A Sensitive and Specific Competitive Enzyme-Linked Immunosorbent Assay for Serodiagnosis of COVID-19 in Animals
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. Experimental Production of Anti-SARS-CoV-2 Antisera by Laboratory Animal Immunization
2.2.1. Viral Strain and Culture
2.2.2. Virus Inactivation and Purification
2.2.3. Animal Immunization
2.2.4. Antibody Detection
2.3. Serum Samples
2.3.1. Sera for Initial Assay Development and Analytical Sensitivity Determination (Panel 1)
2.3.2. COVID-19 Negative Sera for Cut-Off Determination (Panel 2)
2.3.3. Sera from Animals Immunized with Inactive SARS-CoV-2 Virion (Panel 3)
2.3.4. Sera from Animals Infected by βCoVs for Determination of Analytical Specificity (Panel 4)
2.3.5. Sera from Animals in an Amusement Park for Determination of Diagnostic Specificity (Panel 5)
2.3.6. Sera from Laboratory Confirmed Human COVID-19 Patients for Comparison with Commercial Indirect ELISA and Determination of Diagnostic Sensitivity (Panel 6)
2.4. Development of the cELISA
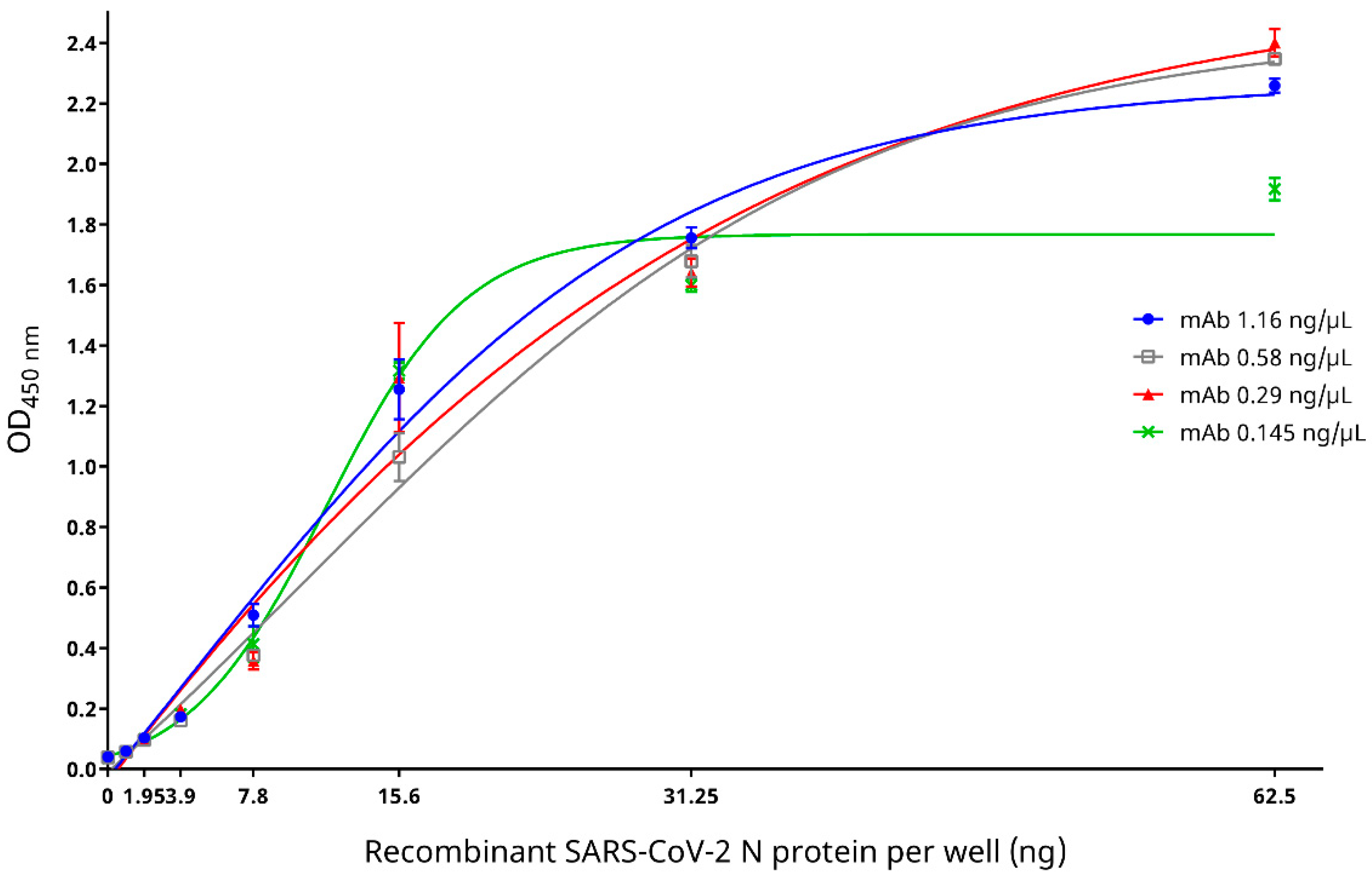
2.4.1. Reactivity and Specificity of Recombinant SARS-CoV-2 N Protein with Mouse Anti-SARS-CoV-2 N Protein mAb
2.4.2. Evaluation of Epitope Blocking by mAb and Determination of Optimal Serum Dilution for the cELISA
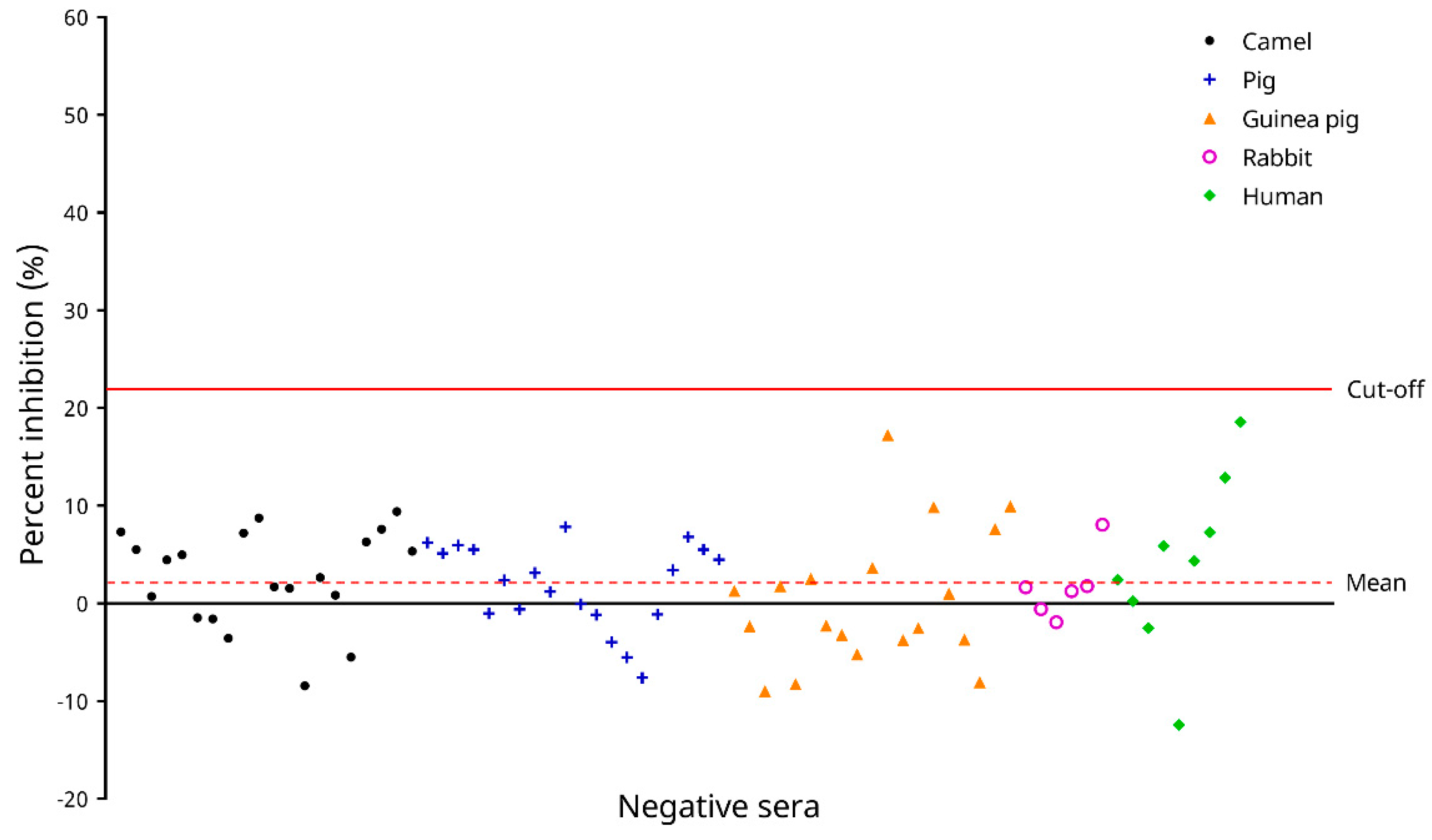
2.4.3. Determination of the cELISA Cut-Off Value
2.4.4. Determination of Analytical Sensitivity of the cELISA
2.4.5. Cross Reactivities to Serum Samples Seropositive to βCoVs Other Than Sarbecovirus and Determination of Analytical Specificity of the cELISA
2.4.6. Preliminary Testing on Diagnostic Performance with Sera from Experimentally Infected Animals
2.5. Evaluation of the Diagnostic Performance of the cELISA
2.6. Statistical Analyses
3. Results
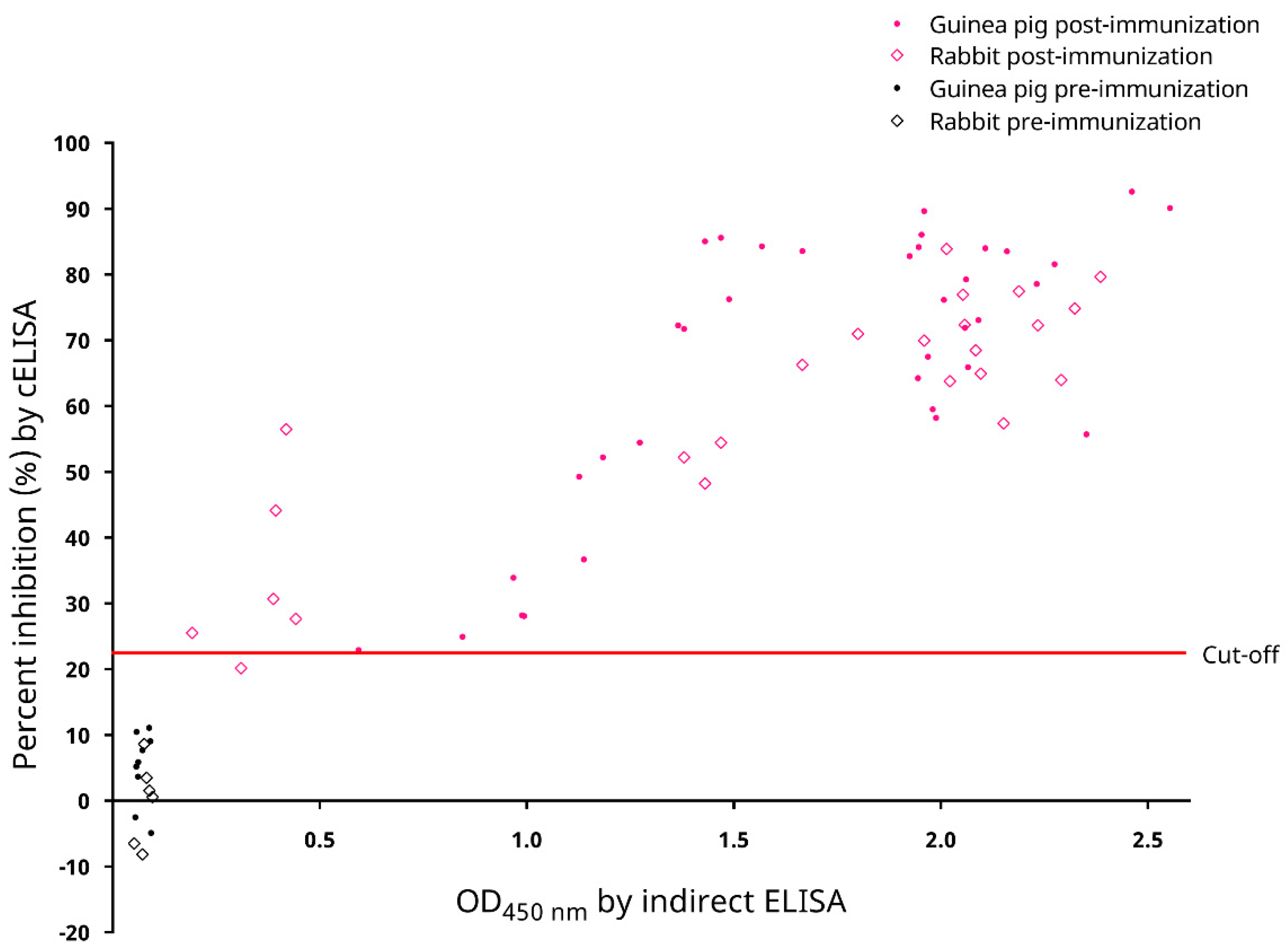
3.1. Experimental Production of Anti-SARS-CoV-2 Antisera by Laboratory Animal Immunization
3.2. Reactivity and Specificity of Recombinant SARS-CoV-2 N Protein with Mouse Anti-SARS-CoV-2 N Protein mAb
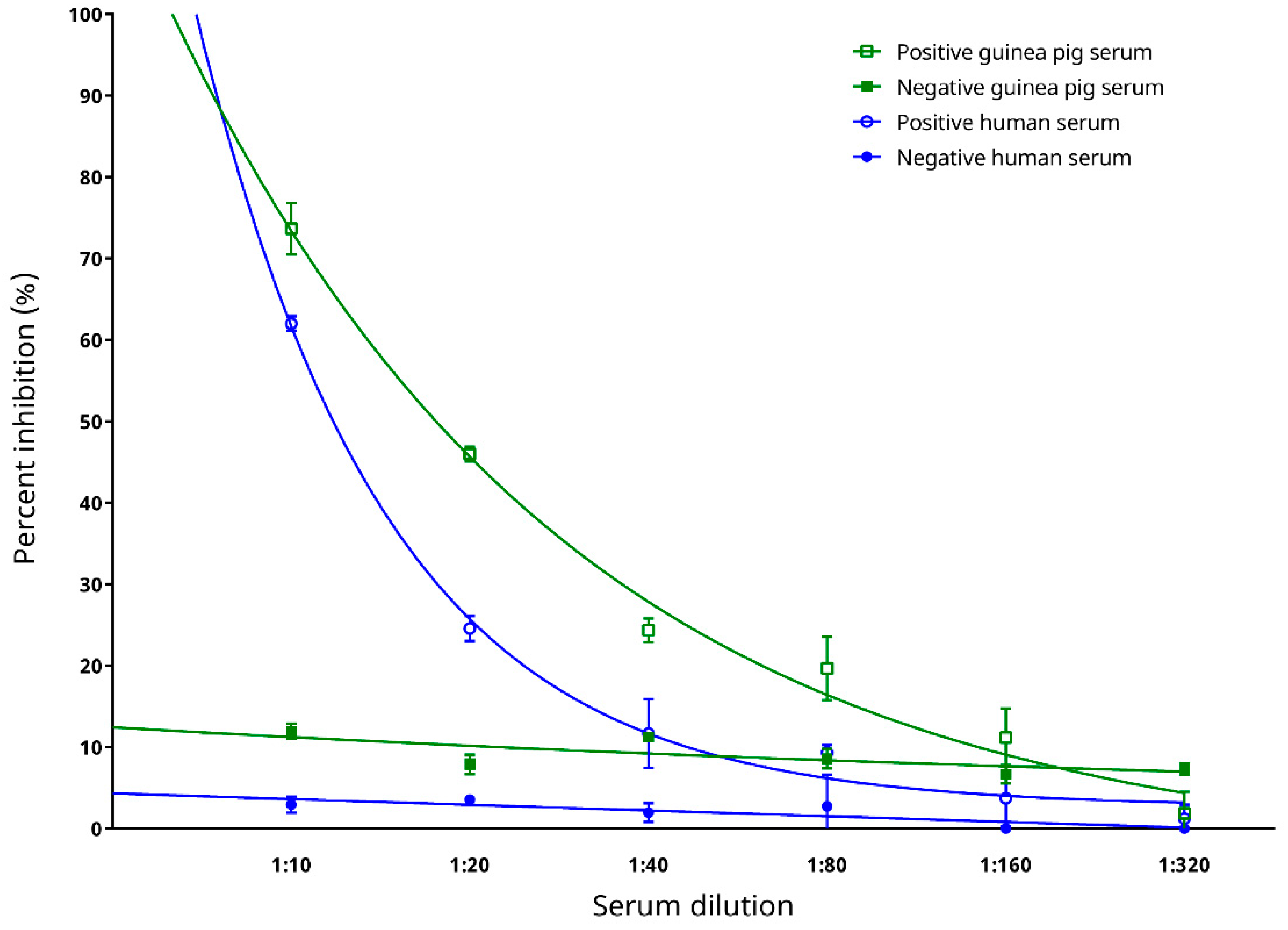
3.3. Epitope Blocking and Determination of Optimal Serum Dilution for the cELISA
3.4. Determination of the Cut-Off Value of the cELISA
3.5. Determination of Analytical Sensitivity
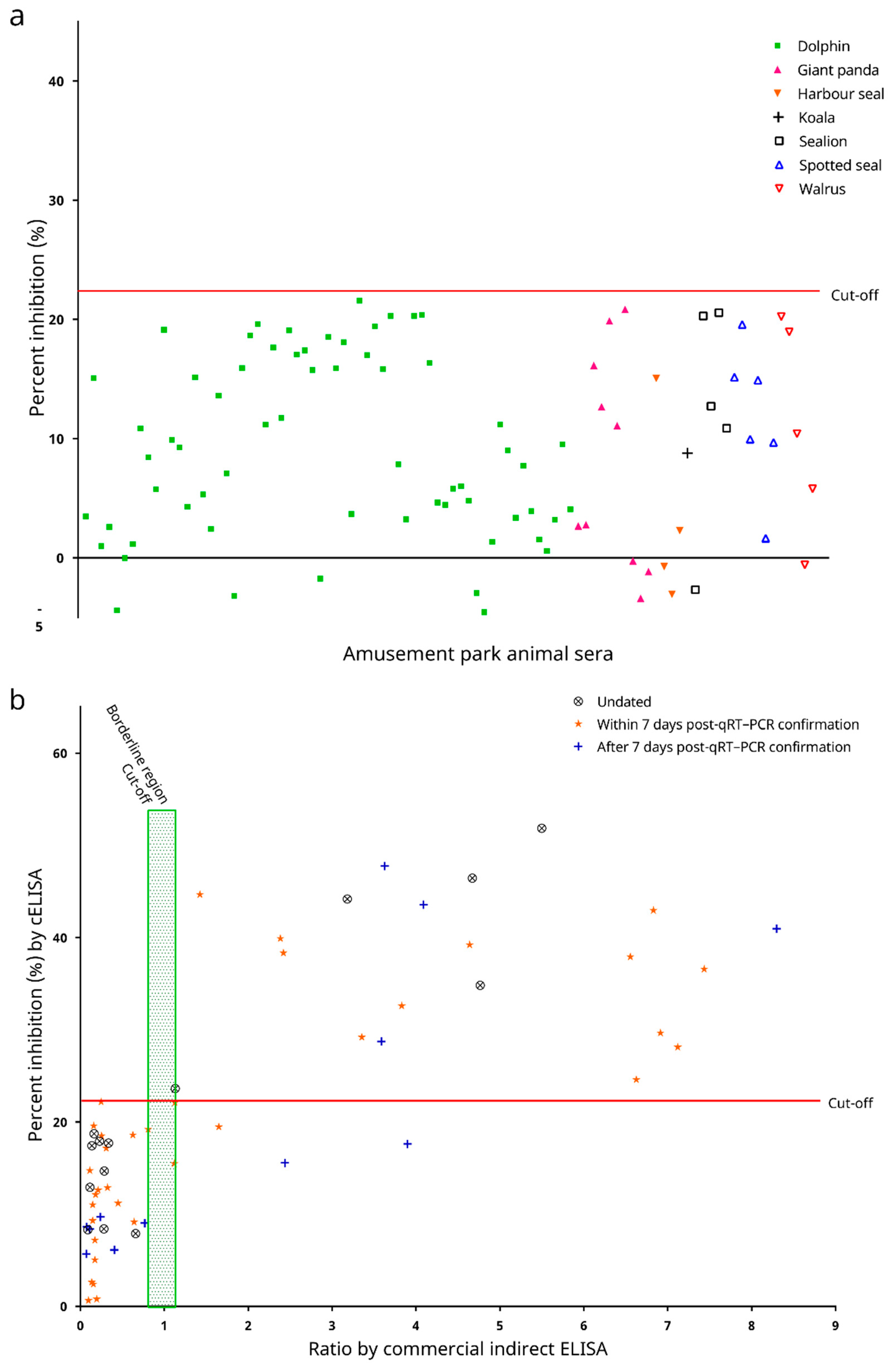
3.6. Cross Reactivities to Serum Samples Seropositive to βCoVs Other Than Sarbecovirus and Analytical Specificity
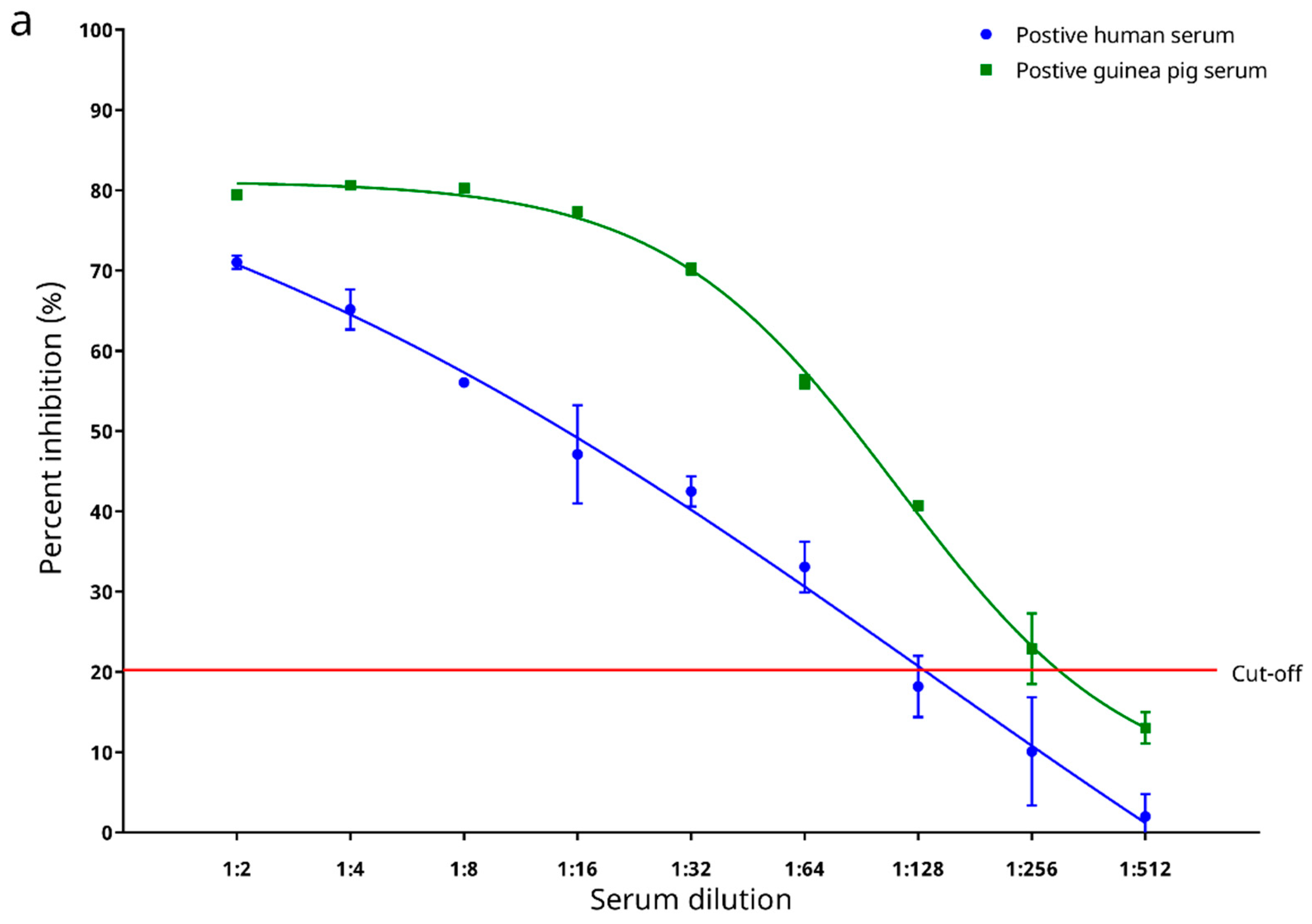
3.7. Preliminary Testing on Diagnostic Performance with Sera from Experimentally Infected Animals
3.8. Diagnostic Performance of the cELISA
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Weekly Operational Update on COVID-19. 03 May 2021; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Chu, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.-Y.; Tsang, J.O.-L.; Huang, X.; et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, e14–e23. [Google Scholar] [CrossRef]
- Abdel-Moneim, A.S.; Abdelwhab, E.M. Evidence for SARS-CoV-2 infection of animal hosts. Pathogens 2020, 9, 529. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Lamers, M.M.; Okba, N.M.A.; Breugem, T.I.; Schipper, D.; van den Doel, P.B.; van Run, P.; van Amerongen, G.; de Waal, L.; Koopmans, M.P.G.; et al. Susceptibility of rabbits to SARS-CoV-2. Emerg. Microbes Infect. 2021, 10, 1–7. [Google Scholar] [CrossRef]
- World Organisation for Animal Health. Covid-19. Available online: https://www.oie.int/en/what-we-offer/emergency-and-resilience/covid-19/ (accessed on 5 May 2021).
- Giner, J.; Villanueva-Saz, S.; Tobajas, A.P.; Pérez, M.D.; González, A.; Verde, M.; Yzuel, A.; García-García, A.; Taleb, V.; Lira-Navarrete, E.; et al. SARS-CoV-2 seroprevalence in household domestic ferrets (Mustela putorius furo). Animals 2021, 11, 667. [Google Scholar] [CrossRef]
- Spada, E.; Vitale, F.; Bruno, F.; Castelli, G.; Reale, S.; Perego, R.; Baggiani, L.; Proverbio, D. A pre- and during pandemic survey of Sars-Cov-2 infection in stray colony and shelter cats from a high endemic area of Northern Italy. Viruses 2021, 13, 618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, H.; Gao, J.; Huang, K.; Yang, Y.; Hui, X.; He, X.; Li, C.; Gong, W.; Zhang, Y.; et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg. Microbes Infect. 2020, 9, 2013–2019. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, P.J.; Hatta, M.; Chiba, S.; Maemura, T.; Fan, S.; Takeda, M.; Kinoshita, N.; Hattori, S.-i.; Sakai-Tagawa, Y.; Iwatsuki-Horimoto, K.; et al. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020, 383, 592–594. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Trujillo, J.D.; Carossino, M.; Meekins, D.A.; Morozov, I.; Madden, D.W.; Indran, S.V.; Bold, D.; Balaraman, V.; Kwon, T.; et al. SARS-CoV-2 infection, disease and transmission in domestic cats. Emerg. Microbes Infect. 2020, 9, 2322–2332. [Google Scholar] [CrossRef]
- Polansek, T. Coronavirus Kills 15,000 U.S. Mink, as Denmark Prepares for Nationwide Cull. Available online: https://www.reuters.com/article/us-health-coronavirus-usa-minks-idINKBN27Q35V (accessed on 13 April 2021).
- Utah Department of Agriculture and Food. Ongoing One Health Response and Investigation on Utah Mink Farms Yields Encouraging Results. Available online: https://ag.utah.gov/wp-content/uploads/2020/12/Press-Release-Ongoing-One-Health-Response-and-Investigation-on-Utah-Mink-Farms-Yields-Encouraging-Results.pdf (accessed on 13 April 2021).
- Gronholt-Pedersen, J. Denmark to Dig Up Millions of Mink From Mass Graves. Available online: https://www.reuters.com/article/idUSL1N2J00ET (accessed on 13 April 2021).
- Oude Munnink, B.B.; Sikkema, R.S.; Nieuwenhuijse, D.F.; Molenaar, R.J.; Munger, E.; Molenkamp, R.; van der Spek, A.; Tolsma, P.; Rietveld, A.; Brouwer, M.; et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 2021, 371, 172–177. [Google Scholar] [CrossRef]
- Wernike, K.; Aebischer, A.; Michelitsch, A.; Hoffmann, D.; Freuling, C.; Balkema-Buschmann, A.; Graaf, A.; Müller, T.; Osterrieder, N.; Rissmann, M.; et al. Multi-species ELISA for the detection of antibodies against SARS-CoV-2 in animals. Transbound. Emerg. Dis. 2020, in press. [Google Scholar] [CrossRef]
- Innovative Diagnostics. ID Screen® SARS-CoV-2 Double Antigen Multi-Species ELISA. Available online: https://www.id-vet.com/produit/id-screen-sars-cov-2-double-antigen-multi-species/ (accessed on 5 May 2021).
- Innovative Diagnostics. Frequently Asked Questions (FAQ). Available online: https://www.id-vet.com/frequently-asked-questions-faq/ (accessed on 5 May 2021).
- Lau, S.K.P.; Luk, H.K.H.; Wong, A.C.P.; Li, K.S.M.; Zhu, L.; He, Z.; Fung, J.; Chan, T.T.Y.; Fung, K.S.C.; Woo, P.C.Y. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020, 26, 1542–1547. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Fan, R.Y.Y.; Lau, C.C.Y.; Wong, E.Y.M.; Joseph, S.; Tsang, A.K.L.; Wernery, R.; Yip, C.C.Y.; Tsang, C.-C.; et al. Isolation and characterization of dromedary camel coronavirus UAE-HKU23 from dromedaries of the Middle East: Minimal serological cross-reactivity between mers coronavirus and dromedary camel coronavirus UAE-HKU23. Int. J. Mol. Sci. 2016, 17, 691. [Google Scholar] [CrossRef]
- Lau, S.K.; Poon, R.W.; Wong, B.H.; Wang, M.; Huang, Y.; Xu, H.; Guo, R.; Li, K.S.; Gao, K.; Chan, K.H.; et al. Coexistence of different genotypes in the same bat and serological characterization of Rousettus bat coronavirus HKU9 belonging to a novel Betacoronavirus subgroup. J. Virol. 2010, 84, 11385–11394. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Fan, R.Y.Y.; Huang, Y.; Wang, M.; Guo, R.; Lam, C.S.F.; Tsang, A.K.L.; Lai, K.K.Y.; et al. Isolation and characterization of a novel betacoronavirus subgroup a coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J. Virol. 2012, 86, 5481–5496. [Google Scholar] [CrossRef] [PubMed]
- Centre for Clinical Research and Biostatistics of The Chinese University of Hong Kong. C.I. Calculator: Diagnostic Statistics. Available online: https://www2.ccrb.cuhk.edu.hk/stat/confidence%20interval/Diagnostic%20Statistic.htm (accessed on 13 April 2021).
- Martelli, P.; Teng, J.L.L.; Lee, F.-K.; Yeong, K.-Y.; Fong, J.Y.H.; Hui, S.-W.; Chan, K.-H.; Lau, S.K.P.; Woo, P.C.Y. Influenza A(H1N1)pdm09 virus infection in a captive giant panda, Hong Kong. Emerg. Infect. Dis. 2019, 25, 2303–2306. [Google Scholar] [CrossRef] [PubMed]
- Wernery, U.; Tsang, C.-C.; Hebel, C.; Damerau, A.; Kinne, J.; Cai, J.-P.; Küspert, H.; Chan, K.-F.; Joseph, M.; Xue, S.; et al. Serodiagnosis of aspergillosis in falcons (Falco spp.) by an Afmp1p-based enzyme-linked immunosorbent assay. Mycoses 2018, 61, 600–609. [Google Scholar] [CrossRef]
- Wernery, U.; Chan, E.; Raghavan, R.; Teng, J.L.L.; Syriac, G.; Sing-Yung, S.; Joseph, M.; Yeung, M.-L.; Jia, L.; Cai, J.-P.; et al. Development and validation of a sensitive competitive enzyme-linked immunosorbent assay for serodiagnosis of Burkholderia mallei, a tier 1 select agent. PLoS Negl. Trop. Dis. 2021, in press. [Google Scholar]
- Khan, I.; Wieler, L.H.; Melzer, F.; Elschner, M.C.; Muhammad, G.; Ali, S.; Sprague, L.D.; Neubauer, H.; Saqib, M. Glanders in animals: A review on epidemiology, clinical presentation, diagnosis and countermeasures. Transbound. Emerg. Dis. 2013, 60, 204–221. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Tsoi, H.-W.; Chan, K.-H.; Wong, B.H.L.; Che, X.-Y.; Tam, V.K.P.; Tam, S.C.F.; Cheng, V.C.C.; Hung, I.F.N.; et al. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 2004, 363, 841–845. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Tsoi, H.-W.; Fung, A.M.Y.; Kao, R.Y.T.; Chan, K.-h.; Peiris, J.S.M.; Yuen, K.-Y. Differential sensitivities of severe acute respiratory syndrome (SARS) coronavirus spike polypeptide enzyme-linked immunosorbent assay (ELISA) and SARS coronavirus nucleocapsid protein ELISA for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2005, 43, 3054–3058. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C.; Rawlings, S.; Smith, D.; Das, S.; Strich, J.R.; Chertow, D.S.; Davey, R.T., Jr.; Cohen, J.I. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020, 222, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Tsoi, H.-W.; Fung, A.M.Y.; Chan, K.-H.; Tam, V.K.P.; Peiris, J.S.M.; Yuen, K.-Y. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 2004, 42, 2306–2309. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Wong, B.H.L.; Chan, K.-H.; Hui, W.-T.; Kwan, G.S.W.; Peiris, J.S.M.; Couch, R.B.; Yuen, K.-Y. False-positive results in a recombinant severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nucleocapsid enzyme-linked immunosorbent assay due to HCoV-OC43 and HCoV-229E rectified by western blotting with recombinant SARS-CoV spike polypeptide. J. Clin. Microbiol. 2004, 42, 5885–5888. [Google Scholar] [PubMed]
- Chow, F.W.-N.; Chan, T.T.-Y.; Tam, A.R.; Zhao, S.; Yao, W.; Fung, J.; Cheng, F.K.-K.; Lo, G.C.-S.; Chu, S.; Aw-Yong, K.L.; et al. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 2020, 21, 5380. [Google Scholar] [CrossRef] [PubMed]
- Fung, J.; Lau, S.K.P.; Woo, P.C.Y. Antigen capture enzyme-linked immunosorbent assay for detecting Middle East respiratory syndrome coronavirus in humans. In MERS Coronavirus: Methods and Protocols; Vijay, R., Ed.; Springer US: New York, NY, USA, 2020; pp. 89–97. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lau, S.K.P.; He, Z.; Tsang, C.-C.; Chan, T.T.Y.; Luk, H.K.H.; Chan, E.; Li, K.S.M.; Fung, J.; Chow, F.W.N.; Tam, A.R.; et al. A Sensitive and Specific Competitive Enzyme-Linked Immunosorbent Assay for Serodiagnosis of COVID-19 in Animals. Microorganisms 2021, 9, 1019. https://doi.org/10.3390/microorganisms9051019
Lau SKP, He Z, Tsang C-C, Chan TTY, Luk HKH, Chan E, Li KSM, Fung J, Chow FWN, Tam AR, et al. A Sensitive and Specific Competitive Enzyme-Linked Immunosorbent Assay for Serodiagnosis of COVID-19 in Animals. Microorganisms. 2021; 9(5):1019. https://doi.org/10.3390/microorganisms9051019
Chicago/Turabian StyleLau, Susanna K. P., Zirong He, Chi-Ching Tsang, Tony T. Y. Chan, Hayes K. H. Luk, Elaine Chan, Kenneth S. M. Li, Joshua Fung, Franklin W. N. Chow, Anthony R. Tam, and et al. 2021. "A Sensitive and Specific Competitive Enzyme-Linked Immunosorbent Assay for Serodiagnosis of COVID-19 in Animals" Microorganisms 9, no. 5: 1019. https://doi.org/10.3390/microorganisms9051019
APA StyleLau, S. K. P., He, Z., Tsang, C.-C., Chan, T. T. Y., Luk, H. K. H., Chan, E., Li, K. S. M., Fung, J., Chow, F. W. N., Tam, A. R., Chung, T. W. H., Wong, S. C. Y., Que, T.-L., Fung, K. S. C., Lung, D. C., Wu, A. K. L., Hung, I. F. N., Teng, J. L. L., Wernery, U., ... Woo, P. C. Y. (2021). A Sensitive and Specific Competitive Enzyme-Linked Immunosorbent Assay for Serodiagnosis of COVID-19 in Animals. Microorganisms, 9(5), 1019. https://doi.org/10.3390/microorganisms9051019











