.F.; Ashton, A.W.; Fu, W.; Mani, K.; Chin, S.F.; et al. Regulation of p53 tetramerization and nuclear export by ARC. Proc. Natl. Acad. Sci. USA 2007, 104, 20826–20831. [Google Scholar] [CrossRef]
- Cao, L.; Petrusca, D.N.; Satpathy, M.; Nakshatri, H.; Petrache, I.; Matei, D. Tissue transglutaminase protects epithelial ovarian cancer cells from cisplatin-induced apoptosis by promoting cell survival signaling. Carcinogenesis 2008, 29, 1893–1900. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Song, X.; Zhuo, W.; Fu, Y.; Shi, H.; Liang, Y.; Tong, M.; Chang, G.; Luo, Y. The regulatory mechanism of Hsp90alpha secretion and its function in tumor malignancy. Proc. Natl. Acad. Sci. USA 2009, 106, 21288–21293. [Google Scholar] [CrossRef]
- Wang, Y.; Trepel, J.B.; Neckers, L.M.; Giaccone, G. STA-9090, a small-molecule Hsp90 inhibitor for the potential treatment of cancer. Curr. Opin. Investig. Drugs 2010, 11, 1466–1476. [Google Scholar]
- Jhaveri, K.; Taldone, T.; Modi, S.; Chiosis, G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta 2012, 1823, 742–755. [Google Scholar] [CrossRef]
- Zheng, G.; Ma, Y.; Zou, Y.; Yin, A.; Li, W.; Dong, D. HCMDB: The human cancer metastasis database. Nucleic Acids Res. 2018, 46, D950–D955. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Vizin, T.; Kos, J. Gamma-enolase: A well-known tumour marker, with a less-known role in cancer. Radiol. Oncol. 2015, 49, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Paolillo, M.; Schinelli, S. Extracellular Matrix Alterations in Metastatic Processes. Int. J. Mol. Sci. 2019, 20, 4847. [Google Scholar] [CrossRef]
- Vaca, D.J.; Thibau, A.; Schutz, M.; Kraiczy, P.; Happonen, L.; Malmstrom, J.; Kempf, V.A.J. Interaction with the host: The role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med. Microbiol. Immunol. 2020, 209, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Troester, M.A.; Lee, M.H.; Carter, M.; Fan, C.; Cowan, D.W.; Perez, E.R.; Pirone, J.R.; Perou, C.M.; Jerry, D.J.; Schneider, S.S. Activation of host wound responses in breast cancer microenvironment. Clin. Cancer Res. 2009, 15, 7020–7028. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Bendas, G.; Borsig, L. Heparanase in Cancer Metastasis—Heparin as a Potential Inhibitor of Cell Adhesion Molecules. Adv. Exp. Med. Biol. 2020, 1221, 309–329. [Google Scholar] [CrossRef] [PubMed]


| Sequence | Function | Distribution |
|---|---|---|
| FYDKERKVY | Binding to plasminogen [39] | Only Streptococcus |
| KK | Binding to plasminogen, according to published papers [40] | Pathogenic and non-pathogenic species |
| KxxK | Binding to plasminogen, according to published papers [40] | Pathogenic and non-pathogenic species |
| Y[LIV]E[LIV] Y[LIV]ED[PLIV] | Binding to PLCgamma –> Blood coagulation and interactionwith PTPN11 (Predicted) | Mostly in pathogenic species of different evolutive pathways |
| YTAV | Binding to PTPN11, a phosphatase related to blood coagulation and platelet formation (interaction with fibrinogen, fibrin,…). Noonan síndrome –> Inbalance in fibrynolitic components. (Predicted) | Mostly in pathogenic species of different evolutive pathways |
| GO Number | Function | % Human Proteome | % Moonlighting Proteins | p-Value |
|---|---|---|---|---|
| 001968 | Binding to Fibronectin | 0.39 | 3.06 | 6.87 × 10−34 |
| 0005518 | Binding to Collagen | 0.22 | 1.96 | 1.99 × 10−26 |
| 0050840 | Binding to ECM | 0.23 | 1.89 | 2.20 × 10−16 |
| 0043236 | Binding to Laminin | 0.15 | 1.32 | 3.01 × 10−17 |
| 0002020 | Protease activity | 0.72 | 5.12 | 2.20 × 10−16 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franco-Serrano, L.; Sánchez-Redondo, D.; Nájar-García, A.; Hernández, S.; Amela, I.; Perez-Pons, J.A.; Piñol, J.; Mozo-Villarias, A.; Cedano, J.; Querol, E. Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors. Microorganisms 2021, 9, 1300. https://doi.org/10.3390/microorganisms9061300
Franco-Serrano L, Sánchez-Redondo D, Nájar-García A, Hernández S, Amela I, Perez-Pons JA, Piñol J, Mozo-Villarias A, Cedano J, Querol E. Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors. Microorganisms. 2021; 9(6):1300. https://doi.org/10.3390/microorganisms9061300
Chicago/Turabian StyleFranco-Serrano, Luis, David Sánchez-Redondo, Araceli Nájar-García, Sergio Hernández, Isaac Amela, Josep Antoni Perez-Pons, Jaume Piñol, Angel Mozo-Villarias, Juan Cedano, and Enrique Querol. 2021. "Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors" Microorganisms 9, no. 6: 1300. https://doi.org/10.3390/microorganisms9061300
APA StyleFranco-Serrano, L., Sánchez-Redondo, D., Nájar-García, A., Hernández, S., Amela, I., Perez-Pons, J. A., Piñol, J., Mozo-Villarias, A., Cedano, J., & Querol, E. (2021). Pathogen Moonlighting Proteins: From Ancestral Key Metabolic Enzymes to Virulence Factors. Microorganisms, 9(6), 1300. https://doi.org/10.3390/microorganisms9061300






