Moderate Seasonal Dynamics Indicate an Important Role for Lysogeny in the Red Sea
Abstract
1. Introduction
2. Materials and Methods
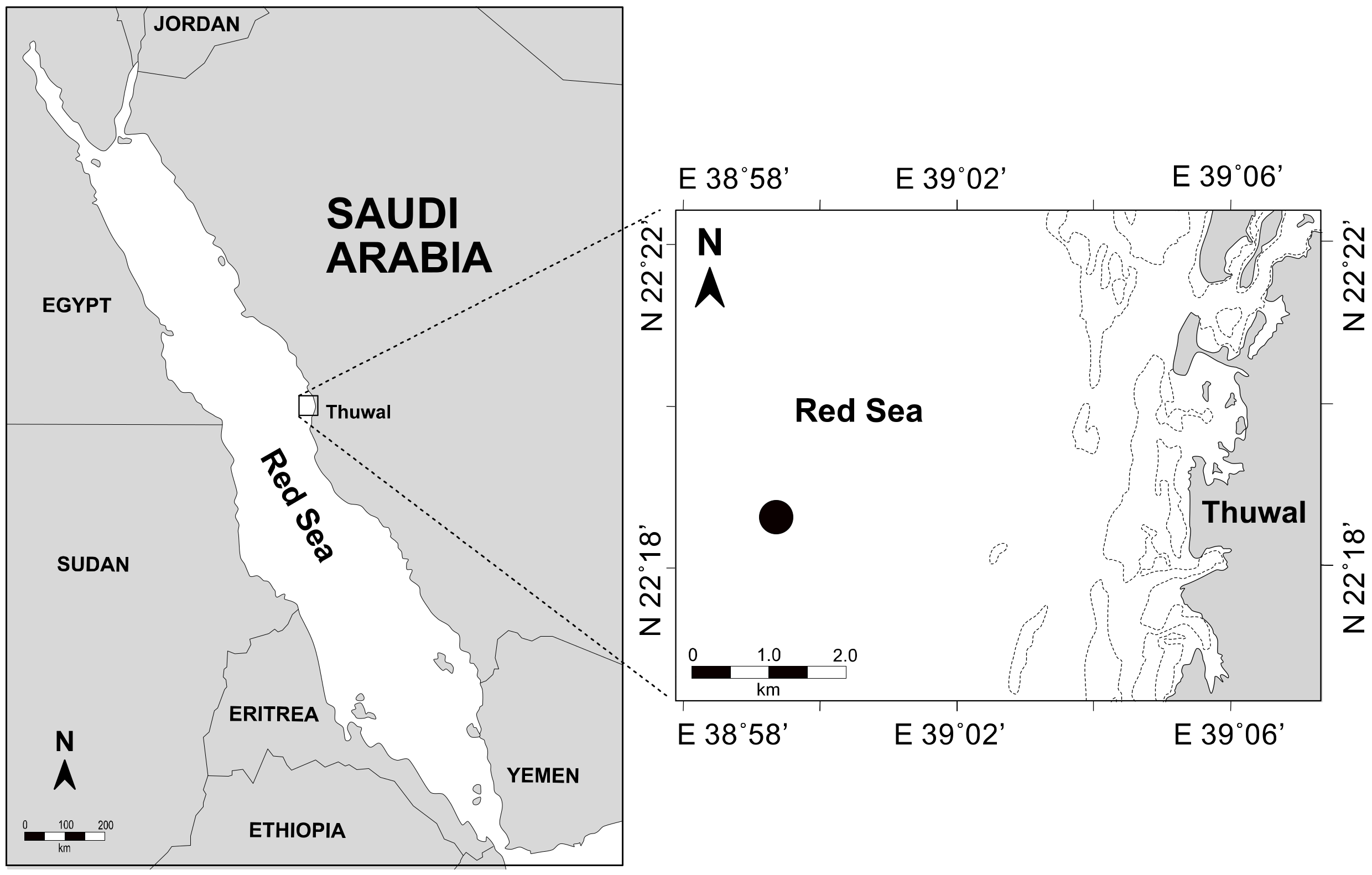
2.1. Sampling
2.2. Virus-Free Water
2.3. Incubations for Measurements of Viral Production (Lytic) and Induction from Lysogenic Phase
2.3.1. Viral Production Rates
2.3.2. Identification of Viral and Heterotrophic Bacterial Populations by Flow Cytometry (FCM)
2.3.3. Burst Size and the Percentage of Lysogenic Heterotrophic Bacteria
2.3.4. Data Analysis
3. Results
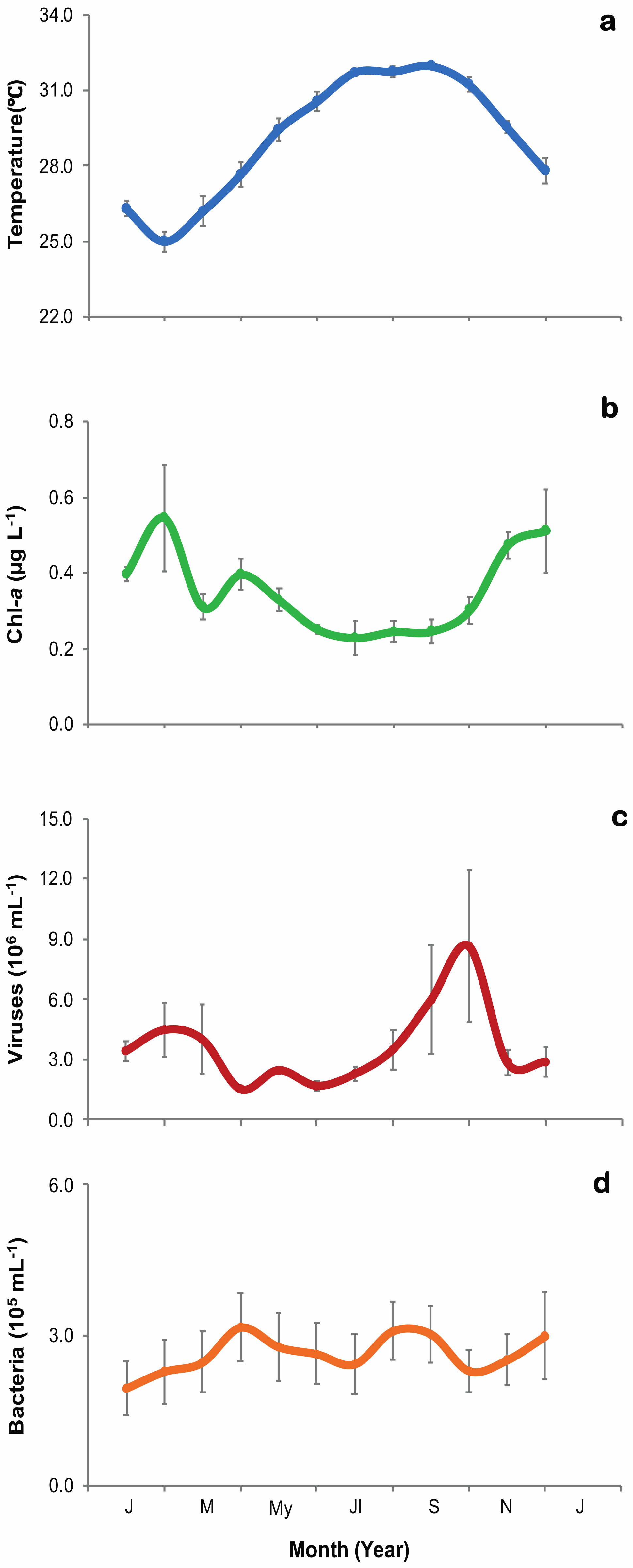
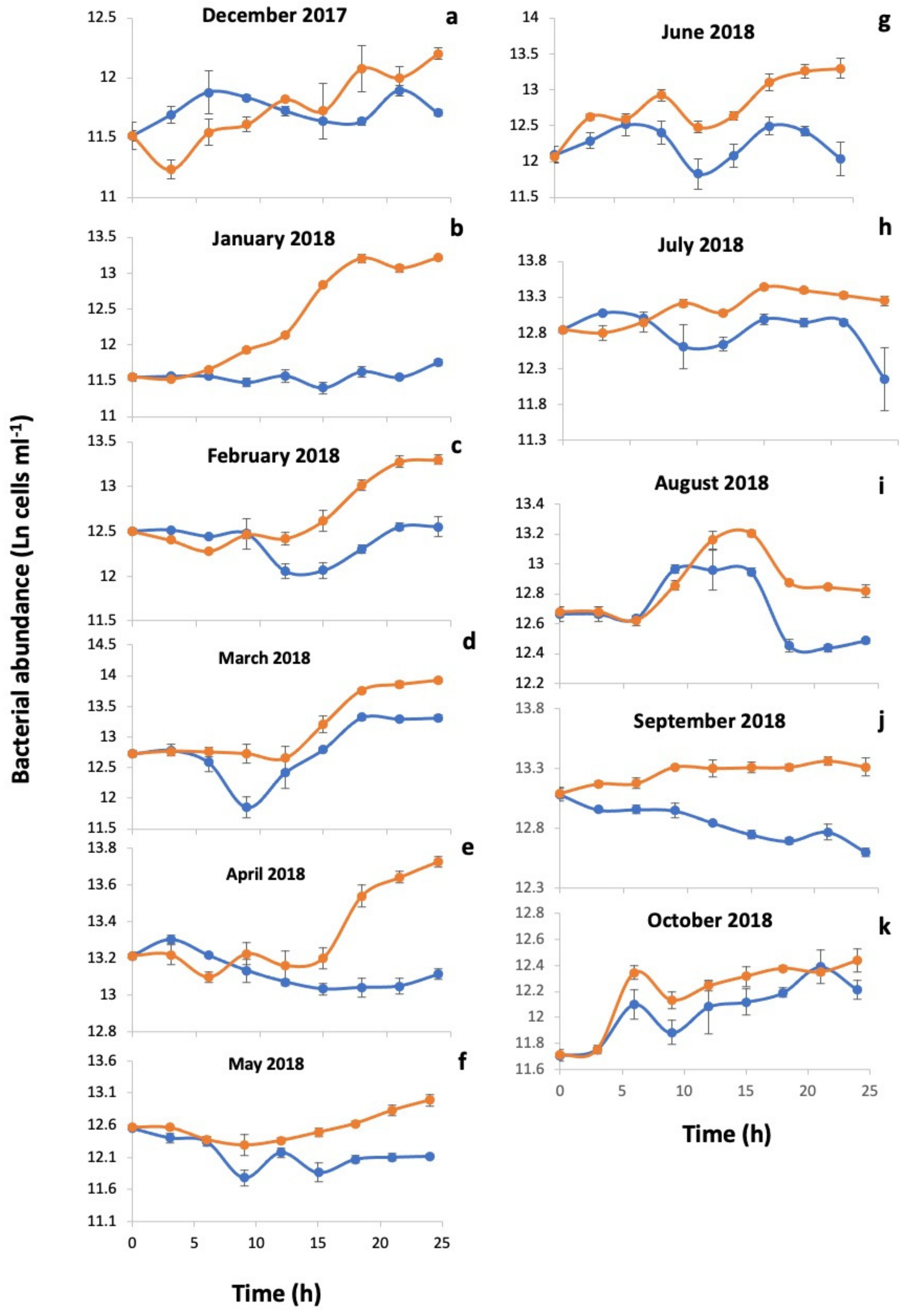
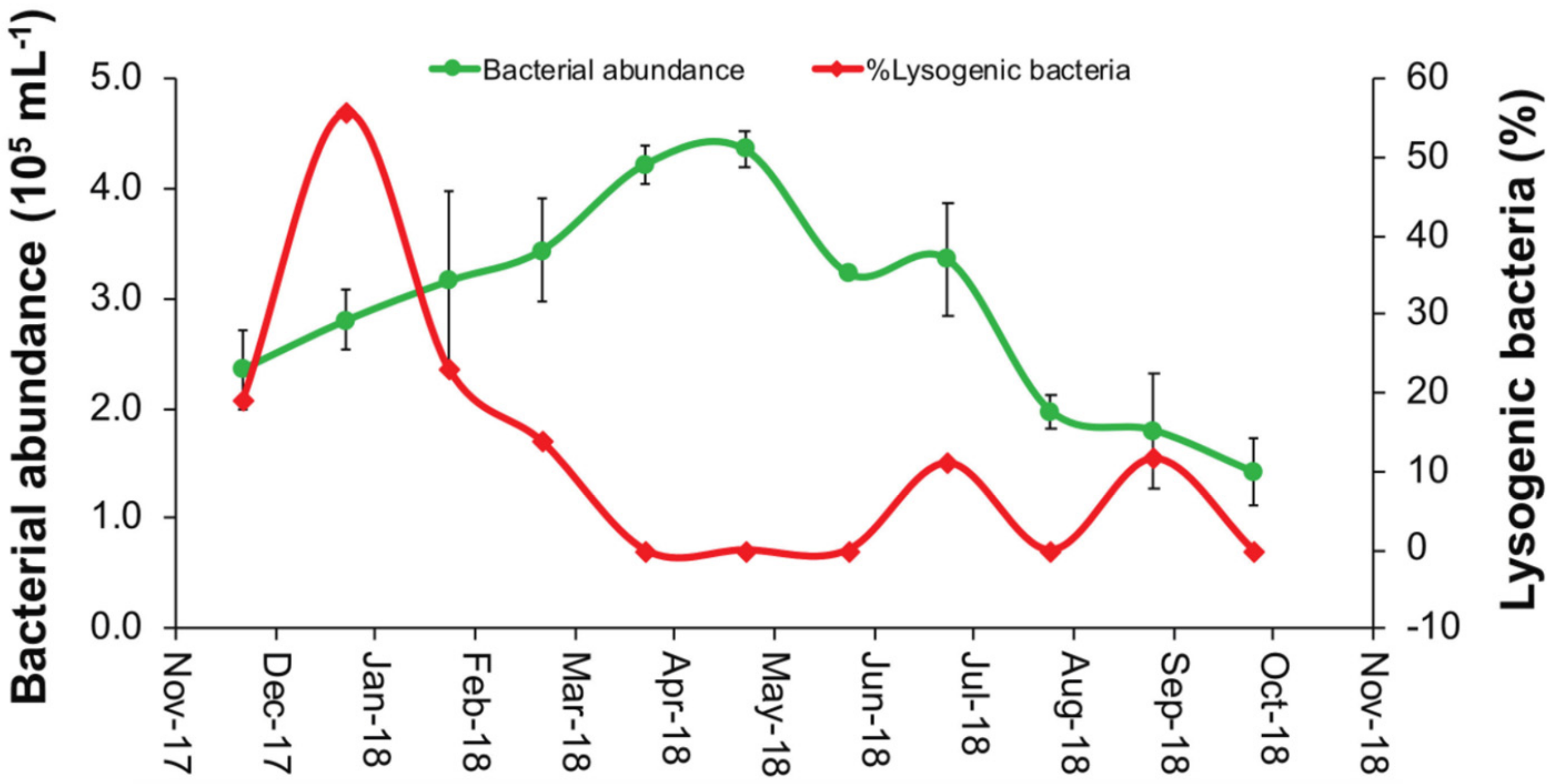
3.1. Environmental Parameters and Viral and Heterotrophic Bacterial Abundances
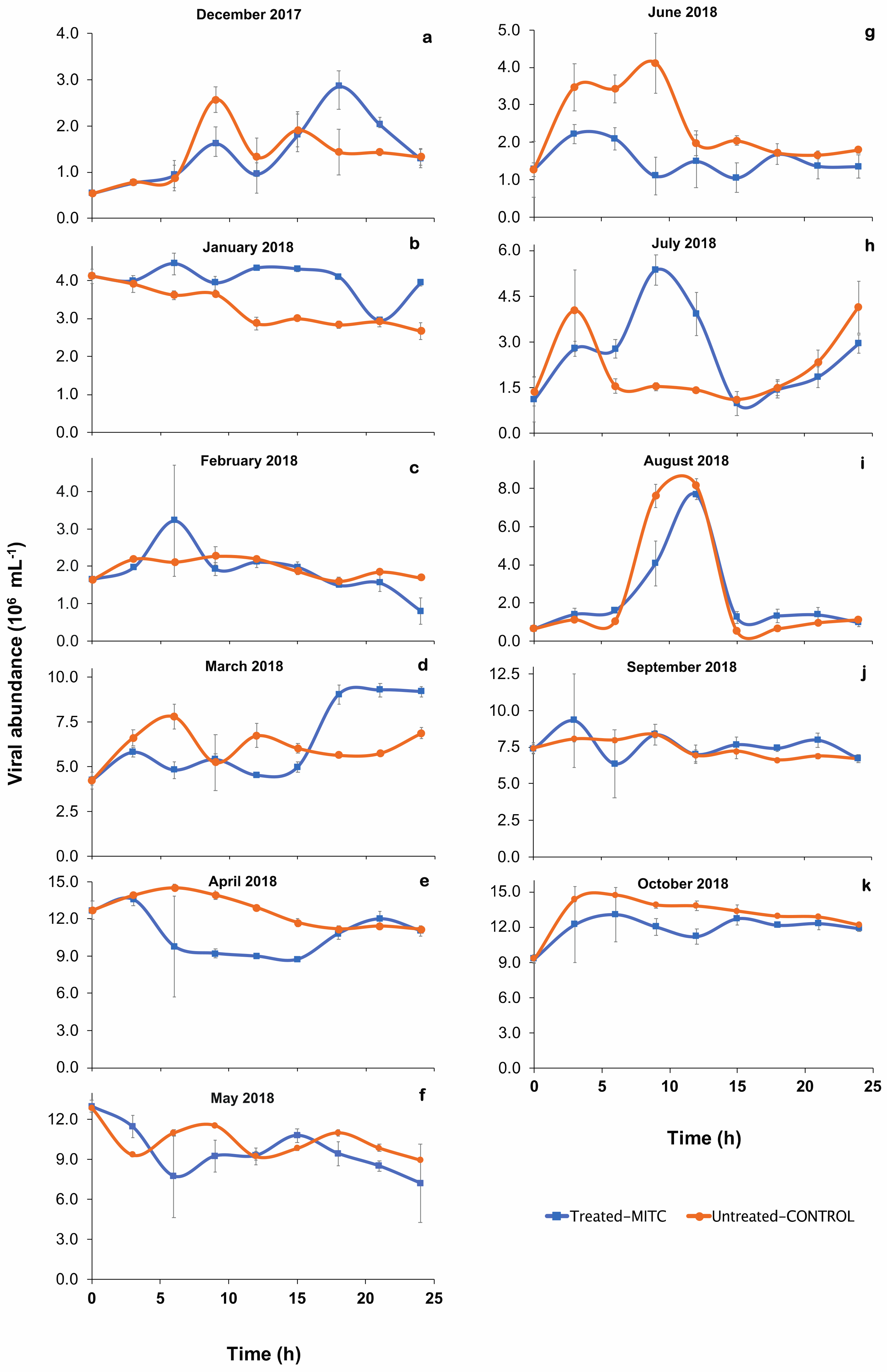
3.2. Percentage of Inducible Lysogens and Viral Production
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breitbart, M. Marine viruses: Truth or dare. Ann. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef]
- Lara, E.; Vaque, D.; Sa, E.L.; Boras, J.A.; Gomes, A.; Borrull, E.; Diez-Vives, C.; Teira, E.; Pernice, M.C.; Garcia, F.C.; et al. Unveiling the role and life strategies of viruses from the surface to the dark ocean. Sci. Adv. 2017, 3, e1602565. [Google Scholar] [CrossRef] [PubMed]
- Boras, J.A.; Sala, M.M.; Arrieta, J.M.; Sa, E.L.; Felipe, J.; Agustí, S.; Duarte, C.M.; Vaqué, D. Effect of ice melting on bacterial carbon fluxes channelled by viruses and protists in the Arctic Ocean. Polar Biol. 2010, 33, 1695–1707. [Google Scholar] [CrossRef]
- Suttle, C.A. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 1994, 28, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Suttle, C.A. Viruses and Nutrient Cycles in the Sea—Viruses play critical roles in the structure and function of aquatic food webs. Bioscience 1999, 49, 781–788. [Google Scholar] [CrossRef]
- Paul, J.H. Prophages in marine bacteria: Dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008, 2, 579–589. [Google Scholar] [CrossRef]
- Kutter, E.; Sulakvelidze, A. Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2004. [Google Scholar]
- Guttman, B.; Raya, R.; Kutter, E. Basic phage biology. In Bacteriophages: Biology and Applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, FL, USA, 2005; Volume 4, pp. 30–63. [Google Scholar]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Weitz, J.S.; Beckett, S.J.; Brum, J.R.; Cael, B.B.; Dushoff, J. Lysis, lysogeny and virus–microbe ratios. Nature 2017, 549, E1. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Williamson, S.J.; Houchin, L.A.; McDaniel, L.; Paul, J.H. Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl. Environ. Microb. 2002, 68, 4307–4314. [Google Scholar] [CrossRef]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [PubMed]
- Payet, J.P.; Suttle, C.A. To kill or not to kill: The balance between lytic and lysogenic viral infection is driven by trophic status. Limnol. Oceanogr. 2013, 58, 465–474. [Google Scholar] [CrossRef]
- Brum, J.R.; Hurwitz, B.L.; Schofield, O.; Ducklow, H.W.; Sullivan, M.B. Seasonal time bombs: Dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J. 2016, 10, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Danovaro, R.; Corinaldesi, C.; Dell’anno, A.; Fuhrman, J.A.; Middelburg, J.J.; Noble, R.T.; Suttle, C.A. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef]
- Stewart, F.M.; Levin, B.R. The Population Biology of Bacterial-Viruses—Why Be Temperate. Popul. Biol. 1984, 26, 93–117. [Google Scholar] [CrossRef]
- Knowles, B.; Silveira, C.; Bailey, B.; Barott, K.; Cantu, V.; Cobián-Güemes, A.; Coutinho, F.; Dinsdale, E.; Felts, B.; Furby, K. Lytic to temperate switching of viral communities. Nature 2016, 531, 466. [Google Scholar] [CrossRef]
- Rasul, N.M.; Stewart, I.C.; Nawab, Z.A. Introduction to the Red Sea: Its origin, structure, and environment. In The Red Sea; Springer: Berlin/Heidelberg, Germany, 2015; pp. 1–28. [Google Scholar]
- Weikert, H. Plankton and the pelagic environment. In Red Sea; Edwards, A.J., Head, S.M., Eds.; 1 November 1987; Pergamon Press: Oxford, UK, 1987; pp. 90–111. [Google Scholar]
- Raitsos, D.E.; Pradhan, Y.; Brewin, R.J.; Stenchikov, G.; Hoteit, I. Remote sensing the phytoplankton seasonal succession of the Red Sea. PLoS ONE 2013, 8, e64909. [Google Scholar] [CrossRef]
- Chaidez, V.; Dreano, D.; Agusti, S.; Duarte, C.M.; Hoteit, I. Decadal trends in Red Sea maximum surface temperature. Sci. Rep. 2017, 7, 8144. [Google Scholar] [CrossRef]
- Al-Aidaroos, A.M.; El-Sherbiny, M.M.O.; Satheesh, S.; Mantha, G.; Agusti, S.; Carreja, B.; Duarte, C.M. Strong Sensitivity of Red Sea Zooplankton to UV-B Radiation. Estuaries Coast 2015, 38, 846–853. [Google Scholar] [CrossRef]
- Abdulrahman Ashy, R.; Agustí, S. Low Host Abundance and High Temperature Determine Switching from Lytic to Lysogenic Cycles in Planktonic Microbial Communities in a Tropical Sea (Red Sea). Viruses 2020, 12, 761. [Google Scholar] [CrossRef]
- Weisse, T. The microbial loop in the Red Sea: Dynamics of pelagic bacteria and heterotrophic nanoflagellates. Mar. Ecol. Prog. Ser. 1989, 55, 241–250. [Google Scholar] [CrossRef]
- Pearman, J.K.; Ellis, J.; Irigoien, X.; Sarma, Y.V.B.; Jones, B.H.; Carvalho, S. Microbial planktonic communities in the Red Sea: High levels of spatial and temporal variability shaped by nutrient availability and turbulence. Sci. Rep. 2017, 7, 6611. [Google Scholar] [CrossRef]
- Silva, L.; Calleja, M.L.; Huete-Stauffer, T.M.; Ivetic, S.; Ansari, M.I.; Viegas, M.; Moran, X.A.G. Low Abundances but High Growth Rates of Coastal Heterotrophic Bacteria in the Red Sea. Front. Microbiol. 2018, 9, 3244. [Google Scholar] [CrossRef]
- Hadas, E.; Marie, D.; Shpigel, M.; Ilan, M. Virus predation by sponges is a new nutrient-flow pathway in coral reef food webs. Limnol. Oceanogr. 2006, 51, 1548–1550. [Google Scholar] [CrossRef]
- Danovaro, R.; Bongiorni, L.; Corinaldesi, C.; Giovannelli, D.; Damiani, E.; Astolfi, P.; Greci, L.; Pusceddu, A. Sunscreens cause coral bleaching by promoting viral infections. Environ. Health Perspect. 2008, 116, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, E.I.; Huete-Stauffer, T.M.; Calleja, M.L.L.; Silva, L.; Viegas, M.; Morán, X.A.G. Weekly variations of viruses and heterotrophic nanoflagellates and their potential impact on bacterioplankton in shallow waters of the central Red Sea. FEMS Microbiol. Ecol. 2020, 96, fiaa033. [Google Scholar] [CrossRef]
- Hevroni, G.; Flores-Uribe, J.; Béjà, O.; Philosof, A. Seasonal and diel patterns of abundance and activity of viruses in the Red Sea. Proc. Natl. Acad. Sci. USA 2020, 117, 29738. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Brigden, S.M.; Suttle, C.A. A dilution technique for the direct measurement of viral production: A comparison in stratified and tidally mixed coastal waters. Microb. Ecol. 2002, 43, 168–173. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Rowe, J.M.; Wilhelm, S. Determining rates of virus production in aquatic systems by the virus reduction approach. In Manual of Aquatic Viral Ecology; Wilhelm, S.W., Weinbauer, M.G., Suttle, C.A., Eds.; Online; ASLO: Waco, TX, USA, 2010; pp. 1–8. [Google Scholar]
- Rowe, J.M.; DeBruyn, J.M.; Poorvin, L.; LeCleir, G.R.; Johnson, Z.I.; Zinser, E.R.; Wilhelm, S.W. Viral and bacterial abundance and production in the Western Pacific Ocean and the relation to other oceanic realms. FEMS Microbiol. Ecol. 2012, 79, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G.; Suttle, C.A. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the gulf of Mexico. Appl. Environ. Microb. 1996, 62, 4374–4380. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Suttle, C.A. Lysogeny and prophage induction in coastal and offshore bacterial communities. Aquat. Microb. Ecol. 1999, 18, 217–225. [Google Scholar] [CrossRef]
- Cochran, P.K.; Paul, J.H. Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl. Environ. Microb. 1998, 64, 2308–2312. [Google Scholar] [CrossRef]
- Magiopoulos, I.; Pitta, P. Viruses in a deep oligotrophic sea: Seasonal distribution of marine viruses in the epi-, meso- and bathypelagic waters of the Eastern Mediterranean Sea. Deep Sea Res. Part II 2012, 66, 1–10. [Google Scholar] [CrossRef]
- Boras, J.A.; Sala, M.M.; Vazquez-Dominguez, E.; Weinbauer, M.G.; Vaque, D. Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ. Microbiol. 2009, 11, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Brussaard, C.P.; Payet, J.P.; Winter, C.; Weinbauer, M.G. Quantification of aquatic viruses by flow cytometry. In Manual of Aquatic Viral Ecology; Wilhelm, S.W., Weinbauer, M.G., Suttle, C.A., Eds.; ASLO: Waco, TX, USA, 2010; Volume 11, pp. 102–107. [Google Scholar]
- Gasol, J.M.; Morán, X.A.G. Flow cytometric determination of microbial abundances and its use to obtain indices of community structure and relative activity. In Hydrocarbon and Lipid Microbiology Protocols; Springer: Berlin/Heidelberg, Germany, 2015; pp. 159–187. [Google Scholar]
- Smith, R.J.; Paterson, J.S.; Sibley, C.A.; Hutson, J.L.; Mitchell, J.G. Putative Effect of Aquifer Recharge on the Abundance and Taxonomic Composition of Endemic Microbial Communities. PLoS ONE 2015, 10, e0129004. [Google Scholar] [CrossRef] [PubMed]
- Antunes, A.; Alam, I.; Simoes, M.F.; Daniels, C.; Ferreira, A.J.S.; Siam, R.; El-Dorry, H.; Bajic, V.B. First Insights into the Viral Communities of the Deep-sea Anoxic Brines of the Red Sea. Genom. Proteom. Bioinf. 2015, 13, 304–309. [Google Scholar] [CrossRef]
- Evans, C.; Brussaard, C.P. Regional variation in lytic and lysogenic viral infection in the Southern Ocean and its contribution to biogeochemical cycling. Appl. Environ. Microb. 2012, 78, 6741–6748. [Google Scholar] [CrossRef]
- Jiang, S.C.; Paul, J.H. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar. Ecol. Prog. Ser. 1996, 142, 27–38. [Google Scholar] [CrossRef]
- Wigington, C.H.; Sonderegger, D.; Brussaard, C.P.D.; Buchan, A.; Finke, J.F.; Fuhrman, J.A.; Lennon, J.T.; Middelboe, M.; Suttle, C.A.; Stock, C.; et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 2016, 1, 15024. [Google Scholar] [CrossRef]
- Jiang, S.C.; Paul, J.H. Seasonal and Diel Abundance of Viruses and Occurrence of Lysogeny/Bacteriocinogeny in the Marine-Environment. Mar. Ecol. Prog. Ser. 1994, 104, 163–172. [Google Scholar] [CrossRef]
- Winget, D.M.; Helton, R.R.; Williamson, K.E.; Bench, S.R.; Williamson, S.J.; Wommack, K.E. Repeating patterns of virioplankton production within an estuarine ecosystem. Proc. Natl. Acad. Sci. USA 2011, 108, 11506–11511. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.M.; Saxton, M.A.; Cottrell, M.T.; DeBruyn, J.M.; Berg, G.M.; Kirchman, D.L.; Hutchins, D.A.; Wilhelm, S.W. Constraints on viral production in the Sargasso Sea and North Atlantic. Aquat. Microb. Ecol. 2008, 52, 233–244. [Google Scholar] [CrossRef]
- Tapper, M.A.; Hicks, R.E. Temperate viruses and lysogeny in Lake Superior bacterioplankton. Limnol. Oceanogr. 1998, 43, 95–103. [Google Scholar] [CrossRef]
- Ortmann, A.C.; Lawrence, J.E.; Suttle, C.A. Lysogeny and lytic viral production during a bloom of the cyanobacterium Synechococcus spp. Microb. Ecol. 2002, 43, 225–231. [Google Scholar] [CrossRef]
- Long, A.; McDaniel, L.D.; Mobberley, J.; Paul, J.H. Comparison of lysogeny (prophage induction) in heterotrophic bacterial and Synechococcus populations in the Gulf of Mexico and Mississippi River plume. ISME J. 2008, 2, 132–144. [Google Scholar] [CrossRef]
- Parada, V.; Herndl, G.J.; Weinbauer, M.G. Viral burst size of heterotrophic prokaryotes in aquatic systems. J. Mar. Biol. Assoc. UK 2006, 86, 613–621. [Google Scholar] [CrossRef]
- Wilcox, R.M.; Fuhrman, J.A. Bacterial-Viruses in Coastal Seawater—Lytic Rather Than Lysogenic Production. Mar. Ecol. Prog. Ser. 1994, 114, 35–45. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Brettar, I.; Höfle, M.G. Lysogeny and virus-induced mortality of bacterioplankton in surface, deep, and anoxic marine waters. Limnol. Oceanogr. 2003, 48, 1457–1465. [Google Scholar] [CrossRef]
- Laybourn-Parry, J.; Marshall, W.A.; Madan, N.J. Viral dynamics and patterns of lysogeny in saline Antarctic lakes. Polar Biol. 2007, 30, 351–358. [Google Scholar] [CrossRef]
- McDaniel, L.; Houchin, L.A.; Williamson, S.J.; Paul, J.H. Lysogeny in marine Synechococcus. Nature 2002, 415, 496. [Google Scholar] [CrossRef]
- Cochran, P.K.; Kellogg, C.A.; Paul, J.H. Prophage induction of indigenous marine lysogenic bacteria by environmental pollutants. Mar. Ecol. Prog. Ser. 1998, 164, 125–133. [Google Scholar] [CrossRef]
- Williamson, S.; Paul, J.H. Nutrient stimulation of lytic phage production in bacterial populations of the Gulf of Mexico. Aquat. Microb. Ecol. 2004, 36, 9–17. [Google Scholar] [CrossRef]
- Winter, C.; Köstner, N.; Kruspe, C.P.; Urban, D.; Muck, S.; Reinthaler, T.; Herndl, G.J. Mixing alters the lytic activity of viruses in the dark ocean. Ecology 2018, 99, 700–713. [Google Scholar] [CrossRef]
- Yoshida-Takashima, Y.; Takaki, Y.; Shimamura, S.; Nunoura, T.; Takai, K. Genome sequence of a novel deep-sea vent epsilonproteobacterial phage provides new insight into the co-evolution of Epsilonproteobacteria and their phages. Extremophiles 2013, 17, 405–419. [Google Scholar] [CrossRef] [PubMed]
- Rastelli, E.; Corinaldesi, C.; Dell’Anno, A.; Martire, M.L.; Greco, S.; Facchini, M.C.; Rinaldi, M.; O’Dowd, C.; Ceburnis, D.; Danovaro, R. Transfer of labile organic matter and microbes from the ocean surface to the marine aerosol: An experimental approach. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chénard, C.; Wirth, J.F.; Suttle, C.A. Viruses Infecting a Freshwater Filamentous Cyanobacterium (Nostoc sp.) Encode a Functional CRISPR Array and a Proteobacterial DNA Polymerase B. mBio 2016, 7, e00667-16. [Google Scholar] [CrossRef]
- Obeng, N.; Pratama, A.A.; Elsas, J.D.V. The Significance of Mutualistic Phages for Bacterial Ecology and Evolution. Trends Microbiol. 2016, 24, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Köstner, N.; Jürgens, K.; Labrenz, M.; Herndl, G.J.; Winter, C. Uneven host cell growth causes lysogenic virus induction in the Baltic Sea. PLoS ONE 2019, 14, e0220716. [Google Scholar] [CrossRef]
| Sample | Viral Abundance (106 mL−1) (Manual) | Viral Abundance (106 mL−1) (Automatic) | HB Abundance (105 mL−1) (Manual) | HB Abundance (105 mL−1) (Automatic) |
|---|---|---|---|---|
| Sample (1) | 6.3 ± 0.1 | 5.5 ± 0.8 | 3.5 ± 0.09 | 3.3 ± 0.2 |
| Sample (2) | 6.9 ± 0.4 | 6.5 ± 0.2 | 5.1 ± 0.1 | 4.9 ± 0.1 |
| Sampling | LVP (mL−1 h−1) | Lysogenic HB (%) | Lysogenic HB * (%) | Burst Size (Phage Bacteria−1) |
|---|---|---|---|---|
| December-17 | 2.1 × 105 ± 4.7 × 104 | 19.2 ± 20.5 | 14.7 | N/A 2 |
| January-18 | N/D 1 | 55.8 ± 8 | 66.5 | N/A |
| February-18 | 6.0 × 104 ± 2.7 × 104 | 23.1 ± 8.7 | 39.4 | 0.8 ± 2.3 |
| March-18 | 6.0 × 105 ± 1.2 × 105 | 13.8 ± 1.7 | 4.2 | 35.1 ± 22 |
| April-18 | 3.1 × 105 ± 1.7 × 105 | N/D | N/D | N/A |
| May-18 | N/D | N/D | N/D | 3.9 ± 2 |
| June-18 | 2.8 × 105 ± 4.0 × 105 | N/D | N/D | N/A |
| July-18 | 3.2 × 104 ± 3.4 × 104 | 11.1 ± 5.1 | 11.0 | 4.9 ± 1.7 |
| August-18 | 7.0 × 105 ± 1.8 × 104 | N/D | N/D | 25.2 ± 7.6 |
| September-18 | 9.0 × 104 ± 1.4 × 105 | 11.7 ± 1.3 | 34.5 | N/A |
| October-18 | 9.0 × 105 ± 5.0 × 104 | N/D | N/D | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdulrahman Ashy, R.; Suttle, C.A.; Agustí, S. Moderate Seasonal Dynamics Indicate an Important Role for Lysogeny in the Red Sea. Microorganisms 2021, 9, 1269. https://doi.org/10.3390/microorganisms9061269
Abdulrahman Ashy R, Suttle CA, Agustí S. Moderate Seasonal Dynamics Indicate an Important Role for Lysogeny in the Red Sea. Microorganisms. 2021; 9(6):1269. https://doi.org/10.3390/microorganisms9061269
Chicago/Turabian StyleAbdulrahman Ashy, Ruba, Curtis A. Suttle, and Susana Agustí. 2021. "Moderate Seasonal Dynamics Indicate an Important Role for Lysogeny in the Red Sea" Microorganisms 9, no. 6: 1269. https://doi.org/10.3390/microorganisms9061269
APA StyleAbdulrahman Ashy, R., Suttle, C. A., & Agustí, S. (2021). Moderate Seasonal Dynamics Indicate an Important Role for Lysogeny in the Red Sea. Microorganisms, 9(6), 1269. https://doi.org/10.3390/microorganisms9061269






