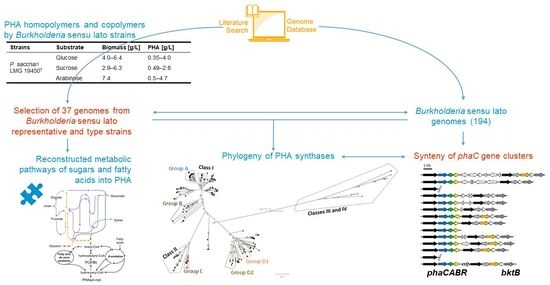
Genome-Wide Metabolic Reconstruction of the Synthesis of Polyhydroxyalkanoates from Sugars and Fatty Acids by Burkholderia Sensu Lato Species
Abstract
1. Introduction
2. Synthesis of PHAs by Burkholderia Sensu Lato Strains
2.1. PHB Homopolymer Synthesis by Burkholderia Sensu Lato
| Strain | Substrate | CDW (g/L) | PHA Type | PHA Concentration (g/L) | Limitation | YPHA/S (g/g) | Reference |
|---|---|---|---|---|---|---|---|
| Paraburkholderia sacchari LMG 19450T (IPT 101, DSM 17165, LFM 101, CCT 6971) | Glu | 5.0–6.4 | PHB | 0.35–4.0 | Nitrogen | 0.25–0.29 | [14,41,47] |
| Xyl | 2.9–6.3 | PHB | 0.49–2.8 | Nitrogen | 0.05–0.26 | [8,14,41,47,48] | |
| Ara | 7.4 | PHB | 0.5–4.7 | Nitrogen | 0.24 | [14,47] | |
| Man | 6.9 | PHB | 4.2 | Nitrogen | 0.21 | [49] | |
| Gal | 4.9 | PHB | 2.2 | Nitrogen | 0.11 | [49] | |
| Scr | 6.14 | PHB | 4.2 | Nitrogen | 0.29 | [50] | |
| Glu/Fatty acids | 1.25–2.4 | P(3HB-co-3HV) | 0.4–0.9 | - | - | [44] | |
| Glu/GBL, 4HBA | 1.8–6.6 | P(3HB-co-4HB) | 0.4–3.1 | Nitrogen | 0.01–0.1 ** | [44,51,52] | |
| Glu/HxA | 2.1 | P(3HB-co-3HHx) | 1.1 | Nitrogen | - | [44] | |
| Paraburkholderia xenovorans LB400T | Glu | - | PHB | (40% w/w) | Nitrogen | - | [40,53] |
| Xyl | - | PHB | NR | Nitrogen | - | [54] | |
| Man | - | PHB | NR | Nitrogen | - | ||
| Burkholderia cepacia ATCC 17759 (DSM 50181) | Glu | 2.6 | PHB | 1.5 | Nitrogen | - | [55] |
| Fru | 5 | PHB | 2 | Nitrogen | 0.07–0.174 | [55,56] | |
| Xyl | 2.6 | PHB | 1.5 | Nitrogen | 0.11 | [55] | |
| Scr | 4.2 | PHB | 2.1 | Nitrogen | 0.18 | [50] | |
| Xyl/LaA | 5.3 | P(3HB-co-3HV) | 2.4 | - | - | [57] | |
| Glu/PA | 1.6–1.8 | P(3HB-co-3HV) | 0.2–1.0 | - | - | [56] | |
| Burkholderia thailandensis E264T | UCO (fatty acids) | 12.6 | PHB | 7.5 | NL | 0.35 | [43] |
| Burkholderia contaminans Kad1 | Waste glycerol/VA | 5.6 | P(3HB-co-3HV) | 1.96 | - | - | [58] |
| Burkholderia contaminans IPT 553 | Glu/Scr | 2.3–4.9 | P(3HB-co-3HDd) | 0.85–1.176 | - | - | [45] |
| Trinickia caryophylli DSM 50341T | Gnt | - | PHB | (34.2% w/w) | - | - | [46] |
| OA | - * | PHB | 1.2 | - | - | ||
| Trinickia caryophylli AS 1.2741 | Glu | 0.981 | P(3HHx-co-3HO-co-3HD) | 0.013 | - | - | [59] |
| OA | 1.084 | P(3HHx-co-3HO-co-3HD) | 0.26 | - | - | ||
| Glu/OA | 1.159 | P(3HHx-co-3HO-co-3HD) | 0.23 | - | - |
2.2. PHA Copolymer Synthesis by Burkholderia Sensu Lato
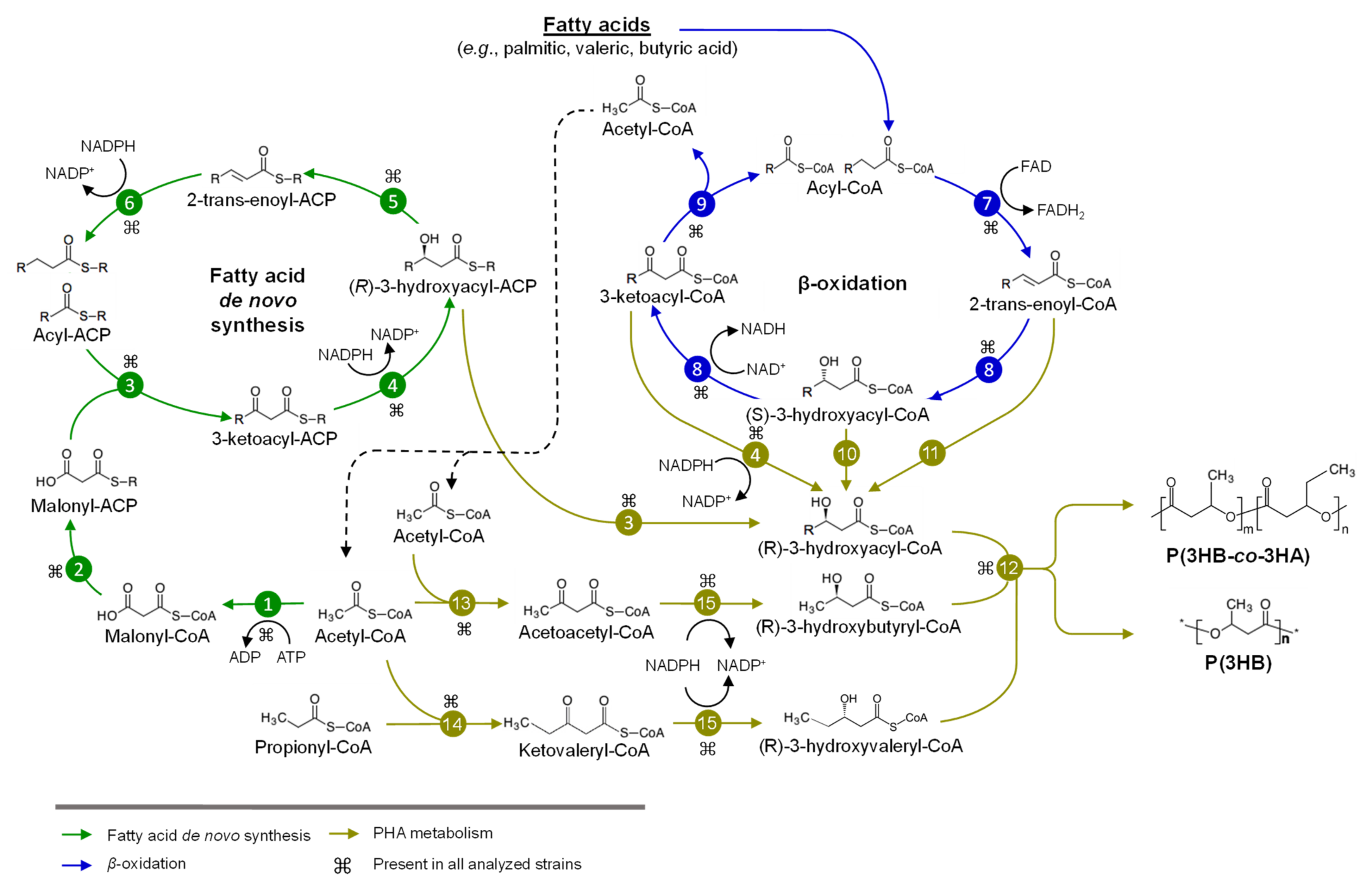
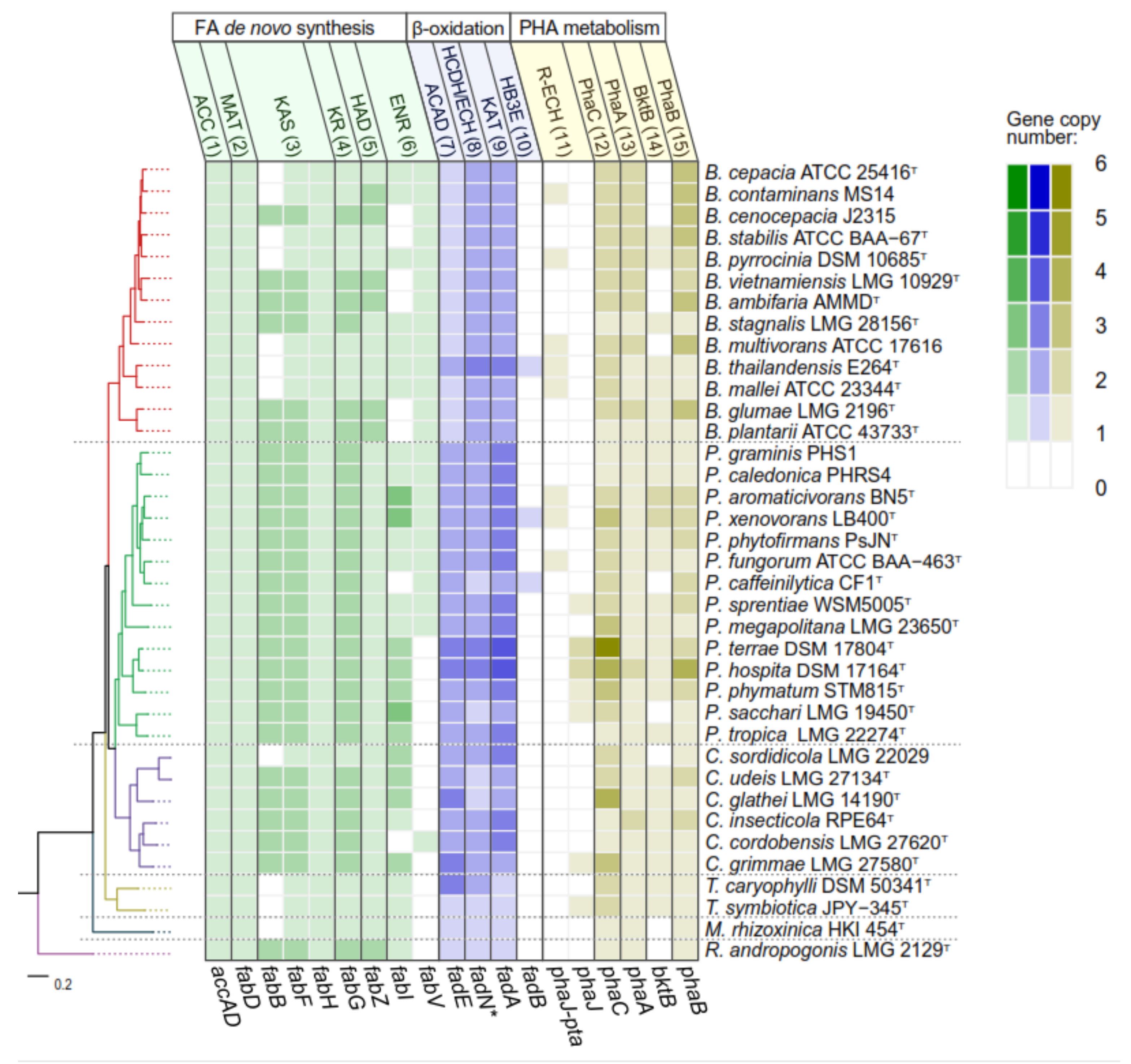
3. Metabolism of Sugars and Fatty Acids in Burkholderia Sensu Lato
3.1. Metabolism of Sugars for PHA Production in Burkholderia Sensu Lato
3.2. Metabolism of Fatty Acids and PHA Synthesis in Burkholderia Sensu Lato
4. PHA Synthases in Burkholderia Sensu Lato Strains
5. Gene Synteny of the phaC Gene Cluster in Burkholderia Sensu Lato
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Geyer, R.; Jambeck, J.; Law, K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017, 3, e1700782. [Google Scholar] [CrossRef]
- Plastics Europe. An Analysis of European Plastics Production, Demand and Waste Data. In Plastics-The Facts 2019. Available online: https:/www.plasticseurope.org/es/resources/publications/1804-plastics-facts-2019 (accessed on 5 December 2019).
- Koller, M. Production of Polyhydroxyalkanoate (PHA) Biopolyesters by Extremophiles? MOJ Polym. Sci. 2017, 1, 1–19. [Google Scholar] [CrossRef]
- Sanhueza, C.; Acevedo, F.; Rocha, S.; Villegas, P.; Seeger, M.; Navia, R. Polyhydroxyalkanoates as biomaterial for electrospun scaffolds. Int. J. Biol. Macromol. 2019, 124, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Sanhueza, C.; Diaz-Rodriguez, P.; Villegas, P.; González, Á.; Seeger, M.; Suárez-González, J.; Concheiro, A.; Alvarez-Lorenzo, C.; Acevedo, F. Influence of the carbon source on the properties of poly-(3)-hydroxybutyrate produced by Paraburkholderia xenovorans LB400 and its electrospun fibers. Int. J. Biol. Macromol. 2020, 152, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Vilchez, A.; Acevedo, F.; Cea, M.; Seeger, M.; Navia, R. Applications of Electrospun Nanofibers with Antioxidant Properties: A Review. Nanomaterials 2020, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, K.; Ismail, I.M.I.; Ali, N.; Rashid, M.I.; Summan, A.S.A.; Kabli, M.R.; Narodoslawsky, M.; Koller, M. LCA, Sustainability and Techno-Economic Studies for PHA Production. In The Handbook of Polyhydroxyalkanoates; CRC Press: Boca Raton, FL, USA, 2020; Volume 2, pp. 455–485. [Google Scholar]
- Silva, L.F.; Taciro, M.K.; Ramos, M.E.M.; Carter, J.M.; Pradella, J.G.C.; Gomez, J.G.C. Poly-3-hydroxybutyrate (P3HB) production by bacteria from xylose, glucose and sugarcane bagasse hydrolysate. J. Ind. Microbiol. Biotechnol. 2004, 31, 245–254. [Google Scholar] [CrossRef]
- Nikel, P.I.; Pettinari, M.; Ramírez, M.; Galvagno, M.A.; Méndez, B.S. Escherichia coli arcA mutants: Metabolic profile characterization of microaerobic cultures using glycerol as a carbon source. J. Mol. Microbiol. Biotechnol. 2008, 15, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tappel, R.C.; Nomura, C.T. Mini-Review: Biosynthesis of poly(hydroxyalkanoates). Polym. Rev. 2009, 49, 226–248. [Google Scholar] [CrossRef]
- Ciesielski, S.; Mozejko-Ciesielska, J.; Pisutpaisal, N. Plant oils as promising substrates for polyhydroxyalkanoates production. J. Clean. Prod. 2015, 106, 408–421. [Google Scholar] [CrossRef]
- Możejko-Ciesielska, J.; Kiewisz, R. Bacterial polyhydroxyalkanoates: Still fabulous? Microbiol. Res. 2016, 192, 271–282. [Google Scholar] [CrossRef]
- Zou, H.; Shi, M.; Zhang, T.; Xian, M.; Li, L. Natural and engineered polyhydroxyalkanoate (PHA) synthase: Key enzyme in biopolyester production. Appl. Microbiol. Biotechnol. 2017, 101, 7417–7426. [Google Scholar] [CrossRef]
- Li, M.; Eskridge, K.M.; Wilkins, M.R. Optimization of polyhydroxybutyrate production by experimental design of combined ternary mixture (glucose, xylose and arabinose) and process variables (sugar concentration, molar C:N ratio). Bioprocess Biosyst. Eng. 2019, 42, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Raza, Z.A.; Abid, S.; Banat, I.M. Polyhydroxyalkanoates: Characteristics, production, recent developments and applications. Int. Biodeterior. Biodegrad. 2018, 126, 45–56. [Google Scholar] [CrossRef]
- Tsuge, T.; Fukui, T.; Matsusaki, H.; Taguchi, S.; Kobayashi, G.; Ishizaki, A.; Doi, Y. Molecular cloning of two (R)-specific enoyl-CoA hydratase genes from Pseudomonas aeruginosa and their use for polyhydroxyalkanoate synthesis. FEMS Microbiol. Lett. 2000, 184, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Nomura, C.T.; Taguchi, K.; Taguchi, S.; Doi, Y. Coexpression of genetically engineered 3-ketoacyl-ACP synthase III (fabH) and polyhydroxyalkanoate synthase (phaC) genes leads to short-chain-length-medium-chain-length polyhydroxyalkanoate copolymer production from glucose in Escherichia coli JM109. Appl. Environ. Microbiol. 2004, 70, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Nomura, C.T. Monitoring differences in gene expression levels and polyhydroxyalkanoate (PHA) production in Pseudomonas putida KT2440 grown on different carbon sources. J. Biosci. Bioeng. 2010, 110, 653–659. [Google Scholar] [CrossRef]
- Madison, L.L.; Huisman, G.W. Metabolic engineering of poly(3-hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 1999, 63, 21–53. [Google Scholar] [CrossRef]
- Qi, Q.; Rehm, B.H.A. Polyhydroxybutyrate biosynthesis in Caulobacter crescentus: Molecular characterization of the polyhydroxybutyrate synthase. Microbiology 2001, 147, 3353–3358. [Google Scholar] [CrossRef] [PubMed]
- Park, J.M.; Kim, T.Y.; Lee, S.Y. Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production. BMC Syst. Biol. 2011, 5, 101. [Google Scholar] [CrossRef]
- Grousseau, E.; Blanchet, E.; Déléris, S.; Albuquerque, M.G.; Paul, E.; Uribelarrea, J.-L. Impact of sustaining a controlled residual growth on polyhydroxybutyrate yield and production kinetics in Cupriavidus necator. Bioresour. Technol. 2013, 148, 30–38. [Google Scholar] [CrossRef]
- Meng, D.-C.; Shen, R.; Yao, H.; Chen, J.-C.; Wu, Q.; Chen, G.-Q. Engineering the diversity of polyesters. Curr. Opin. Biotechnol. 2014, 29, 24–33. [Google Scholar] [CrossRef]
- Sawana, A.; Eadeolu, M.; Gupta, R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 2014, 5, 429. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Samadpour, M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016, 66, 2836–2846. [Google Scholar] [CrossRef] [PubMed]
- Dobritsa, A.P.; Samadpour, M. Reclassification of Burkholderia insecticola as Caballeronia insecticola comb. nov. and reliability of conserved signature indels as molecular synapomorphies. Int. J. Syst. Evol. Microbiol. 2019, 69, 2057–2063. [Google Scholar] [CrossRef]
- Lin, Q.-H.; Lv, Y.-Y.; Gao, Z.-H.; Qiu, L.-H. Pararobbsia silviterrae gen. nov., sp. nov., isolated from forest soil and reclassification of Burkholderia alpina as Pararobbsia alpina comb. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Chain, P.S.G.; Denef, V.J.; Konstantinidis, K.T.; Vergez, L.M.; Agulló, L.; Reyes, V.L.; Hauser, L.; Córdova, M.; Gómez, L.; González, M.; et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc. Natl. Acad. Sci. USA 2006, 103, 15280–15287. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pantoja, D.; Donoso, R.; Agulló, L.; Córdova, M.; Seeger, M.; Pieper, D.H.; González, B. Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environ. Microbiol. 2012, 14, 1091–1117. [Google Scholar] [CrossRef]
- Peeters, C.; Meier-Kolthoff, J.P.; Verheyde, B.; De Brandt, E.; Cooper, V.S.; Vandamme, P. Phylogenomic study of Burkholderia glathei-like organisms, proposal of 13 novel Burkholderia Species and emended descriptions of Burkholderia sordidicola, Burkholderia zhejiangensis, and Burkholderia grimmiae. Front. Microbiol. 2016, 7, 877. [Google Scholar] [CrossRef]
- Agulló, L.; Romero-Silva, M.J.; Domenech, M.; Seeger, M. p-Cymene promotes its catabolism through the p-cymene and the p-cumate pathways, activates a stress response and reduces the biofilm formation in Burkholderia xenovorans LB400. PLoS ONE 2017, 12, e0169544. [Google Scholar] [CrossRef]
- Seeger, M.; Zielinski, M.; Timmis, K.N.; Hofer, B. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 1999, 65, 3614–3621. [Google Scholar] [CrossRef]
- Seeger, M.; González, M.; Cámara, B.; Muñoz, L.; Ponce, E.; Mejías, L.; Mascayano, C.; Vásquez, Y.; Sepúlveda-Boza, S. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 2003, 69, 5045–5050. [Google Scholar] [CrossRef]
- Chirino, B.; Strahsburger, E.; Agulló, L.; González, M.; Seeger, M. Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS ONE 2013, 8, e75746. [Google Scholar] [CrossRef]
- Fuentes, S.; Méndez, V.; Aguila, P.; Seeger, M. Bioremediation of petroleum hydrocarbons: Catabolic genes, microbial communities, and applications. Appl. Microbiol. Biotechnol. 2014, 98, 4781–4794. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Straube, M.J.; Cámara, B.; Tello, M.; Silva, F.M.; Cárdenas, F.; Seeger, M. Genetic and functional analysis of the biosynthesis of a non-ribosomal peptide siderophore in Burkholderia xenovorans LB400. PLoS ONE 2016, 11, e0151273. [Google Scholar] [CrossRef] [PubMed]
- Vega-Celedón, P.; Canchignia, H.; González, M.; Seeger, M. Biosynthesis of indole-3-acetic acid and plant growth promoting by bacteria. Cultiv. Trop. 2016, 37, 31–37. [Google Scholar]
- Donoso, R.; Leiva-Novoa, P.; Zúñiga, A.; Timmermann, T.; Recabarren-Gajardo, G.; González, B. Biochemical and genetic bases of indole-3-acetic acid (auxin phytohormone) degradation by the plant-growth-promoting rhizobacterium Paraburkholderia phytofirmans PsJN. Appl. Environ. Microbiol. 2017, 83, 01991-16. [Google Scholar] [CrossRef]
- Mendonça, T.T.; Tavares, R.R.; Cespedes, L.G.; Rodriguez, R.J.S.; Schripsema, J.; Taciro, M.K.; Gomez, J.G.; Silva, L.F. Combining molecular and bioprocess techniques to produce poly(3-hydroxybutyrate- co -3-hydroxyhexanoate) with controlled monomer composition by Burkholderia sacchari. Int. J. Biol. Macromol. 2017, 98, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Urtuvia, V.; Villegas, P.; Fuentes, S.; González, M.; Seeger, M. Burkholderia xenovorans LB400 possesses a functional polyhydroxyalkanoate anabolic pathway encoded by the pha genes and synthesizes poly(3-hydroxybutyrate) under nitrogen-limiting conditions. Int. Microbiol. 2018, 21, 47–57. [Google Scholar] [CrossRef]
- Lopes, M.S.G.; Rocha, R.C.S.; Zanotto, S.P.; Gomez, J.G.C.; Da Silva, L.F. Screening of bacteria to produce polyhydroxyalkanoates from xylose. World J. Microbiol. Biotechnol. 2009, 25, 1751–1756. [Google Scholar] [CrossRef]
- Pan, W.; Perrotta, J.A.; Stipanovic, A.J.; Nomura, C.T.; Nakas, J.P. Production of polyhydroxyalkanoates by Burkholderia cepacia ATCC 17759 using a detoxified sugar maple hemicellulosic hydrolysate. J. Ind. Microbiol. Biotechnol. 2012, 39, 459–469. [Google Scholar] [CrossRef]
- Kourmentza, C.; Costa, J.; Azevedo, Z.; Servin, C.; Grandfils, C.; Freitas, V.; Reis, M. Burkholderia thailandensis as a microbial cell factory for the bioconversion of used cooking oil to polyhydroxyalkanoates and rhamnolipids. Bioresour. Technol. 2018, 247, 829–837. [Google Scholar] [CrossRef]
- Mendonça, T.; Gomez, J.; Buffoni, E.; Rodriguez, R.J.S.; Schripsema, J.; Lopes, M.; Silva, L. Exploring the potential of Burkholderia sacchari to produce polyhydroxyalkanoates. J. Appl. Microbiol. 2014, 116, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Nahar, S.; Jeong, M.-H.; Hur, J.-S. Lichen-Associated bacterium, a novel bioresource of polyhydroxyalkanoate (pha) production and simultaneous degradation of naphthalene and anthracene. J. Microbiol. Biotechnol. 2019, 29, 79–90. [Google Scholar] [CrossRef]
- Timm, A.; Steinbüchel, A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl. Environ. Microbiol. 1990, 56, 3360–3367. [Google Scholar] [CrossRef] [PubMed]
- Cesário, M.T.; Raposo, R.S.; de Almeida, M.C.M.; van Keulen, F.; Ferreira, B.; da Fonseca, M.M. Enhanced bioproduction of poly-3-hydroxybutyrate from wheat straw lignocellulosic hydrolysates. New Biotechnol. 2014, 31, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Lopes, M.S.; Gomez, J.G.; Silva, L.F. Cloning and overexpression of the xylose isomerase gene from Burkholderia sacchari and production of polyhydroxybutyrate from xylose. Can. J. Microbiol. 2009, 55, 1012–1015. [Google Scholar] [CrossRef]
- Dietrich, K.; Dumont, M.-J.; Schwinghamer, T.; Orsat, V.; Del Rio, L.F. Model study to assess softwood hemicellulose hydrolysates as the carbon source for PHB production in Paraburkholderia sacchari IPT 101. Biomacromolecules 2018, 19, 188–200. [Google Scholar] [CrossRef]
- Gomez, J.G.C.; Rodrigues, M.F.A.; Alli, R.C.P.; Torres, B.B.; Netto, C.L.B.; Oliveira, M.S.; Da Silva, L.F. Evaluation of soil gram-negative bacteria yielding polyhydroxyalkanoic acids from carbohydrates and propionic acid. Appl. Microbiol. Biotechnol. 1996, 45, 785–791. [Google Scholar] [CrossRef]
- Cesário, M.T.; Raposo, R.S.; De Almeida, M.C.M.; Van Keulen, F.; Ferreira, B.; Telo, J.; da Fonseca, M.M. Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) by Burkholderia sacchari using wheat straw hydrolysates and gamma-butyrolactone. Int. J. Biol. Macromol. 2014, 71, 59–67. [Google Scholar] [CrossRef]
- Raposo, R.S.; De Almeida, M.C.M.; Da Fonseca, M.; Cesário, M.T. Feeding strategies for tuning poly (3-hydroxybutyrate-co-4-hydroxybutyrate) monomeric composition and productivity using Burkholderia sacchari. Int. J. Biol. Macromol. 2017, 105, 825–833. [Google Scholar] [CrossRef]
- Urtuvia, V.; Villegas, P.; González, M.; Seeger, M. Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int. J. Biol. Macromol. 2014, 70, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, F.; Villegas, P.; Urtuvia, V.; Hermosilla, J.; Navia, R.; Seeger, M. Bacterial polyhydroxybutyrate for electrospun fiber production. Int. J. Biol. Macromol. 2018, 106, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, J.A.; Hassan, M.-C.A.; Ramsay, B.A. Hemicellulose as a potential substrate for production of poly (β-hydroxyalkanoates). Can. J. Microbiol. 1995, 41, 262–266. [Google Scholar] [CrossRef]
- Ramsay, B.A.; Ramsay, J.A.; Cooper, D.G. Production of poly-β-hydroxyalkanoic acid by Pseudomonas cepacia. Appl. Environ. Microbiol. 1989, 55, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Keenan, T.; Tanenbaum, S.; Stipanovic, A.; Nakas, J. Production and characterization of poly-β-hydroxyalkanoate copolymers from Burkholderia cepacia utilizing xylose and levulinic acid. Biotechnol. Prog. 2004, 20, 1697–1704. [Google Scholar] [CrossRef]
- Al-Kaddo, K.B.; Mohamad, F.; Murugan, P.; Tan, J.S.; Sudesh, K.; Samian, M.R. Production of P(3HB-co-4HB) copolymer with high 4HB molar fraction by Burkholderia contaminans Kad1 PHA synthase. Biochem. Eng. J. 2020, 153, 107394. [Google Scholar] [CrossRef]
- Hang, X.; Zhang, G.; Wang, G.; Zhao, X.; Chen, G.-Q. PCR cloning of polyhydroxyalkanoate biosynthesis genes from Burkholderia caryophylli and their functional expression in recombinant Escherichia coli. FEMS Microbiol. Lett. 2002, 210, 49–54. [Google Scholar] [CrossRef][Green Version]
- Habe, H.; Sato, S.; Morita, T.; Fukuoka, T.; Kirimura, K.; Kitamoto, D. Bacterial production of short-chain organic acids and trehalose from levulinic acid: A potential cellulose-derived building block as a feedstock for microbial production. Bioresour. Technol. 2015, 177, 381–386. [Google Scholar] [CrossRef]
- Ashby, R.D.; Solaiman, D.K.; Nuñez, A.; Strahan, G.D.; Johnston, D.B. Burkholderia sacchari DSM 17165: A source of compositionally-tunable block-copolymeric short-chain poly(hydroxyalkanoates) from xylose and levulinic acid. Bioresour. Technol. 2018, 253, 333–342. [Google Scholar] [CrossRef]
- Volova, T.G.; Vinogradova, O.N.; Zhila, N.O.; Kiselev, E.G.; Peterson, I.V.; Vasilev, A.D.; Sukovatyi, A.G.; Shishatskaya, E. Physicochemical properties of multicomponent polyhydroxyalkanoates: Novel aspects. Polym. Sci. Ser. A 2017, 59, 98–106. [Google Scholar] [CrossRef]
- Jiang, X.; Luo, X.; Zhou, N.-Y. Two polyhydroxyalkanoate synthases from distinct classes from the aromatic degrader Cupriavidus pinatubonensis JMP134 exhibit the same substrate preference. PLoS ONE 2015, 10, e0142332. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.; Escapa, I.F.; Martinez, V.; Dinjaski, N.; Herencias, C.; De La Peña, F.; Tarazona, N.; Revelles, O. A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida. Environ. Microbiol. 2016, 18, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Kihara, T.; Hiroe, A.; Ishii-Hyakutake, M.; Mizuno, K.; Tsuge, T. Bacillus cereus-type polyhydroxyalkanoate biosynthetic gene cluster contains R-specific enoyl-CoA hydratase gene. Biosci. Biotechnol. Biochem. 2017, 81, 1627–1635. [Google Scholar] [CrossRef]
- Álvarez-Santullano, N.; Villegas, P.; Sepúlveda, M.; Vilchez, A.; Donoso, R.; Pérez-Pantoja, D.; Navia, R.; Acevedo, F.; Seeger, M. Production of polyhydroxyalkanoates by Paraburkholderia and Burkholderia species: A journey from the genes through metabolic routes. In The Handbook of Polyhydroxyalkanoates; Koller, M., Ed.; CRC Press: Boca Raton, FL, USA, 2020; Volume 1, pp. 115–136. [Google Scholar]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with standing in nomenclature (lpsn) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Mendler, K.; Chen, H.; Parks, D.H.; Lobb, B.; Hug, L.; Doxey, A.C. AnnoTree: Visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res. 2019, 47, 4442–4448. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; A Fulcher, C.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Latendresse, M.; E Midford, P.; Ong, Q.; Ong, W.K.; et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018, 46, D633–D639. [Google Scholar] [CrossRef]
- Segata, N.; Börnigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Pratama, A.A.; Jiménez, D.J.; Chen, Q.; Bunk, B.; Spröer, C.; Overmann, J.; Van Elsas, J.D. Delineation of a subgroup of the genus Paraburkholderia, including P. terrae DSM 17804T, P. hospita DSM 17164T, and four soil-isolated fungiphiles, reveals remarkable genomic and ecological features—proposal for the definition of a P. hospita species cluster. Genome Biol. Evol. 2020, 12, 325–344. [Google Scholar] [CrossRef]
- Feng, Y.; Cronan, J.E. Escherichia coli unsaturated fatty acid synthesis. J. Biol. Chem. 2009, 284, 29526–29535. [Google Scholar] [CrossRef] [PubMed]
- Kutralam-Muniasamy, G.; Marsch, R.; Pérez-Guevara, F. Investigation on the evolutionary relation of diverse polyhydroxyalkanoate gene clusters in betaproteobacteria. J. Mol. Evol. 2018, 86, 470–483. [Google Scholar] [CrossRef] [PubMed]
- Urakami, T.; Ito-Yoshida, C.; Araki, H.; Kijima, T.; Suzuki, K.-I.; Komagata, K. Transfer of Pseudomonas plantarii and Pseudomonas glumae to Burkholderia as Burkholderia spp. and description of Burkholderia vandii sp. nov. Int. J. Syst. Bacteriol. 1994, 44, 235–245. [Google Scholar] [CrossRef]
- Vandamme, P.; Holmes, B.; Vancanneyt, M.; Coenye, T.; Hoste, B.; Coopman, R.; Revets, H.; Lauwers, S.; Gillis, M.; Kersters, K.; et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 1997, 47, 1188–1200. [Google Scholar] [CrossRef]
- Seth-Smith, H.M.; Casanova, C.; Sommerstein, R.; Meinel, D.M.; Abdelbary, M.M.; Blanc, D.S.; Droz, S.; Führer, U.; Lienhard, R.; Lang, C.; et al. Phenotypic and genomic analyses of Burkholderia stabilis clinical contamination, Switzerland. Emerg. Infect. Dis. 2019, 25, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Brämer, C.; Vandamme, P.; Da Silva, L.F.; Gomez, J.G.C.; Steinbüchel, A. Polyhydroxyalkanoate-accumulating bacterium isolated from soil of a sugar-cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 2001, 51, 1709–1713. [Google Scholar] [CrossRef]
- Gillis, M.; Van Van, T.; Bardin, R.; Goor, M.; Hebbar, P.; Willems, A.; Segers, P.; Kersters, K.; Heulin, T.; Fernandez-Fernandez, M.P. Polyphasic taxonomy in the genus Burkholderia Leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for n2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 1995, 45, 274–289. [Google Scholar] [CrossRef]
- Coenye, T.; Laevens, S.; Willems, A.; Olén, M.; Hannat, W.; Govan, J.; Gillis, M.; Falsen, E.; Vandamme, P. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 2001, 51, 1099–1107. [Google Scholar] [CrossRef]
- De Smet, B.; Mayo, M.; Peeters, C.; Zlosnik, J.; Spilker, T.; Hird, T.J.; Lipuma, J.J.; Kidd, T.; Kaestli, M.; Ginther, J.L.; et al. Burkholderia stagnalis sp. nov. and Burkholderia territorii sp. nov., two novel Burkholderia cepacia complex species from environmental and human sources. Int. J. Syst. Evol. Microbiol. 2015, 65, 2265–2271. [Google Scholar] [CrossRef]
- Vandamme, P.; Opelt, K.; Knöchel, N.; Berg, C.; Schönmann, S.; De Brandt, E.; Eberl, L.; Falsen, E.; Berg, G. Burkholderia bryophila sp. nov. and Burkholderia megapolitana sp. nov., moss-associated species with antifungal and plant-growth-promoting properties. Int. J. Syst. Evol. Microbiol. 2007, 57, 2228–2235. [Google Scholar] [CrossRef]
- Brett, P.J.; DeShazer, D.; Woods, D.E. Note: Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 1998, 48, 317–320. [Google Scholar] [CrossRef]
- Victor, I.U.; Kwiencien, M.; Tripathi, L.; Cobice, D.; McClean, S.; Marchant, R.; Banat, I.M. Quorum sensing as a potential target for increased production of rhamnolipid biosurfactant in Burkholderia thailandensis E264. Appl. Microbiol. Biotechnol. 2019, 103, 6505–6517. [Google Scholar] [CrossRef] [PubMed]
- Yabuuchi, E.; Kosako, Y.; Oyaizu, H.; Yano, I.; Hotta, H.; Hashimoto, Y.; Ezaki, T.; Arakawa, M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group ii to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992, 36, 1251–1275. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, C.O. Paraburkholderia aromaticivorans sp. nov., an aromatic hydrocarbon-degrading bacterium, isolated from gasoline-contaminated soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 1251–1257. [Google Scholar] [CrossRef] [PubMed]
- Barberan, A. International Journal of Systematic and Evolutionary Microbiology (IJSEM) Phenotypic Database. Figshare Dataset. 2016. Available online: https://figshare.com/articles/dataset/International_Journal_of_Systematic_and_Evolutionary_Microbiology_IJSEM_phenotypic_database/4272392 (accessed on 20 January 2021). [CrossRef]
- Sessitsch, A.; Coenye, T.; Sturz, A.V.; Vandamme, P.; Barka, E.A.; Salles, J.F.; Van Elsas, J.D.; Faure, D.; Reiter, B.; Glick, B.R.; et al. Burkholderia phytofirmans sp. nov., a novel plant-associated bacterium with plant-beneficial properties. Int. J. Syst. Evol. Microbiol. 2005, 55, 1187–1192. [Google Scholar] [CrossRef]
- Gao, Z.; Yuan, Y.; Xu, L.; Liu, R.; Chen, M.; Zhang, C. Paraburkholderia caffeinilytica sp. nov., isolated from the soil of a tea plantation. Int. J. Syst. Evol. Microbiol. 2016, 66, 4185–4190. [Google Scholar] [CrossRef]
- Yang, H.-C.; Im, W.-T.; Kim, K.K.; An, D.-S.; Lee, S.-T. Burkholderia terrae sp. nov., isolated from a forest soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 453–457. [Google Scholar] [CrossRef]
- Vandamme, P.; De Brandt, E.; Houf, K.; Salles, J.F.; Van Elsas, J.D.; Spilker, T.; LiPuma, J.J. Burkholderia humi sp. nov., Burkholderia choica sp. nov., Burkholderia telluris sp. nov., Burkholderia terrestris sp. nov. and Burkholderia udeis sp. nov.: Burkholderia glathei-like bacteria from soil and rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 4707–4718. [Google Scholar] [CrossRef]
- Takeshita, K.; Tamaki, H.; Ohbayashi, T.; Meng, X.-Y.; Sone, T.; Mitani, Y.; Peeters, C.; Kikuchi, Y.; Vandamme, P. Burkholderia insecticola sp. nov., a gut symbiotic bacterium of the bean bug Riptortus pedestris. Int. J. Syst. Evol. Microbiol. 2018, 68, 2370–2374. [Google Scholar] [CrossRef]
- Draghi, W.O.; Peeters, C.; Cnockaert, M.; Snauwaert, C.; Wall, L.G.; Zorreguieta, A.; Vandamme, P. Burkholderia cordobensis sp. nov., from agricultural soils. Int. J. Syst. Evol. Microbiol. 2014, 64, 2003–2008. [Google Scholar] [CrossRef]
- Santos, P.E.-D.L.; Palmer, M.; Chávez-Ramírez, B.; Beukes, C.; Steenkamp, E.T.; Briscoe, L.; Khan, N.; Maluk, M.; Lafos, M.; Humm, E.; et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 2018, 9, 389. [Google Scholar] [CrossRef]
- Lopes-Santos, L.; Castro, D.B.A.; Ferreira-Tonin, M.; Corrêa, D.B.A.; Weir, B.S.; Park, D.; Ottoboni, L.M.M.; Neto, J.R.; Destéfano, S.A.L. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie van Leeuwenhoek 2017, 110, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Klemke, F.; Beyer, G.; Sawade, L.; Saitov, A.; Korte, T.; Maldener, I.; Lockau, W.; Nürnberg, D.; Volkmer, T. All1371 is a polyphosphate-dependent glucokinase in Anabaena sp. PCC 7120. Microbiology 2014, 160, 2807–2819. [Google Scholar] [CrossRef] [PubMed]
- Francke, C.; Postma, P.; Westerhoff, H.; Blom, J.; Peletier, M. Why the phosphotransferase system of Escherichia coli escapes diffusion limitation? Biophys. J. 2013, 85, 612–622. [Google Scholar] [CrossRef]
- Jeckelmann, J.-M.; Harder, D.; Mari, S.A.; Meury, M.; Ucurum, Z.; Muller, D.J.; Erni, B.; Fotiadis, D. Structure and function of the glucose PTS transporter from Escherichia coli. J. Struct. Biol. 2011, 176, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, R.A.; Mendonca, C.M.; Aristilde, L. A Cyclic Metabolic Network in Pseudomonas protegens Pf-5 prioritizes the Entner-Doudoroff pathway and exhibits substrate hierarchy during carbohydrate co-utilization. Appl. Environ. Microbiol. 2018, 85, e02084-18. [Google Scholar] [CrossRef] [PubMed]
- Solhtalab, M.; Karbalaei-Heidari, H.R.; Absalan, G. Tuning of hydrophilic ionic liquids concentration: A way to prevent enzyme instability. J. Mol. Catal. B Enzym. 2015, 122, 125–130. [Google Scholar] [CrossRef]
- Pastor, J.M.; Borges, N.; Pagán, J.P.; Castaño-Cerezo, S.; Csonka, L.N.; Goodner, B.W.; Reynolds, K.A.; Gonçalves, L.G.; Argandoña, M.; Nieto, J.J.; et al. Fructose metabolism in Chromohalobacter salexigens: Interplay between the Embden–Meyerhof–Parnas and Entner–Doudoroff pathways. Microb. Cell Fact. 2019, 18, 1–15. [Google Scholar] [CrossRef]
- Fuhrer, T.; Fischer, E.; Sauer, U. Experimental Identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 2005, 187, 1581–1590. [Google Scholar] [CrossRef]
- Klingner, A.; Bartsch, A.; Dogs, M.; Wagner-Döbler, I.; Jahn, D.; Simon, M.; Brinkhoff, T.; Becker, J.; Wittmann, C. Large-scale 13C flux profiling reveals conservation of the Entner-Doudoroff Pathway as a glycolytic strategy among marine bacteria that use glucose. Appl. Environ. Microbiol. 2015, 81, 2408–2422. [Google Scholar] [CrossRef]
- Jyoti, P.; Shree, M.; Joshi, C.; Prakash, T.; Ray, S.K.; Satapathy, S.S.; Masakapalli, S.K. The Entner-Doudoroff and nonoxidative pentose phosphate pathways bypass glycolysis and the oxidative pentose phosphate pathway in Ralstonia solanacearum. mSystems 2020, 5, 00091-20. [Google Scholar] [CrossRef]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Greuning, N.-M.; Krueger, A.; Alam, M.T.; et al. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; De Lorenzo, V. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef]
- Chavarría, M.; Nikel, P.I.; Pérez-Pantoja, D.; De Lorenzo, V. The Entner-Doudoroff pathway empowers Pseudomonas putida KT2440 with a high tolerance to oxidative stress. Environ. Microbiol. 2013, 15, 1772–1785. [Google Scholar] [CrossRef] [PubMed]
- Lemire, J.; AlHasawi, A.; Appanna, V.; Tharmalingam, S. Metabolic defense against oxidative stress: The road less travelled so far. J. Appl. Microbiol. 2017, 123, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Obruca, S.; Sedlacek, P.; Koller, M.; Kucera, D.; Pernicová, I. Involvement of polyhydroxyalkanoates in stress resistance of microbial cells: Biotechnological consequences and applications. Biotechnol. Adv. 2018, 36, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Ponce, B.L.; Latorre, V.K.; González, M.; Seeger, M. Antioxidant compounds improved PCB-degradation by Burkholderia xenovorans strain LB400. Enzym. Microb. Technol. 2011, 49, 509–516. [Google Scholar] [CrossRef]
- Kivisaar, M. The Effect of cellular redox status on the evolvability of new catabolic pathways. mBio 2018, 9, e01981-18. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Castro, L.; Méndez, V.; Durán, R.E.; Seeger, M. Long-chain flavodoxin FldX1 improves Paraburkholderia xenovorans LB400 tolerance to oxidative stress caused by paraquat and H2O2. PLoS ONE 2019, 14, e0221881. [Google Scholar] [CrossRef]
- Sacomboio, E.N.M.; Kim, E.Y.S.; Correa, H.L.R.; Bonato, P.; Pedrosa, F.D.O.; De Souza, E.M.; Chubatsu, L.S.; Müller-Santos, M. The transcriptional regulator NtrC controls glucose-6-phosphate dehydrogenase expression and polyhydroxybutyrate synthesis through NADPH availability in Herbaspirillum seropedicae. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Orellana, R.; Macaya, C.; Bravo, G.; Dorochesi, F.; Cumsille, A.; Valencia, R.; Rojas, C.; Seeger, M. Living at the frontiers of life: Extremophiles in chile and their potential for bioremediation. Front. Microbiol. 2018, 9, 2309. [Google Scholar] [CrossRef]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef]
- Zhao, J.; Binns, A.N. Characterization of the mmsAB-araD1 (gguABC) Genes of Agrobacterium tumefaciens. J. Bacteriol. 2011, 193, 6586–6596. [Google Scholar] [CrossRef]
- Watanabe, S.; Shimada, N.; Tajima, K.; Kodaki, T.; Makino, K. Identification and characterization of L-arabonate dehydratase, l-2-keto-3-deoxyarabonate dehydratase, and l-arabinolactonase involved in an alternative pathway of L-arabinose metabolism: Novel evolutionary insight into sugar metabolism. J. Biol. Chem. 2006, 281, 33521–33536. [Google Scholar] [CrossRef]
- Watanabe, S.; Yamada, M.; Ohtsu, I.; Makino, K. α-Ketoglutaric Semialdehyde Dehydrogenase isozymes involved in metabolic pathways of d-glucarate, d-galactarate, and hydroxy-l-proline. J. Biol. Chem. 2007, 282, 6685–6695. [Google Scholar] [CrossRef]
- Ribeiro, P.L.L.; da Silva, A.C.M.S.; Filho, J.A.M.; Druzian, J.I. Impact of different by-products from the biodiesel industry and bacterial strains on the production, composition, and properties of novel polyhydroxyalkanoates containing achiral building blocks. Ind. Crop. Prod. 2015, 69, 212–223. [Google Scholar] [CrossRef]
- Sacco, L.P.; Castellane, T.C.L.; Lopes, E.; Lemos, E.G.D.M.; Alves, L.M.C. Properties of polyhydroxyalkanoate granules and bioemulsifiers from Pseudomonas sp. and Burkholderia sp. isolates growing on glucose. Appl. Biochem. Biotechnol. 2015, 178, 990–1001. [Google Scholar] [CrossRef]
- Nomura, C.T.; Taguchi, K.; Gan, Z.; Kuwabara, K.; Tanaka, T.; Takase, K.; Doi, Y. Expression of 3-ketoacyl-acyl carrier protein Reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl. Environ. Microbiol. 2005, 71, 4297–4306. [Google Scholar] [CrossRef] [PubMed]
- Scarsdale, J.N.; Kazanina, G.; He, X.; Reynolds, K.A.; Wright, H.T. Crystal Structure of the Mycobacterium tuberculosis β-ketoacyl-acyl carrier protein synthase III. J. Biol. Chem. 2001, 276, 20516–20522. [Google Scholar] [CrossRef] [PubMed]
- Röttig, A.; Steinbüchel, A. Acyltransferases in bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 277–321. [Google Scholar] [CrossRef]
- Rehm, B.H.A.; Mitsky, T.A.; Steinbuchel, A. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by Pseudomonads: Establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl. Environ. Microbiol. 2001, 67, 3102–3109. [Google Scholar] [CrossRef]
- Zheng, Z.; Chen, J.-C.; Tian, H.-L.; Bei, F.-F.; Chen, G.-Q. Specific identification of (R)-3-hydroxyacyl-ACP: CoA transacylase gene from Pseudomonas and Burkholderia strains by polymerase chain reaction. Sheng Wu Gong Cheng Xue Bao 2005, 21, 19–24. [Google Scholar] [PubMed]
- Davis, R.; Chandrashekar, A.; Shamala, T.R. Role of (R)-specific enoyl coenzyme A hydratases of Pseudomonas sp in the production of polyhydroxyalkanoates. Antonie van Leeuwenhoek 2007, 93, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Taguchi, S.; Seiichi, T.; Doi, Y. Molecular characterization and properties of (R)-specific enoyl-CoA hydratases from Pseudomonas aeruginosa: Metabolic tools for synthesis of polyhydroxyalkanoate via fatty acid beta-oxidation. Int. J. Biol. Macromol. 2003, 31, 195–205. [Google Scholar] [CrossRef]
- Rehm, B.H.A. Genetics and biochemistry of polyhydroxyalkanoate granule self-assembly: The key role of polyester synthases. Biotechnol. Lett. 2006, 28, 207–213. [Google Scholar] [CrossRef]
- Snell, K.; Feng, F.; Zhong, L.; Martin, D.; Madison, L. YfcX enables medium-chain-length poly(3-hydroxyalkanoate) formation from fatty acids in recombinant Escherichia coli fadB strains. J. Bacteriol. 2002, 184, 5696–5705. [Google Scholar] [CrossRef]
- Fujita, Y.; Matsuoka, H.; Hirooka, K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef]
- Riedel, S.L.; Lu, J.; Stahl, U.; Brigham, C.J. Lipid and fatty acid metabolism in Ralstonia eutropha: Relevance for the biotechnological production of value-added products. Appl. Microbiol. Biotechnol. 2014, 98, 1469–1483. [Google Scholar] [CrossRef]
- Slater, S.; Houmiel, K.L.; Tran, M.; Mitsky, T.A.; Taylor, N.B.; Padgette, S.R.; Gruys, K.J. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J. Bacteriol. 1998, 180, 1979–1987. [Google Scholar] [CrossRef]
- Rand, J.M.; Pisithkul, T.; Clark, R.L.; Thiede, J.M.; Mehrer, C.R.; Agnew, D.E.; Campbell, C.E.; Markley, A.L.; Price, M.N.; Ray, J.; et al. A metabolic pathway for catabolizing levulinic acid in bacteria. Nat. Microbiol. 2017, 2, 1624–1634. [Google Scholar] [CrossRef]
- Chee, J.-Y.; Lau, N.-S.; Samian, M.-R.; Tsuge, T.; Sudesh, K. Expression of Aeromonas caviae polyhydroxyalkanoate synthase gene in Burkholderia sp. USM (JCM15050) enables the biosynthesis of SCL-MCL PHA from palm oil products. J. Appl. Microbiol. 2011, 112, 45–54. [Google Scholar] [CrossRef]
- Mezzolla, V.; D’Urso, O.F.; Poltronieri, P. Role of PhaC type i and type ii enzymes during PHA biosynthesis. Polymers 2018, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Wittenborn, E.C.; Jost, M.; Wei, Y.; Stubbe, J.; Drennan, C.L. Structure of the catalytic domain of the class i polyhydroxybutyrate synthase from Cupriavidus necator. J. Biol. Chem. 2016, 291, 25264–25277. [Google Scholar] [CrossRef] [PubMed]
- Rehm, B.H.A. Polyester synthases: Natural catalysts for plastics. Biochem. J. 2003, 376, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Gradíssimo, D.G.; Xavier, L.P.; Santos, A.V. Cyanobacterial Polyhydroxyalkanoates: A sustainable alternative in circular economy. Molecules 2020, 25, 4331. [Google Scholar] [CrossRef] [PubMed]
- Jendrossek, D.; Pfeiffer, D. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ. Microbiol. 2014, 16, 2357–2373. [Google Scholar] [CrossRef]
- Tsuge, T.; Hyakutake, M.; Mizuno, K. Class IV polyhydroxyalkanoate (PHA) synthases and PHA-producing Bacillus. Appl. Microbiol. Biotechnol. 2015, 99, 6231–6240. [Google Scholar] [CrossRef]
- Rodrigues, M.F.D.A.; Vicente, E.J.; Steinbüchel, A. Studies on polyhydroxyalkanoate (PHA) accumulation in a PHA synthase I-negative mutant of Burkholderia cepacia generated by homogenotization. FEMS Microbiol. Lett. 2000, 193, 179–185. [Google Scholar] [CrossRef][Green Version]
- A Chen, I.-M.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R.; et al. IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019, 47, D666–D677. [Google Scholar] [CrossRef]
- Tan, I.K.P.; Foong, C.P.; Tan, H.T.; Lim, H.; Zain, N.-A.A.; Tan, Y.C.; Hoh, C.C.; Sudesh, K. Polyhydroxyalkanoate (PHA) synthase genes and PHA-associated gene clusters in Pseudomonas spp. and Janthinobacterium spp. isolated from Antarctica. J. Biotechnol. 2020, 313, 18–28. [Google Scholar] [CrossRef]
- Kim, J.; Kim, Y.-J.; Choi, S.Y.; Lee, S.Y.; Kim, K.-J. Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol. J. 2017, 12, 27808482. [Google Scholar] [CrossRef]
- Hiroe, A.; Tsuge, K.; Nomura, C.T.; Itaya, M.; Tsuge, T. Rearrangement of gene order in the phaCAB operon leads to effective production of ultrahigh-molecular-weight poly[(R)-3-hydroxybutyrate] in genetically engineered Escherichia coli. Appl. Environ. Microbiol. 2012, 78, 3177–3184. [Google Scholar] [CrossRef]
- Mezzina, M.P.; Pettinari, M.J. Phasins, Multifaceted polyhydroxyalkanoate granule-associated proteins. Appl. Environ. Microbiol. 2016, 82, 5060–5067. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, S.Y. Identification and characterization of a new enoyl coenzyme a hydratase involved in biosynthesis of medium-chain-length polyhydroxyalkanoates in recombinant Escherichia coli. J. Bacteriol. 2003, 185, 5391–5397. [Google Scholar] [CrossRef]
- Wolfe, A.J. The Acetate Switch. Microbiol. Mol. Biol. Rev. 2005, 69, 12–50. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Zhang, Z.-Z.; Tan, T.-W.; Li, Z.-J. Metabolic engineering of Escherichia coli for the synthesis of polyhydroxyalkanoates using acetate as a main carbon source. Microb. Cell Fact. 2018, 17, 1–12. [Google Scholar] [CrossRef]
- Summers, M.L.; Denton, M.C.; McDermott, T.R. Genes coding for phosphotransacetylase and acetate kinase in Sinorhizobium meliloti are in an operon that is inducible by phosphate stress and controlled by phob. J. Bacteriol. 1999, 181, 2217–2224. [Google Scholar] [CrossRef]
- Morris, J.; Fane, A.; Rush, C.; Govan, B.; Mayo, M.; Currie, B.J.; Ketheesan, N. Neurotropic threat characterization of Burkholderia pseudomallei strains. Emerg. Infect. Dis. 2015, 21, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Price, E.P.; MacHunter, B.; Spratt, B.G.; Wagner, D.M.; Currie, B.J.; Sarovich, D.S. Improved multilocus sequence typing of Burkholderia pseudomallei and closely related species. J. Med. Microbiol. 2016, 65, 992–997. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Zwahlen, R.D.; Yang, P.; Van Elsas, J.D. The response of Paraburkholderia terrae strains to two soil fungi and the potential role of oxalate. Front. Microbiol. 2018, 9, 989. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Dejonghe, W.; Falsen, E.; De Clerck, E.; Geeraerts, B.; Willems, A.; Top, E.M.; Vandamme, P.; De Vos, P. Diversity of transconjugants that acquired plasmid pjp4 or pEMT1 after Inoculation of a donor strain in the a- and b-horizon of an agricultural soil and description of Burkholderia hospita sp. nov. and Burkholderia terricola sp. nov. Syst. Appl. Microbiol. 2002, 25, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Li, C.-X.; Luo, X.-J.; Lai, Q.-L.; Xu, J.-H. Burkholderia jiangsuensis sp. nov., a methyl parathion degrading bacterium, isolated from methyl parathion contaminated soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 3247–3253. [Google Scholar] [CrossRef] [PubMed]
- Baek, I.; Seo, B.; Lee, I.; Yi, H.; Chun, J. Burkholderia monticola sp. nov., isolated from mountain soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 504–509. [Google Scholar] [CrossRef]
- Pötter, M.; Madkour, M.H.; Mayer, F.; Steinbüchel, A. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 2002, 148, 2413–2426. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for ac-curate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
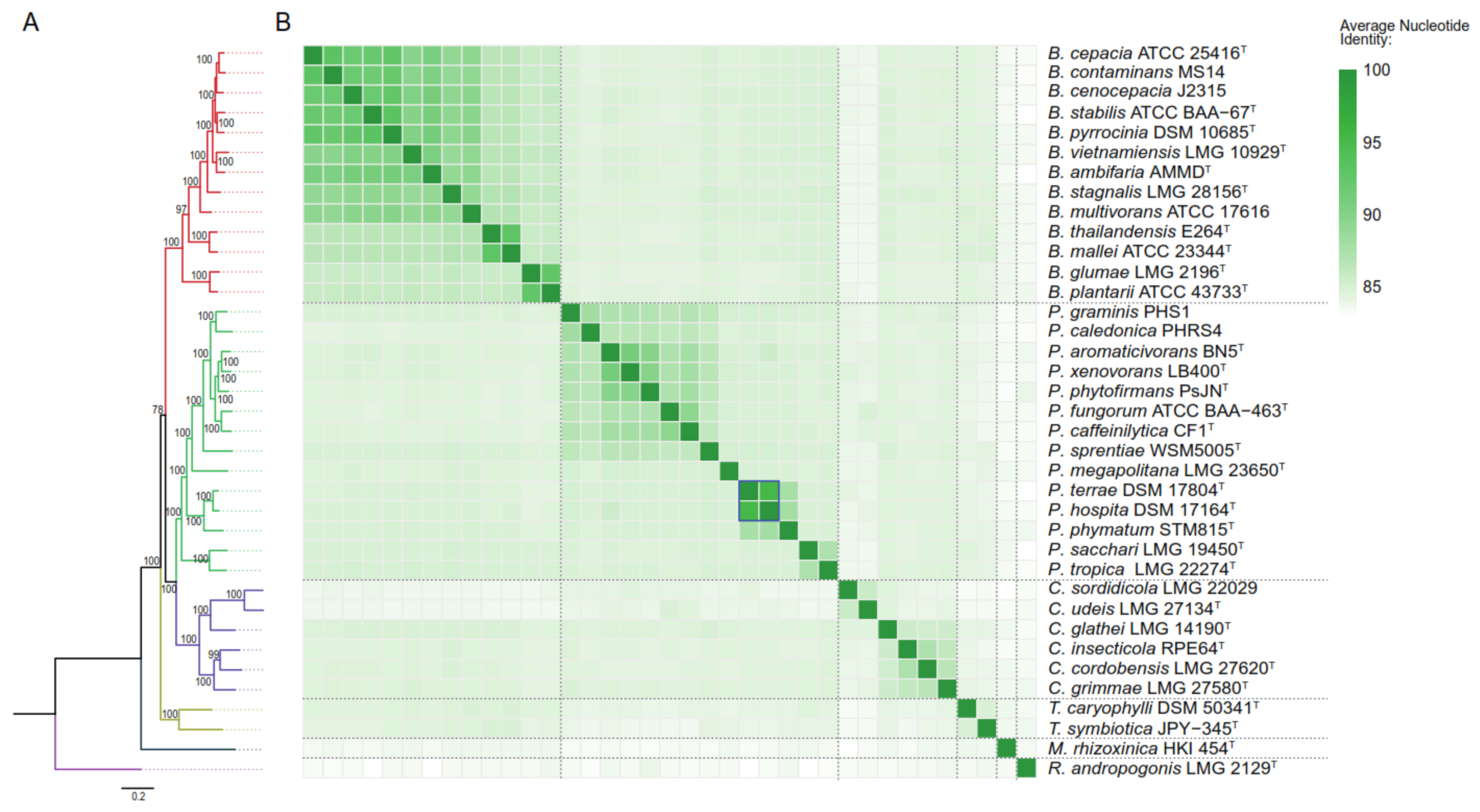





| Strain | Accession Number | Chr | Plasmids | Size (Mbp) | Contigs | CDS | G+C Content (mol%) |
|---|---|---|---|---|---|---|---|
| Burkholderia cepacia ATCC 25416T | GCA_006094315 | 3 | 2 | 8574 | 5 | 7619 | 66.59 |
| Burkholderia contaminans MS14 | GCA_001029145 | 3 | 0 | 8509 | 3 | 7494 | 66.38 |
| Burkholderia cenocepacia J2315T | GCA_902830575 | - | - | 7911 | 89 | 7105 | 66.99 |
| Burkholderia stabilis ATCC BAA-67T | GCA_001742165 | 3 | 0 | 8528 | 3 | 7552 | 66.42 |
| Burkholderia pyrrocinia DSM 10685T | GCA_001028665 | 3 | 1 | 7961 | 4 | 6920 | 66.46 |
| Burkholderia vietnamiensis LMG 10929T | GCA_902830295 | - | - | 6876 | 65 | 5397 | 66.89 |
| Burkholderia ambifaria AMMDT | GCA_000959545 | 3 | 1 | 7528 | 4 | 6548 | 66.77 |
| Burkholderia stagnalis LMG 28156T | GCA_902830275 | - | - | 8032 | 149 | 7039 | 67.23 |
| Burkholderia multivorans ATCC 17616 | GCA_000010545 | 3 | 1 | 7009 | 4 | 6262 | 66.69 |
| Burkholderia thailandensis E264T | GCA_000012365 | 2 | 0 | 6724 | 2 | 5656 | 67.73 |
| Burkholderia mallei ATCC 23344T | GCA_000011705 | 2 | 0 | 5836 | 2 | 4820 | 68.49 |
| Burkholderia glumae LMG 2196T | GCA_902832765 | - | - | 6662 | 142 | 5623 | 68.34 |
| Burkholderia plantarii ATCC 43733T | GCA_001411805 | 2 | 1 | 8081 | 3 | 6715 | 68.55 |
| Paraburkholderia graminis PHS1 | GCA_003330785 | 2 | 1 | 7508 | 3 | 6510 | 62.83 |
| Paraburkholderia caledonica PHRS4 | GCA_003330745 | 2 | 1 | 7305 | 3 | 6042 | 61.93 |
| Paraburkholderia aromaticivorans BN5T | GCA_002278075 | 2 | 6 | 8908 | 8 | 7753 | 62.94 |
| Paraburkholderia xenovorans LB400T | GCA_000756045 | 2 | 1 | 9703 | 3 | 8321 | 62.63 |
| Paraburkholderia phytofirmans PsJNT | GCA_000020125 | 2 | 1 | 8215 | 3 | 7175 | 62.29 |
| Paraburkholderia fungorum ATCC BAA-463T | GCA_000961515 | 3 | 1 | 9059 | 4 | 7898 | 61.75 |
| Paraburkholderia caffeinilytica CF1T | GCA_003368325 | 2 | 1 | 8324 | 3 | 7142 | 62.21 |
| Paraburkholderia sprentiae WSM5005T | GCA_001865575 | 2 | 3 | 7829 | 5 | 6699 | 63.21 |
| Paraburkholderia megapolitana LMG 23650T | GCA_900113825 | - | - | 7607 | 32 | 6571 | 62.07 |
| Paraburkholderia terrae DSM 17804T | GCA_002902925 | 4 | 0 | 10,062 | 4 | 8754 | 61.92 |
| Paraburkholderia hospita DSM 17164T | GCA_002902965 | 5 | 1 | 11,528 | 6 | 9975 | 61.79 |
| Paraburkholderia phymatum STM 815T | GCA_000020045 | 2 | 2 | 8676 | 4 | 7405 | 62.29 |
| Paraburkholderia sacchari LMG 19450T | GCA_000785435 * | - | - | 7318 | 21 | 6341 | 64.01 |
| Paraburkholderia tropica LMG 22274T | GCA_902833865 | - | - | 8598 | 53 | 7619 | 64.77 |
| Caballeronia sordidicola LMG 22029 | GCA_001544455 * | - | - | 6874 | 72 | 6002 | 60.15 |
| Caballeronia udeis LMG 27134T | GCA_001544555 * | - | - | 10,052 | 242 | 8774 | 60.04 |
| Caballeronia glathei LMG 14190T | GCA_902833485 | - | - | 8637 | 356 | 7660 | 64.41 |
| Caballeronia insecticola RPE64T | GCA_000402035 | 3 | 2 | 6964 | 5 | 6266 | 63.15 |
| Caballeronia cordobensis LMG 27620T | GCA_001544575 * | - | - | 8208 | 74 | 7428 | 63.69 |
| Caballeronia grimmiae LMG 27580T | GCA_000698555 | - | - | 6704 | 160 | 6024 | 63.02 |
| Trinickia caryophylli DSM 50341T | GCA_002879875 | - | - | 6581 | 158 | 5626 | 64.62 |
| Trinickia symbiotica JPY-345T | GCA_002934455 | - | - | 6714 | 62 | 5823 | 63.00 |
| Mycetohabitans rhizoxinica HKI 454T | GCA_000198775 | 1 | 2 | 3750 | 3 | 2875 | 60.71 |
| Robbsia andropogonis LMG 2129T | GCA_902833845 | - | - | 6.33 | 77 | 5183 | 58.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez-Santullano, N.; Villegas, P.; Mardones, M.S.; Durán, R.E.; Donoso, R.; González, A.; Sanhueza, C.; Navia, R.; Acevedo, F.; Pérez-Pantoja, D.; et al. Genome-Wide Metabolic Reconstruction of the Synthesis of Polyhydroxyalkanoates from Sugars and Fatty Acids by Burkholderia Sensu Lato Species. Microorganisms 2021, 9, 1290. https://doi.org/10.3390/microorganisms9061290
Alvarez-Santullano N, Villegas P, Mardones MS, Durán RE, Donoso R, González A, Sanhueza C, Navia R, Acevedo F, Pérez-Pantoja D, et al. Genome-Wide Metabolic Reconstruction of the Synthesis of Polyhydroxyalkanoates from Sugars and Fatty Acids by Burkholderia Sensu Lato Species. Microorganisms. 2021; 9(6):1290. https://doi.org/10.3390/microorganisms9061290
Chicago/Turabian StyleAlvarez-Santullano, Natalia, Pamela Villegas, Mario Sepúlveda Mardones, Roberto E. Durán, Raúl Donoso, Angela González, Claudia Sanhueza, Rodrigo Navia, Francisca Acevedo, Danilo Pérez-Pantoja, and et al. 2021. "Genome-Wide Metabolic Reconstruction of the Synthesis of Polyhydroxyalkanoates from Sugars and Fatty Acids by Burkholderia Sensu Lato Species" Microorganisms 9, no. 6: 1290. https://doi.org/10.3390/microorganisms9061290
APA StyleAlvarez-Santullano, N., Villegas, P., Mardones, M. S., Durán, R. E., Donoso, R., González, A., Sanhueza, C., Navia, R., Acevedo, F., Pérez-Pantoja, D., & Seeger, M. (2021). Genome-Wide Metabolic Reconstruction of the Synthesis of Polyhydroxyalkanoates from Sugars and Fatty Acids by Burkholderia Sensu Lato Species. Microorganisms, 9(6), 1290. https://doi.org/10.3390/microorganisms9061290