From the Farms to the Dining Table: The Distribution and Molecular Characteristics of Antibiotic-Resistant Enterococcus spp. in Intensive Pig Farming in South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Clearance
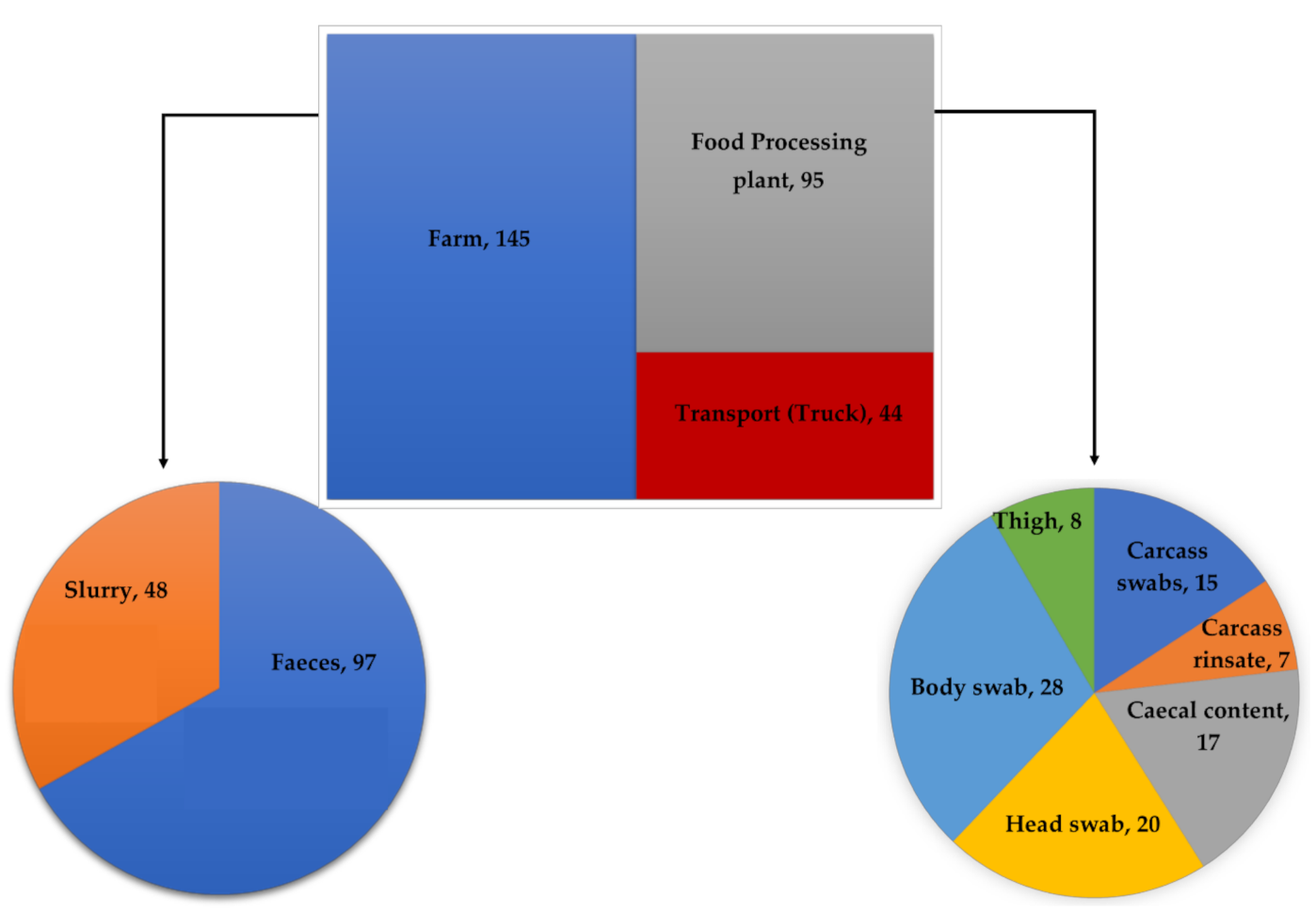
2.2. Study Population and Sampling Strategy
2.3. Isolation and Purification of Enterococcus spp.
2.4. Molecular Confirmation and Speciation of Enterococcus spp.
2.5. Antimicrobial Susceptibility Testing
2.6. Molecular Detection of Antibiotic Resistance and Virulence Genes
2.7. Clonality
3. Results
3.1. Prevalence of Enterococcus spp.
3.2. Antibiotic Susceptibility Profile of Isolates
3.3. Prevalence of Antibiotic Resistance Genes
3.4. Detection of Virulence of Factors
3.5. Clonal Relatedness
4. Discussion
4.1. Isolation and Identification of Enterococcus spp.
4.2. Antibiotic Susceptibility Profiles and Detection of Antibiotic Resistance Genes
4.3. Determination of Virulence Potentials of Enterococcus spp.
4.4. Clonal Relatedness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 18 February 2021).
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef] [PubMed]
- FAO. The FAO Action Plan on Antimicrobial Resistance; Food and Agricultural Organization: Rome, Italy, 2016. [Google Scholar]
- Doyle, M.P.; Busta, F.; Cords, B.R.; Davidson, P.M.; Hawke, J.; Hurd, H.S.; Isaacson, R.E.; Matthews, K.; Maurer, J.; Meng, J.; et al. Antimicrobial resistance: Implications for the food system: An expert report, funded by the IFT Foundation. Compr. Rev. Food Sci. Food Saf. 2006, 5, 71–137. [Google Scholar]
- You, Y.; Silbergeld, E.K. Learning from agriculture: Understanding low-dose antimicrobials as drivers of resistome expansion. Front. Microbiol. 2014, 5, 284. [Google Scholar] [PubMed]
- Landers, T.F.; Cohen, B.; Wittum, T.E.; Larson, E.L. A review of antibiotic use in food animals: Perspective, policy, and potential. Public Health Rep. 2012, 127, 4–22. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Fisher, K.; Phillips, C. The ecology, epidemiology and virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef] [PubMed]
- WHO. Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach: Guidance from the WHO Advisory Group on Integrated Surveillanec of Antimicrobial Resistance (AGISAR); World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Kaye, K.S.; Engemann, J.J.; Fraimow, H.S.; Abrutyn, E.; Kaye, K.S. Pathogens resistant to antimicrobial agents: Epidemiology, molecular mechanisms, and clinical management. Infect. Dis. Clin. N. Am. 2004, 18, 467–511. [Google Scholar] [CrossRef]
- Torres, C.; Alonso, C.A.; Ruiz-Lipa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Quiloan, M.L.G.; Vu, J.; Carvalho, J. Enterococcus faecalis can be distinguished from Enterococcus faecium via differential susceptibility to antibiotics and growth and fermentation characteristics on mannitol salt agar. Front. Biol. 2012, 7, 167–177. [Google Scholar] [CrossRef]
- Abdalla, S.E.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From Farm-to-Fork: E. coli from an intensive pig production system in South Africa shows high resistance to critically important antibiotics for human and animal use. Antibiotics 2021, 10, 178. [Google Scholar] [CrossRef]
- Abia, A.L.K.; Ubomba-Jaswa, E.; Momba, M.N.B. Impact of seasonal variation on Escherichia coli concentrations in the riverbed sediments in the Apies River, South Africa. Sci. Total Environ. 2015, 537, 462–469. [Google Scholar] [CrossRef]
- Molechan, C.; Amoako, D.G.; Abia, A.L.K.; Somboro, A.M.; Bester, L.A.; Essack, S.Y. Molecular epidemiology of antibiotic-resistant Enterococcus spp. from the farm-to-fork continuum in intensive poultry production in KwaZulu-Natal, South Africa. Sci. Total Environ. 2019, 692, 868–878. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing, Breakpoint tables for interpretation of MICs and zone diameters. Version 8. 2017. Available online: www.eucast.org/clinical_breakpoints/ (accessed on 4 February 2018).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 27th Edition Informational Supplement M100-S127; Clinical and Laboratory Standards Institute: Wayne, NJ, USA, 2017. [Google Scholar]
- Aarestrup, F.M.; Agerso, Y.; Gerner-Smidt, P.; Madsen, M.; Jensen, L.B. Comparison of antimicrobial resistance phenotypes and resistance genes in Enterococcus faecalis and Enterococcus faecium from humans in the community, broilers, and pigs in Denmark. Diagn. Microbiol. Infect. Dis. 2000, 37, 127–137. [Google Scholar] [CrossRef]
- Šeputiene, V.; Bogdaite, A.; Ružauskas, M.; Sužiedeliene, E. Antibiotic resistance genes and virulence factors in Enterococcus faecium and Enterococcus faecalis from diseased farm animals: Pigs, cattle and poultry. Pol. J. Vet. Sci. 2012, 15, 431–438. [Google Scholar]
- Tan, S.C.; Chong, C.W.; Teh, C.S.J.; Ooi, P.T.; Thong, K.L. Occurrence of virulent multidrug-resistant Enterococcus faecalis and Enterococcus faecium in the pigs, farmers and farm environments in Malaysia. PeerJ 2018, 6, e5353. [Google Scholar] [CrossRef]
- de Jong, A.; Simjee, S.; Rose, M.; Moyaert, H.; El Garch, F.; Youala, M.; Marion, O.; Lin, D.; Filip, B.; Mireille, B.; et al. Antimicrobial resistance monitoring in commensal enterococci from healthy cattle, pigs and chickens across Europe during 2004–14 (EASSA Study). J. Antimicrob. Chemother. 2019, 74, 921–930. [Google Scholar] [CrossRef]
- Hwang, I.Y.; Lim, S.K.; Ku, H.O.; Park, C.K.; Jung, S.C.; Park, Y.H.; Nam, H.M. Occurrence of virulence determinants in fecal Enterococcus faecalis isolated from pigs and chickens in Korea. J. Microbiol. Biotechnol. 2011, 21, 1352–1355. [Google Scholar] [CrossRef]
- Novais, C.; Freitas, A.R.; Silveira, E.; Antunes, P.; Silva, R.; Coque, T.M.; Peixe, L. Spread of multidrug-resistant Enterococcus to animals and humans: An underestimated role for the pig farm environment. J. Antimicrob. Chemother. 2013, 68, 2746–2754. [Google Scholar] [CrossRef]
- de Jong, A.; Simjee, S.; Garch, F.E.; Moyaert, H.; Rose, M.; Youala, M.; Dry, M. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics. Vet. Microbiol. 2018, 216, 168–175. [Google Scholar] [CrossRef]
- Beshiru, A.; Igbinosa, I.H.; Omeje, F.I.; Ogofure, A.G.; Eyong, M.M.; Igbinosa, E.O. Multi-antibiotic resistant and putative virulence gene signatures in Enterococcus species isolated from pig farms environment. Microb. Pathog. 2017, 104, 90–96. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Virulence and antimicrobial resistance factors of Enterococcus spp. isolated from fecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol. 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and acquired resistance mechanisms in Enterococcus. Virulence 2012, 3, 421–569. [Google Scholar] [CrossRef]
- Eagar, H.; Swan, G.; van Vuuren, M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J. S. Afr. Vet. Assoc. 2012, 83, 16. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, H.; Wang, Z. Distribution of acquired antibiotic resistance genes among Enterococcus spp. isolated from a hospital in Baotou, China. BMC Res. Notes 2019, 12, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Ayeni, F.A.; Odumosu, B.T.; Oluseyi, A.E.; Ruppitsch, W. Identification and prevalence of tetracycline resistance in enterococci isolated from poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 2016, 8, 69–73. [Google Scholar] [CrossRef]
- Kim, Y.B.; Seo, K.W.; Jeon, H.Y.; Lim, S.K.; Sung, H.W.; Lee, Y.J. Molecular characterization of erythromycin and tetracycline-resistant Enterococcus faecalis isolated from retail chicken meats. Poult. Sci. 2019, 98, 977–983. [Google Scholar] [CrossRef]
- Van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal infection—treatment and antibiotic resistance. In Enterococci from Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Isnard, C.; Malbruny, B.; Leclercq, R.; Cattoira, V. Genetic basis for in vitro and in vivo resistance to lincosamides, streptogramins a, and pleuromutilins (LSAP Phenotype) in Enterococcus faecium. Antimicrob. Agents Chemother. 2013, 57, 4463–4469. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO List of Critically Important Antimicrobials for Human Medicine (WHO CIA List); World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Hasannejad Bibalan, M.; Eshaghi, M.; Sadeghi, J.; Asadian, M.; Narimani, T.; Talebi, M. Clonal diversity in multi drug resistant (MDR) enterococci isolated from fecal normal flora. Int. J. Mol. Cell. Med. 2015, 4, 240–244. [Google Scholar] [PubMed]
- Diarra, M.S.; Rempel, H.; Champagne, J.; Masson, L.; Pritchard, J.; Topp, E. Distribution of antimicrobial resistance and virulence genes in Enterococcus spp. and characterization of isolates from broiler chickens. Appl. Environ. Microbiol. 2010, 76, 8033–8043. [Google Scholar] [CrossRef]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence factors of Enterococcus spp. presented in food. LWT Food Sci. Technol. 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Zou, L.K.; Wang, H.N.; Zeng, B.; Li, J.N.; Li, X.T.; Zhang, A.Y.; Zhou, Y.S.; Yang, X.; Xu, C.W.; Xia, Q.Q. Erythromycin resistance and virulence genes in Enterococcus faecdlis from swine in china. New Microbiol. 2011, 34, 73–80. [Google Scholar]



| Antibiotics | E. faecalis (n = 225) | E. faecium (n = 19) | E. casseliflavus (n = 7) | E. gallinarum (n = 1) | Other Enterococcus spp. (n = 32) | Total (n = 284) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |
| CIP | 204 (90.7) | 0 (0) | 21 (9.3) | 16 (84.2) | 0 (0) | 3 (15.8) | 7 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 30 (93.8) | 0 (0) | 2 (6.3) | 258 (90.8) | 0 (0) | 26 (9.2) |
| GEN | 191 (84.9) | 0 (0) | 34 (15.1) | 19 (100) | 0 (0) | 0 (0) | 6 (85.7) | 0 (0) | 1 (14.3) | 1 (100) | 0 (0) | 0 (0) | 28 (87.5) | 0 (0) | 4 (12.5) | 245 (86.3) | 0 (0) | 39 (13.7) |
| STR | 68 (30.2) | 0 (0) | 157 (69.8) | 5 (26.3) | 0 (0) | 14 (73.7) | 2 (28.6) | 0 (0) | 5 (71.4) | 0 (0) | 0 (0) | 1 (100) | 5 (15.6) | 0 (0) | 27 (84.4) | 80 (28.2) | 0 (0) | 204 (71.8) |
| Q-D * | 4 (21.1) | 0 (0) | 15 (78.9) | 4 (21.1) | 0 (0) | 15 (78.9) | ||||||||||||
| NIT | 218 (96.9) | 0 (0) | 7 (3.1) | 17 (89.5) | 0 (0) | 2 (10.5) | 7 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 32 (100) | 0 (0) | 0 (0) | 275 (96.8) | 0 (0) | 9 (3.2) |
| SXT | 50 (22.2) | 0 (0) | 175 (77.8) | 3 (15.8) | 0 (0) | 16 (84.2) | 2 (28.6) | 0 (0) | 5 (71.4) | 1 (100) | 0 (0) | 0 (0) | 13 (40.6) | 0 (0) | 19 (59.4) | 69 (24.3) | 0 (0) | 215 (75.7) |
| ERY | 8 (3.6) | 56 (24.9) | 161 (71.6) | 0 (0) | 6 (31.6) | 13 (68.4) | 0 (0) | 3 (42.9) | 4 (57.1) | 0 (0) | 0 (0) | 1 (100) | 1 (3.1) | 5 (15.6) | 26 (81.3) | 9 (3.2) | 70 (24.6) | 205 (72.2) |
| TET | 36 (16) | 10 (4.4) | 179 (79.6) | 7 (36.8) | 1 (5.3) | 11 (57.9) | 1 (14.3) | 0 (0) | 6 (85.7) | 0 (0) | 0 (0) | 1 (100) | 1 (3.1) | 0 (0) | 31 (96.9) | 45 (15.8) | 11 (3.9) | 228 (80.3) |
| CHL | 126 (56) | 42 (18.7) | 57 (25.3) | 13 (68.4) | 4 (21.1) | 2 (10.5) | 4 (57.1) | 1 (14.3) | 2 (28.6) | 0 (0) | 0 (0) | 1 (100) | 17 (53.1) | 7 (21.9) | 8 (25) | 160 (56.3) | 54 (19) | 70 (24.6) |
| LEV | 215 (95.6) | 0 (0) | 10 (4.4) | 19 (100) | 0 (0) | 0 (0) | 7 (100) | 0 (0) | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 31 (96.9) | 0 (0) | 1 (3.1) | 273 (96.1) | 0 (0) | 11 (3.9) |
| Enterococcus spp. | Antibiotic Resistance Genes | ||||
|---|---|---|---|---|---|
| ermB (n = 205) | aph(3′)-IIIa (n = 204) | tetK (n = 228) | tetM (n = 228) | aac(6′)-Ie-aph(2″)-Ia (n = 39) | |
| E. faecalis | 139 (67.8%) | 155 (76.0%) | 35 (15.4%) | 177 (77.6%) | 2 (5.1%) |
| E. faecium | 13 (6.3%) | 11 (5.4%) | 1 (0.4%) | 11 (4.8%) | 0 (0.0%) |
| E. casseliflavus | 2 (1.0%) | 4 (2.0%) | 0 (0.0%) | 6 (2.6%) | 0 (0.0%) |
| E. gallinarum | 1 (0.5%) | 0 (0.0%) | 0 (0.0%) | 1 (0.4%) | 0 (0.0%) |
| Other Enterococcus spp. | 26 (12.7%) | 26 (12.7%) | 3 (1.3%) | 31 (13.6%) | 0 (0.0%) |
| Total | 181 (88.3%) | 196 (96.1%) | 39 (17.1%) | 226 (99.1%) | 2 (5.1%) |
| Enterococcus spp. | Virulence Genes | |||||
|---|---|---|---|---|---|---|
| efaAfs | gelE | cpd | cylB | cylA | efaAfm | |
| E. faecalis (n = 225) | 201 (89.3%) | 208 (92.4%) | 186 (82.7%) | 80 (35.6%) | 49 (21.8%) | 1 (0.4%) |
| E. faecium (n = 19) | 0 (0.0%) | 13 (68.4%) | 10 (52.6%) | 1 (5.3%) | 0 (0.0%) | 9 (47.4%) |
| E. casseliflavus (n = 7) | 3 (42.9%) | 5 (71.4%) | 2 (28.6%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| E. gallinarum (n = 1) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Other Enterococcus spp. (n = 32) | 19 (59.4%) | 27 (84.4%) | 21 (65.6%) | 8 (25.0%) | 3 (9.4%) | 0 (0.0%) |
| Total (n = 284) | 223 (78.5%) | 253 (89.1%) | 219 (77.1%) | 89 (31.3%) | 52 (18.3%) | 10 (3.5%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badul, S.; Abia, A.L.K.; Amoako, D.G.; Perrett, K.; Bester, L.A.; Essack, S.Y. From the Farms to the Dining Table: The Distribution and Molecular Characteristics of Antibiotic-Resistant Enterococcus spp. in Intensive Pig Farming in South Africa. Microorganisms 2021, 9, 882. https://doi.org/10.3390/microorganisms9050882
Badul S, Abia ALK, Amoako DG, Perrett K, Bester LA, Essack SY. From the Farms to the Dining Table: The Distribution and Molecular Characteristics of Antibiotic-Resistant Enterococcus spp. in Intensive Pig Farming in South Africa. Microorganisms. 2021; 9(5):882. https://doi.org/10.3390/microorganisms9050882
Chicago/Turabian StyleBadul, Sasha, Akebe L. K. Abia, Daniel G. Amoako, Keith Perrett, Linda A. Bester, and Sabiha Y. Essack. 2021. "From the Farms to the Dining Table: The Distribution and Molecular Characteristics of Antibiotic-Resistant Enterococcus spp. in Intensive Pig Farming in South Africa" Microorganisms 9, no. 5: 882. https://doi.org/10.3390/microorganisms9050882
APA StyleBadul, S., Abia, A. L. K., Amoako, D. G., Perrett, K., Bester, L. A., & Essack, S. Y. (2021). From the Farms to the Dining Table: The Distribution and Molecular Characteristics of Antibiotic-Resistant Enterococcus spp. in Intensive Pig Farming in South Africa. Microorganisms, 9(5), 882. https://doi.org/10.3390/microorganisms9050882










