Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Bacterial Pathogens from Bovine and Caprine Mastitis in Northern Lebanon
Abstract
1. Introduction
2. Materials and Methods
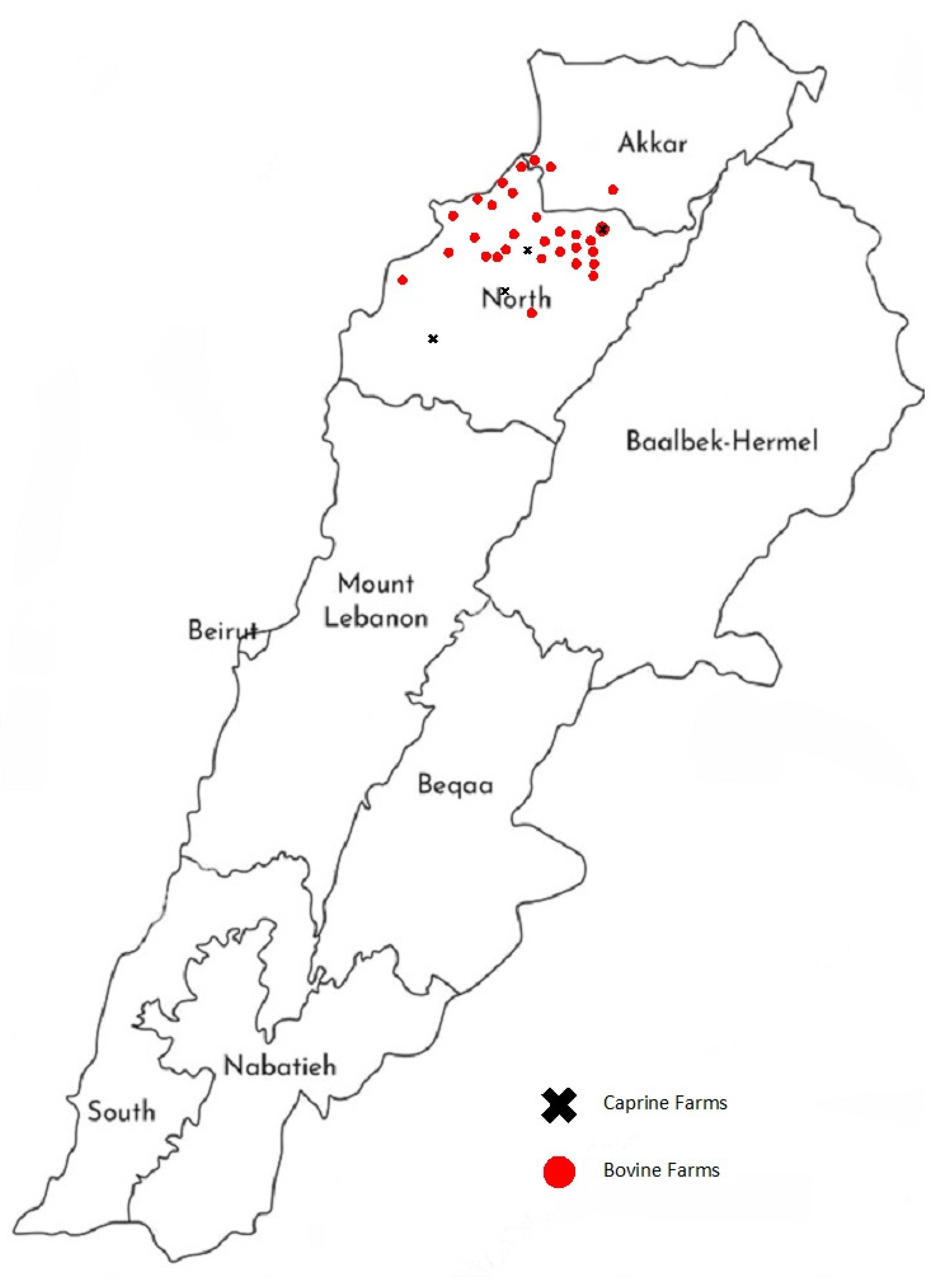
2.1. Survey Design
2.2. Sample Collection
2.3. Culturing Techniques and Species Identification
2.4. Antimicrobial Susceptibility Testing
2.5. Detection of AMR Genes
2.6. Determination of Phylogenetic Groups
2.7. Detection of Biofilm and Virulence Associated Genes of Staphylococcus Spp.
3. Results
3.1. Main Findings of the Questionnaire-Based Survey
3.2. Distribution of Pathogens
3.3. Antimicrobial Resistance
3.4. Molecular Characterization of Clinical Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Banos, G.; Bramis, G.; Bush, S.J.; Clark, E.L.; McCulloch, M.E.B.; Smith, J.; Schulze, G.; Arsenos, G.; Hume, D.A.; Psifidi, A. The genomic architecture of mastitis resistance in dairy sheep. BMC Genom. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Aghamohammadi, M.; Haine, D.; Kelton, D.F.; Barkema, H.W.; Hogeveen, H.; Keefe, G.P.; Dufour, S. Herd-Level mastitis-associated costs on canadian dairy farms. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef]
- Benić, M.; Maćešić, N.; Cvetnić, L.; Habrun, B.; Cvetnić, Ž.; Turk, R.; Đuričić, D.; Lojkić, M.; Dobranić, V.; Valpotić, H.; et al. Bovine mastitis: A persistent and evolving problem requiring novel approaches for its control—A review. Vet. Arh. 2018, 88, 535–557. [Google Scholar] [CrossRef]
- Mekonnen, S.A.; Koop, G.; Lam, T.J.G.M.; Hogeveen, H. The intention of North-Western Ethiopian dairy farmers to control mastitis. PLoS ONE 2017, 12. [Google Scholar] [CrossRef]
- Oliver, S.P.; Murinda, S.E. Antimicrobial resistance of mastitis pathogens. Vet. Clin. N. Am. Food Anim. Pract. 2012, 28, 165–185. [Google Scholar] [CrossRef]
- Lima, M.C.; de Barros, M.; Scatamburlo, T.M.; Polveiro, R.C.; de Castro, L.K.; Guimarães, S.H.S.; da Costa, S.L.; da Costa, M.M.; Moreira, M.A.S. Profiles of Staphyloccocus aureus isolated from goat persistent mastitis before and after treatment with enrofloxacin. BMC Microbiol. 2020, 20. [Google Scholar] [CrossRef]
- Merz, A.; Stephan, R.; Johler, S. Staphylococcus aureus isolates from goat and sheep milk seem to be closely related and differ from isolates detected from bovine milk. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Food and Agriculture Organization. Recovery and Rehabilitation of the Dairy Sector in Lebanon. 2016. Available online: http://www.fao.org/3/i5745e/i5745e.pdf (accessed on 29 April 2021).
- Osman, M.; Al Mir, H.; Rafei, R.; Dabboussi, F.; Madec, J.-Y.; Haenni, M.; Hamze, M. Epidemiology of antimicrobial resistance in Lebanese extra-hospital settings: An overview. J. Glob. Antimicrob. Resist. 2019, 17, 123–129. [Google Scholar] [CrossRef]
- Kassem, I.I.; Hijazi, M.A.; Saab, R. On a collision course: The availability and use of colistin-containing drugs in human therapeutics and food-animal farming in Lebanon. J. Glob. Antimicrob. Resist. 2019, 16, 162–164. [Google Scholar] [CrossRef]
- Higham, L.E.; Deakin, A.; Tivey, E.; Porteus, V.; Ridgway, S.; Rayner, A.C. A survey of dairy cow farmers in the United Kingdom: Knowledge, attitudes and practices surrounding antimicrobial use and resistance. Vet. Rec. 2018, 183, 746. [Google Scholar] [CrossRef]
- Palma, E.; Tilocca, B.; Roncada, P. Antimicrobial resistance in veterinary medicine: An overview. Int. J. Mol. Sci. 2020, 21, 1914. [Google Scholar] [CrossRef] [PubMed]
- Saleh, I.; Zouhairi, O.; Alwan, N.; Hawi, A.; Barbour, E.; Harakeh, S. Antimicrobial resistance and pathogenicity of Escherichia coli isolated from common dairy products in the Lebanon. Ann. Trop. Med. Parasitol. 2009, 103, 39–52. [Google Scholar] [CrossRef] [PubMed]
- Diab, M.; Hamze, M.; Bonnet, R.; Saras, E.; Madec, J.Y.; Haenni, M. OXA-48 and CTX-M-15 extended-spectrum beta-lactamases in raw milk in Lebanon: Epidemic spread of dominant Klebsiella pneumoniae clones. J. Med. Microbiol. 2017, 66, 1688–1691. [Google Scholar] [CrossRef] [PubMed]
- Diab, M.; Hamze, M.; Madec, J.-Y.; Haenni, M. High Prevalence of Non-ST131 CTX-M-15-Producing Escherichia coli in healthy cattle in Lebanon. Microb. Drug. Resist. 2017, 23, 261–266. [Google Scholar] [CrossRef]
- Zouhairi, O.; Saleh, I.; Alwan, N.; Toufeili, I.; Barbour, E.; Harakeh, S. Antimicrobial resistance of Staphylococcus species isolated from Lebanese dairy-based products. East. Mediterr. Health J. 2012, 16, 1221–1225. [Google Scholar] [CrossRef]
- Al Omari, S.; Al Mir, H.; Wrayde, S.; Merhabi, S.; Dhaybi, I.; Jamal, S.; Chahine, M.; Bayaa, R.; Tourba, F.; Tantawi, H.; et al. First Lebanese antibiotic awareness week campaign: Knowledge, attitudes and practices towards antibiotics. J. Hosp. Infect. 2019, 101, 475–479. [Google Scholar] [CrossRef]
- Al-Mir, H.; Osman, M.; Drapeau, A.; Hamze, M.; Madec, J.-Y.; Haenni, M. WGS analysis of clonal and plasmidic epidemiology of colistin-resistance mediated by mcr genes in the poultry sector in Lebanon. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Al-Mir, H.; Osman, M.; Azar, N.; Madec, J.Y.; Hamze, M.; Haenni, M. Emergence of clinical mcr-1-positive Escherichia coli in Lebanon. J. Glob. Antimicrob. Resist. 2019, 19, 83–84. [Google Scholar] [CrossRef]
- Kim, J.; Jeon, S.; Rhie, H.; Lee, B.; Park, M.; Lee, H.; Lee, J.; Kim, S. Rapid detection of extended spectrum β-lactamase (ESBL) for Enterobacteriaceae by use of a multiplex PCR-based method. Infect. Chemother. 2009, 41, 181–184. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups: A new E. coli phylo-typing method. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Notcovich, S.; DeNicolo, G.; Flint, S.H.; Williamson, N.B.; Gedye, K.; Grinberg, A.; Lopez-Villalobos, N. Biofilm-forming potential of Staphylococcus aureus isolated from clinical mastitis cases in New Zealand. Vet. Sci. 2018, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Cucarella, C.; Tormo, M.Á.; Úbeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, Í.; Penadés, J.R. Role of Biofilm-associated protein Bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.; Gaglio, S.; Galluzzo, P.; Cascone, G.; Piraino, C.; Di Marco Lo Presti, V.; Alduina, R. Antibiotic resistance profiling, analysis of virulence aspects and molecular genotyping of Staphylococcus aureus isolated in Sicily, Italy. Foodborne. Pathog. Dis. 2018, 15, 177–185. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- El Achkar, S.; Demanche, C.; Osman, M.; Rafei, R.; Ismail, M.B.; Yaacoub, H.; Pinçon, C.; Duthoy, S.; De Matos, F.; Gaudin, C.; et al. Drug-resistant tuberculosis, Lebanon, 2016–2017. Emerg. Infect. Dis. 2019, 25, 564–568. [Google Scholar] [CrossRef]
- Osman, M.; Al Bikai, A.; Rafei, R.; Mallat, H.; Dabboussi, F.; Hamze, M. Species distribution and antifungal susceptibility patterns of clinical Candida isolates in North Lebanon: A pilot cross-sectional multicentric study. J. Mycol. Med. 2020, 30, 100986. [Google Scholar] [CrossRef]
- Osman, M.; Bidon, B.; Abboud, C.; Zakaria, A.; Hamze, B.; Achcar, M.E.; Mallat, H.; Dannaoui, E.; Dabboussi, F.; Papon, N.; et al. Species distribution and antifungal susceptibility of Aspergillus clinical isolates in Lebanon. Future Microbiol. 2021, 16, 13–26. [Google Scholar] [CrossRef]
- Bendary, M.M.; Solyman, S.M.; Azab, M.M.; Mahmoud, N.F.; Hanora, A.M. Characterization of Methicillin Resistant Staphylococcus aureus isolated from human and animal samples in Egypt. Cell. Mol. Biol. 2016, 62, 94–100. [Google Scholar]
- El-Ashker, M.; Gwida, M.; Monecke, S.; El-Gohary, F.; Ehricht, R.; Elsayed, M.; Akinduti, P.; El-Fateh, M.; Maurischat, S. Antimicrobial resistance pattern and virulence profile of S. aureus isolated from household cattle and buffalo with mastitis in Egypt. Vet. Microbiol. 2020, 240, 108535. [Google Scholar] [CrossRef] [PubMed]
- Klibi, A.; Maaroufi, A.; Torres, C.; Jouini, A. Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int. J. Antimicrob. Agents 2018, 52, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Ben Said, M.; Abbassi, M.S.; Bianchini, V.; Sghaier, S.; Cremonesi, P.; Romano, A.; Gualdi, V.; Hassen, A.; Luini, M.V. Genetic characterization and antimicrobial resistance of Staphylococcus aureus isolated from bovine milk in Tunisia. Lett. Appl. Microbiol. 2016, 63, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Cvetnić, L.; Samardžija, M.; Habrun, B.; Kompes, G.; Benić, M. Microbiological monitoring of mastitis pathogens in the control of udder health in dairy cows. Slov. Vet. Res. 2016, 53, 131–140. [Google Scholar]
- Burović, J. Isolation of bovine clinical mastitis bacterial pathogens and their antimicrobial susceptibility in the Zenica region in 2017. Vet. Stn. 2020, 51, 47–52. [Google Scholar] [CrossRef]
- Saidi, R.; Cantekin, Z.; Mimoune, N.; Ergun, Y.; Solmaz, H.; Khelef, D.; Kaidi, R. Investigation of the presence of slime production, VanA gene and antiseptic resistance genes in Staphylococci isolated from bovine mastitis in Algeria. Vet. Stn. 2021, 52. [Google Scholar] [CrossRef]
- Marzoli, F.; Turchi, B.; Pedonese, F.; Torracca, B.; Bertelloni, F.; Cilia, G.; Cerri, D.; Fratini, F. Coagulase negative staphylococci from ovine bulk-tank milk: Effects of the exposure to sub-inhibitory concentrations of disinfectants for teat-dipping. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101656. [Google Scholar] [CrossRef]
- Turchi, B.; Bertelloni, F.; Marzoli, F.; Cerri, D.; Tola, S.; Azara, E.; Longheu, C.M.; Tassi, R.; Schiavo, M.; Cilia, G.; et al. Coagulase negative staphylococci from ovine milk: Genotypic and phenotypic characterization of susceptibility to antibiotics, disinfectants and biofilm production. Small Ruminant. Res. 2020, 183, 106030. [Google Scholar] [CrossRef]
- Zecconi, A.; dell’Orco, F.; Rizzi, N.; Vairani, D.; Cipolla, M.; Pozzi, P.; Zanini, L. Cross-sectional study on the prevalence of contagious pathogens in bulk tank milk and their effects on somatic cell counts and milk yield. Italian J. Anim. Sci. 2020, 19, 66–74. [Google Scholar] [CrossRef]
- Botrel, M.-A.; Haenni, M.; Morignat, E.; Sulpice, P.; Madec, J.-Y.; Calavas, D. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog. Dis. 2010, 7, 479–487. [Google Scholar] [CrossRef]
- Castro, A.; Pereira, J.; Amiama, C.; Lema, J. Mastitis diagnosis in ten Galician dairy herds (NW Spain) with automatic milking systems. Span. J. Agric. Res. 2015, 13, e0504. [Google Scholar] [CrossRef]
- Filioussis, G.; Kachrimanidou, M.; Christodoulopoulos, G.; Kyritsi, M.; Hadjichristodoulou, C.; Adamopoulou, M.; Tzivara, A.; Kritas, S.K.; Grinberg, A. Short communication: Bovine mastitis caused by a multidrug-resistant, mcr-1-positive (colistin-resistant), extended-spectrum β-lactamase-producing Escherichia coli clone on a Greek dairy farm. J. Dairy Sci. 2020, 103, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; de Jong, A.; Moyaert, H.; Simjee, S.; El Garch, F.; Morrissey, I.; Marion, H.; Vallé, M. Antimicrobial susceptibility monitoring of mastitis pathogens isolated from acute cases of clinical mastitis in dairy cows across Europe: VetPath results. Int J. Antimicrob. Agents 2015, 46, 13–20. [Google Scholar] [CrossRef]
- Ghatak, S.; Singha, A.; Sen, A.; Guha, C.; Ahuja, A.; Bhattacharjee, U.; Das, S.; Pradhan, N.R.; Puro, K.; Jana, C.; et al. Detection of New Delhi metallo-beta-lactamase and extended-spectrum beta-lactamase genes in Escherichia coli isolated from mastitic milk samples. Transbound. Emerg. Dis. 2013, 60, 385–389. [Google Scholar] [CrossRef]
- Su, Y.; Yu, C.-Y.; Tsai, Y.; Wang, S.-H.; Lee, C.; Chu, C. Fluoroquinolone-resistant and extended-spectrum β-lactamase-producing Escherichia coli from the milk of cows with clinical mastitis in Southern Taiwan. J. Microbiol. Immunol. Infect. 2016, 49, 892–901. [Google Scholar] [CrossRef] [PubMed]
- El Moujaber, G.; Osman, M.; Rafei, R.; Dabboussi, F.; Hamze, M. Molecular mechanisms and epidemiology of resistance in Streptococcus pneumoniae in the Middle East region. J. Med. Microbiol. 2017, 66, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Reslan, L.; Finianos, M.; Bitar, I.; Moumneh, M.B.; Araj, G.F.; Zaghlout, A.; Boutros, C.; Jisr, T.; Nabulsi, M.; Kara Yaccoub, G.; et al. The emergence of invasive Streptococcus pneumoniae serotype 24F in Lebanon: Complete genome sequencing reveals high virulence and antimicrobial resistance characteristics. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- El Ashkar, S.; Osman, M.; Rafei, R.; Mallat, H.; Achkar, M.; Dabboussi, F.; Hamze, M. Molecular detection of genes responsible for macrolide resistance among Streptococcus pneumoniae isolated in North Lebanon. J. Infect. Public Health 2017, 10, 745–748. [Google Scholar] [CrossRef]
- Tian, X.Y.; Zheng, N.; Han, R.W.; Ho, H.; Wang, J.; Wang, Y.T.; Wang, S.Q.; Li, H.G.; Liu, H.W.; Yu, Z.N. Antimicrobial resistance and virulence genes of Streptococcus isolated from dairy cows with mastitis in China. Microb. Pathog. 2019, 131, 33–39. [Google Scholar] [CrossRef]
- Mileva, R.; Karadaev, M.; Fasulkov, I.; Petkova, T.; Rusenova, N.; Vasilev, N.; Milanova, A. Oxytetracycline Pharmacokinetics After Intramuscular Administration in Cows with Clinical Metritis Associated with Trueperella pyogenes Infection. Antibiotics 2020, 9, 392. [Google Scholar] [CrossRef]
- Denamiel, G.; Llorente, P.; Carabella, M.; Rebuelto, M.; Gentilini, E. Anti-microbial susceptibility of Streptococcus spp. isolated from bovine mastitis in Argentina. J. Vet. Med. B Infect. Dis. Vet. Public Health 2005, 52, 125–128. [Google Scholar] [CrossRef]
- Petrovski, K.R.; Grinberg, A.; Williamson, N.B.; Abdalla, M.E.; Lopez-Villalobos, N.; Parkinson, T.J.; Tucker, I.G.; Rapnicki, P. Susceptibility to antimicrobials of mastitis-causing Staphylococcus aureus, Streptococcus uberis and Str. dysgalactiae from New Zealand and the USA as assessed by the disk diffusion test. Aust. Vet. J. 2015, 93, 227–233. [Google Scholar] [CrossRef]
- Osman, M.; Kamal-Dine, K.; El Omari, K.; Rafei, R.; Dabboussi, F.; Hamze, M. Prevalence of Staphylococcus aureus methicillin-sensitive and methicillin-resistant nasal carriage in food handlers in Lebanon: A potential source of transmission of virulent strains in the community. Access Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Rafei, R.; Hawli, M.; Osman, M.; Dabboussi, F.; Hamze, M. Distribution of emm types and macrolide resistance determinants among group A streptococci in the Middle East and North Africa. J. Glob. Antimicrob. Resist. 2020. [Google Scholar] [CrossRef] [PubMed]
- Rafei, R.; Hawli, M.; Osman, M.; Khelissa, S.; Salloum, T.; Dabboussi, F.; Tokajian, S.; Hamze, M. Molecular epidemiology of nonpharyngeal group A streptococci isolates in northern Lebanon. Future Microbiol. 2020, 15, 1555–1569. [Google Scholar] [CrossRef]
- Moghnieh, R.; Araj, G.F.; Awad, L.; Daoud, Z.; Mokhbat, J.E.; Jisr, T.; Abdallah, D.; Azar, N.; Irani-Hakimeh, N.; Balkis, M.M.; et al. A compilation of antimicrobial susceptibility data from a network of 13 Lebanese hospitals reflecting the national situation during 2015–2016. Antimicrob. Resist. Infect. Control. 2019, 8, 41. [Google Scholar] [CrossRef]
- Cvetnić, L.; Samardžija, M.; Duvnjak, S.; Habrun, B.; Cvetnić, M.; Jaki Tkalec, V.; Đuričić, D.; Benić, M. Multi Locus Sequence Typing and spa typing of Staphylococcus aureus isolated from the milk of cows with subclinical mastitis in Croatia. Microorganisms 2021, 9, 725. [Google Scholar] [CrossRef]
- Elias, L.; Balasubramanyam, A.S.; Ayshpur, O.Y.; Mushtuk, I.U.; Sheremet, N.O.; Gumeniuk, V.V.; Musser, J.M.B.; Rogovskyy, A.S. Antimicrobial susceptibility of Staphylococcus aureus, Streptococcus agalactiae, and Escherichia coli isolated from mastitic dairy cattle in Ukraine. Antibiotics 2020, 9, 469. [Google Scholar] [CrossRef]
- Hmede, Z.; Kassem, I.I. The colistin resistance gene mcr-1 is prevalent in commensal Escherichia coli isolated from preharvest poultry in lebanon. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Coura, F.M.; Diniz Sde, A.; Silva, M.X.; Mussi, J.M.; Barbosa, S.M.; Lage, A.P.; Heinemann, M.B. Phylogenetic Group Determination of Escherichia coli isolated from animals samples. Sci. World J. 2015, 2015, 258424. [Google Scholar] [CrossRef]
- Kempf, F.; Slugocki, C.; Blum, S.E.; Leitner, G.; Germon, P. Genomic comparative study of bovine mastitis Escherichia coli. PLoS ONE 2016, 11, e0147954. [Google Scholar] [CrossRef]
- Ghanbarpour, R.; Oswald, E. Phylogenetic distribution of virulence genes in Escherichia coli isolated from bovine mastitis in Iran. Res. Vet. Sci. 2010, 88, 6–10. [Google Scholar] [CrossRef]
- Tomazi, T.; Coura, F.M.; Gonçalves, J.L.; Heinemann, M.B.; Santos, M.V. Antimicrobial susceptibility patterns of Escherichia coli phylogenetic groups isolated from bovine clinical mastitis. J. Dairy Sci. 2018, 101, 9406–9418. [Google Scholar] [CrossRef]
- Nuesch-Inderbinen, M.; Kappeli, N.; Morach, M.; Eicher, C.; Corti, S.; Stephan, R. Molecular types, virulence profiles and antimicrobial resistance of Escherichia coli causing bovine mastitis. Vet. Rec. Open 2019, 6, e000369. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Z.; Huang, C.; Gao, X.; Wang, Z.; Liu, Y.; Tian, C.; Hong, W.; Niu, S.; Liu, M. The phylogenetic group, antimicrobial susceptibility, and virulence genes of Escherichia coli from clinical bovine mastitis. J. Dairy Sci. 2018, 101, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Todorovic, D.; Velhner, M.; Grego, E.; Vidanovic, D.; Milanov, D.; Krnjaic, D.; Kehrenberg, C. Molecular characterization of multidrug-resistant Escherichia coli isolates from bovine clinical mastitis and pigs in the Vojvodina province, Serbia. Microb. Drug Resist. 2018, 24, 95–103. [Google Scholar] [CrossRef]
- Keane, O.M. Genetic diversity, the virulence gene profile and antimicrobial resistance of clinical mastitis-associated Escherichia coli. Res. Microbiol. 2016, 167, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, S.; Shang, X.; Wang, L.; Li, H.; Wang, X. Characteristics of quinolone-resistant Escherichia coli isolated from bovine mastitis in China. J. Dairy Sci. 2018, 101, 6244–6252. [Google Scholar] [CrossRef] [PubMed]
- Szweda, P.; Schielmann, M.; Milewski, S.; Frankowska, A.; Jakubczak, A. Biofilm production and presence of ica and bap genes in Staphylococcus aureus strains isolated from cows with mastitis in the eastern Poland. Pol. J. Microbiol. 2012, 61, 65–69. [Google Scholar] [CrossRef]
- Prenafeta, A.; Sitjà, M.; Holmes, M.A.; Paterson, G.K. Short communication: Biofilm production characterization of mecA and mecC methicillin-resistant Staphylococcus aureus isolated from bovine milk in Great Britain. J. Dairy Sci. 2014, 97, 4838–4841. [Google Scholar] [CrossRef]
- Fitzgerald, J.R.; Monday, S.R.; Foster, T.J.; Bohach, G.A.; Hartigan, P.J.; Meaney, W.J.; Smyth, C.J. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 2001, 183, 63–70. [Google Scholar] [CrossRef]
| Bacterial Species | Animal Origin | Status of Infection | Percentage (%) | ||
|---|---|---|---|---|---|
| Cow | Goat | Clinical | Subclinical | ||
| Streptococcus uberis | 14 | 14 | 19.2 | ||
| Streptococcus agalactiae | 11 | 11 | 15.1 | ||
| Escherichia coli | 7 | 2 | 9 | 12.3 | |
| Staphylococcus aureus | 7 | 1 | 8 | 11 | |
| Trueperella pyogenes | 5 | 2 | 7 | 9.6 | |
| Aerococcus viridans | 4 | 4 | 5.5 | ||
| Raoultella ornithinolytica | 3 | 3 | 4.1 | ||
| Streptococcus dysgalactiae | 2 | 2 | 2.7 | ||
| Streptococcus pluranimalium | 1 | 1 | 1 | 1 | 2.7 |
| Corynebacterium bovis | 2 | 2 | 2.7 | ||
| Corynebacterium xerosis | 2 | 2 | 2.7 | ||
| Pseudomonas aeruginosa | 2 | 2 | 2.7 | ||
| Staphylococcus caprae | 1 | 1 | 1.4 | ||
| Staphylococcus haemolyticus | 1 | 1 | 1.4 | ||
| Pantoea agglomerans | 1 | 1 | 1.4 | ||
| Pasteurella multocida | 1 | 1 | 1.4 | ||
| Klebsiella oxytoca | 1 | 1 | 1.4 | ||
| Serratia marcescens | 1 | 1 | 1.4 | ||
| Streptococcus pneumoniae | 1 | 1 | 1.4 | ||
| Total | 65 | 8 | 66 | 7 | 100% |
| N | PEN ‡ | AMP ‡ | XNL ‡ | GEN ‡ | ERY ‼ | CLI ‡ | OXY ‡ | CTET ‡ | CHL ‼ | FFN ‡ | ENR ‡ | SXT ‡ | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | 9 | - | 100 | 11.1 | 11.1 | - | - | 77.8 | 88.9 | 0 | 55.6 | 22.2 | 44.4 |
| Klebsiella oxytoca | 1 | - | 100 | 0 | 0 | - | - | 100 | 100 | 0 | 0 | 0 | 0 |
| Pantoea agglomerans | 1 | - | 0 | 0 | 0 | - | - | 0 | 100 | 0 | 0 | 100 | 0 |
| Serratia marcescens | 1 | - | 100 | 0 | 0 | - | - | 100 | 100 | 0 | 100 | 0 | 0 |
| Raoultella ornithinolytica | 3 | - | 100 | 0 | 0 | - | - | 0 | 100 | 0 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 2 | - | - | - | 0 | - | - | - | - | - | - | 50 | - |
| Staphylococcus aureus | 8 | 62.5 | 50 | - | 12.5 | 0 | 0 | 12.5 | 12.5 | 0 | - | 0 | 0 |
| Staphylococcus caprae | 1 | 100 | 100 | - | 0 | 0 | 0 | 0 | 100 | 0 | - | 0 | 0 |
| Staphylococcus haemolyticus | 1 | 100 | 100 | - | 0 | 0 | 0 | 100 | 100 | 0 | - | 0 | 0 |
| Streptococcus uberis | 14 | 14.3 | 28.6 | 14.3 | - | 71.4 | 100 | 92.9 | 92.9 | 42.9 | - | 50 | 0 |
| Streptococcus agalactiae | 10 | 0 | 0 | 0 | - | 81.8 | 100 | 100 | 100 | 18.2 | - | 100 | 0 |
| Streptococcusdysgalactiae | 2 | 0 | 0 | 0 | - | 50 | 50 | 50 | 50 | 50 | - | 0 | 0 |
| Isolate | Enterotoxins | Exfoliative Toxins and Toxic Shock Syndrome Toxin-1 | Methicillin Resistance | Biofilm Formation |
|---|---|---|---|---|
| S. aureus AL 081 | mecA | ica operon | ||
| S. aureus AL 084 | sei, seg | mecA | ica operon | |
| S. aureus AL 085 | ica operon | |||
| S. aureus AL 086 | ica operon | |||
| S. aureus AL 087 | ica operon | |||
| S. aureus AL 088 | ica operon | |||
| S. aureus AL 089 | sec | tsst-1 | ica operon | |
| S. aureus AL 090 | ica operon | |||
| S. haemolyticus AL 091 | sea, sej, sep | mecA | ||
| S. caprae AL 092 | seb |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abboud, Z.; Galuppo, L.; Tolone, M.; Vitale, M.; Puleio, R.; Osman, M.; Loria, G.R.; Hamze, M. Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Bacterial Pathogens from Bovine and Caprine Mastitis in Northern Lebanon. Microorganisms 2021, 9, 1148. https://doi.org/10.3390/microorganisms9061148
Abboud Z, Galuppo L, Tolone M, Vitale M, Puleio R, Osman M, Loria GR, Hamze M. Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Bacterial Pathogens from Bovine and Caprine Mastitis in Northern Lebanon. Microorganisms. 2021; 9(6):1148. https://doi.org/10.3390/microorganisms9061148
Chicago/Turabian StyleAbboud, Zahie, Lucia Galuppo, Marco Tolone, Maria Vitale, Roberto Puleio, Marwan Osman, Guido Ruggero Loria, and Monzer Hamze. 2021. "Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Bacterial Pathogens from Bovine and Caprine Mastitis in Northern Lebanon" Microorganisms 9, no. 6: 1148. https://doi.org/10.3390/microorganisms9061148
APA StyleAbboud, Z., Galuppo, L., Tolone, M., Vitale, M., Puleio, R., Osman, M., Loria, G. R., & Hamze, M. (2021). Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Bacterial Pathogens from Bovine and Caprine Mastitis in Northern Lebanon. Microorganisms, 9(6), 1148. https://doi.org/10.3390/microorganisms9061148









