Presence of Broad-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Zoo Mammals
Abstract
1. Introduction
2. Materials and Methods
2.1. Belgian Zoos
2.2. Selective Isolation from Fecal Samples
2.3. Antimicrobial Susceptibility Testing
2.4. Molecular Mechanisms of Resistance
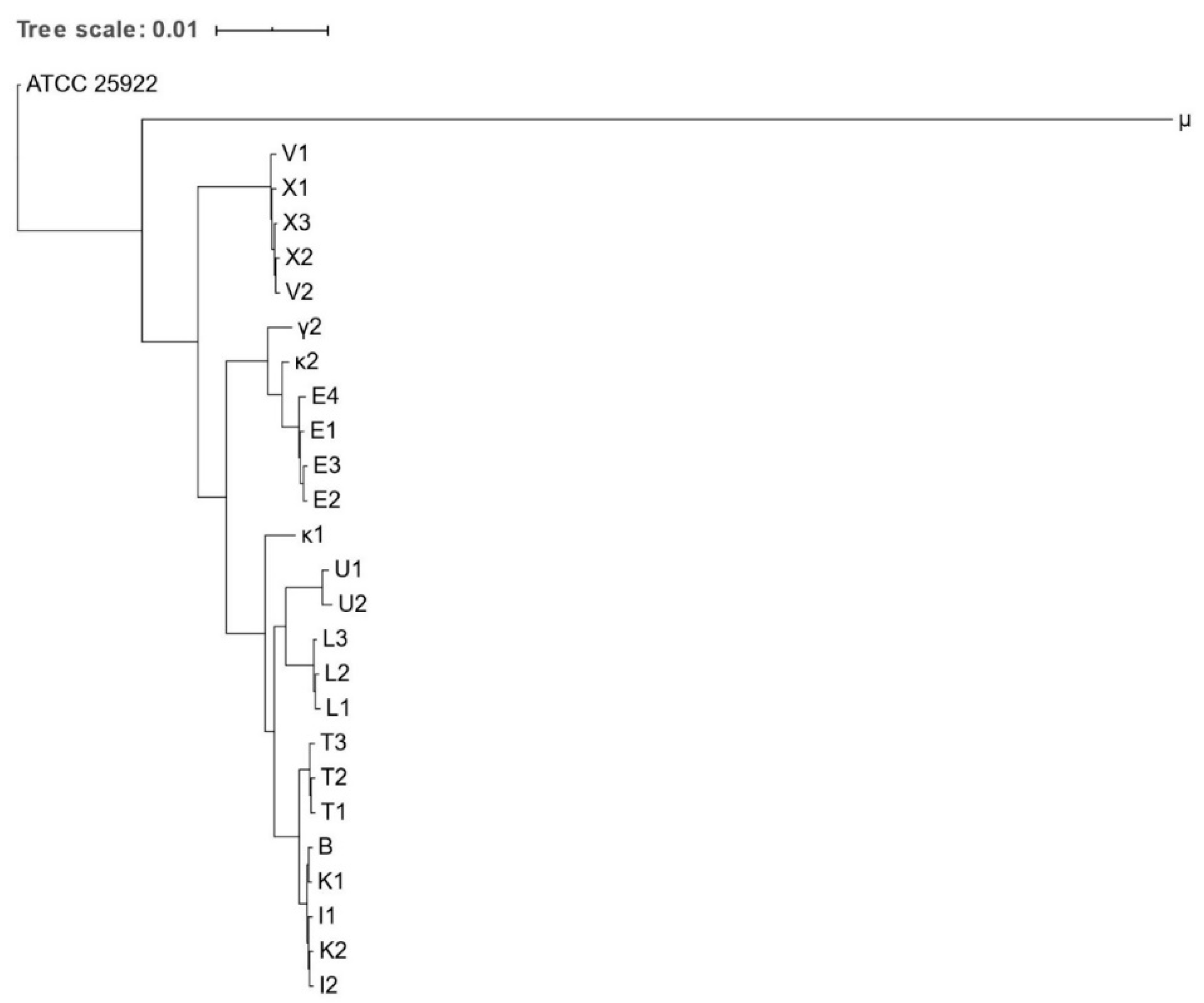
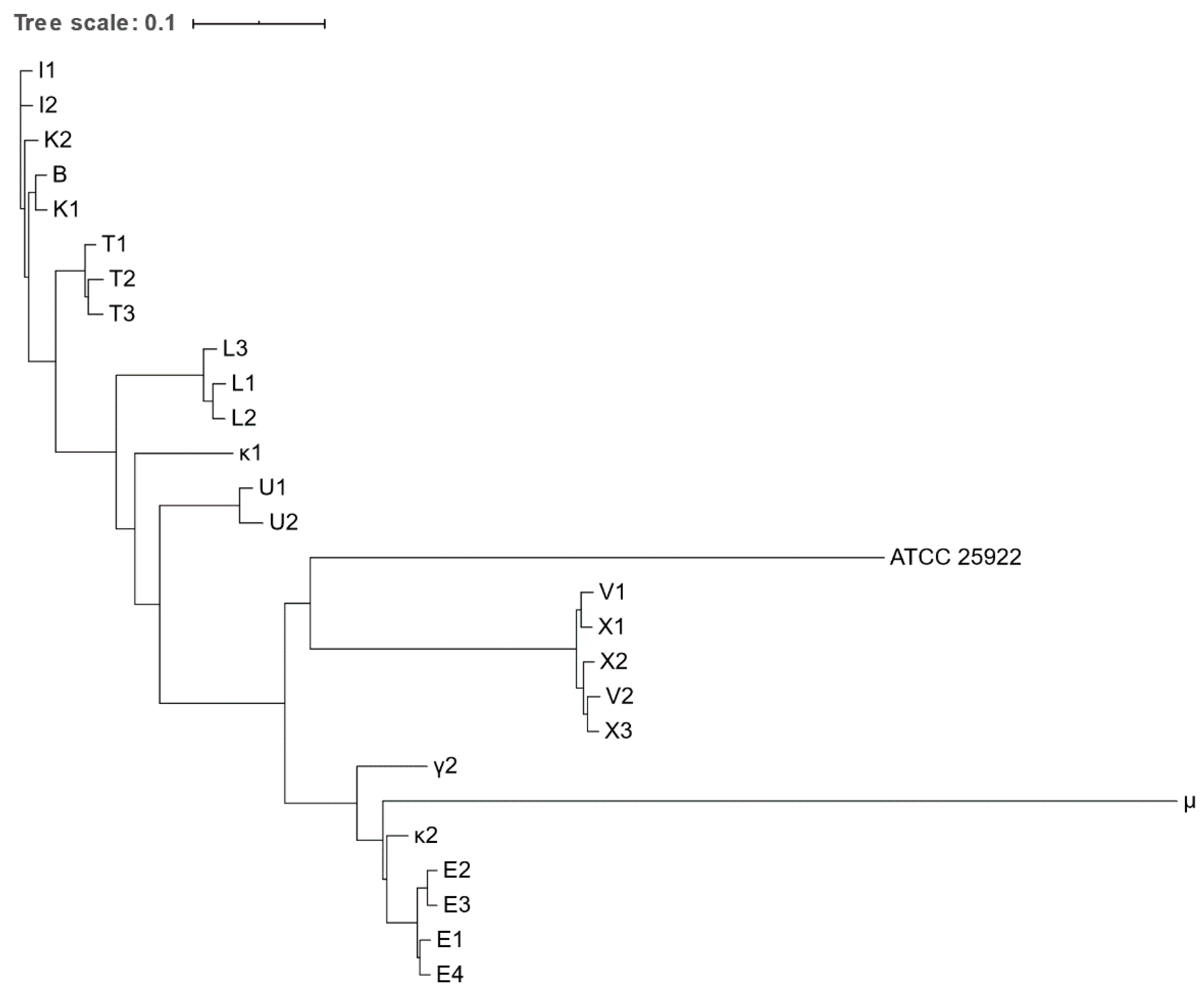
2.5. Phylogenetic Analysis and Strain Typing
3. Results
3.1. Bacterial Isolates and Antimicrobial Susceptibility
3.2. Molecular Mechanisms of Resistance
3.3. WGS and Resistome
3.4. Phylogenetic Analysis and Strain Typing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Herman, L.; Haesebrouck, F.; Butaye, P. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010, 34, 295–316. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Coque, T.M.; Baquero, F.; Canton, R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill. 2008, 13, 19044. [Google Scholar] [CrossRef] [PubMed]
- Dorado-García, A.; Smid, J.H.; van Pelt, W.; Bonten, M.J.M.; Fluit, A.C.; van den Bunt, G.; Wagenaar, J.A.; Hordijk, J.; Dierikx, C.M.; Veldman, K.T.; et al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: A pooled analysis. J. Antimicrob. Chemother. 2018, 73, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Dobiasova, H.; Dolejska, M.; Jamborova, I.; Brhelova, E.; Blazkova, L.; Papousek, I.; Kozlova, M.; Klimes, J.; Cizek, A.; Literak, I. Extended spectrum beta-lactamase and fluoroquinolone resistance genes and plasmids among Escherichia coli isolates from zoo animals, Czech Republic. FEMS Microbiol. Ecol. 2013, 85, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Motoi, Y.; Sato, M.; Maruyama, A.; Watanabe, H.; Fukumoto, Y.; Shimamoto, T. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl. Environ. Microbiol. 2007, 73, 6686–6690. [Google Scholar] [CrossRef]
- Ishihara, K.; Hosokawa, Y.; Makita, K.; Noda, J.; Ueno, H.; Muramatsu, Y.; Ueno, H.; Mukai, T.; Yamamoto, H.; Ito, M.; et al. Factors associated with antimicrobial-resistant Escherichia coli in zoo animals. Res. Vet. Sci. 2012, 93, 574–580. [Google Scholar] [CrossRef]
- Wang, Y.; He, T.; Han, J.; Wang, J.; Foley, S.L.; Yang, G.; Wan, S.; Shen, J.; Wu, C. Prevalence of ESBLs and PMQR genes in fecal Escherichia coli isolated from the non-human primates in six zoos in China. Vet. Microbiol. 2012, 159, 53–59. [Google Scholar] [CrossRef]
- Bender, J.B.; Shulman, S.A.; Animals in Public Contact Subcommittee; National Association of State Public Health Veterinarians. Reports of zoonotic disease outbreaks associated with animal exhibits and availability of recommendations for preventing zoonotic disease transmission from animals to people in such settings. J. Am. Vet. Med. Assoc. 2004, 224, 1105–1109. [Google Scholar] [CrossRef]
- Sala, A.; Taddei, S.; Santospirito, D.; Sandri, C.; Magnone, W.; Cabassi, C.S. Antibiotic resistance in conjunctival and enteric bacterial flora in raptors housed in a zoological garden. Vet. Med. Sci. 2016, 2, 239–245. [Google Scholar] [CrossRef]
- Sherwen, S.L.; Hemsworth, P.H.; Butler, K.L.; Fanson, K.V.; Magrath, M.J.L. Impacts of visitor number on Kangaroos housed in free-range exhibits. Zoo Biol. 2015, 34, 287–295. [Google Scholar] [CrossRef]
- Van Driessche, L.; Bokma, J.; Gille, L.; Ceyssens, P.-J.; Sparbier, K.; Haesebrouck, F.; Deprez, P.; Boyen, F.; Pardon, B. Rapid detection of tetracycline resistance in bovine Pasteurella multocida isolates by MALDI Biotyper antibiotic susceptibility test rapid assay (MBT-ASTRA). Sci. Rep. 2018, 8, 13599. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Catry, B.; Herman, L.; Haesebrouck, F.; Butaye, P. Diversity of extended-spectrum beta-lactamases and class C beta-lactamases among cloacal Escherichia coli Isolates in Belgian broiler farms. Antimicrob. Agents Chemother. 2008, 52, 1238–1243. [Google Scholar] [CrossRef]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Sović, I.; Šikić, M.; Wilm, A.; Fenlon, S.N.; Chen, S.; Nagarajan, N. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 2016, 7, 11307. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019, 48, D517–D525. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Alonso, S.; Rogers, M.R.C.; Braat, J.C.; Verschuuren, T.D.; Top, J.; Corander, J.; Willems, R.J.L.; Schürch, A.C. mlplasmids: A user-friendly tool to predict plasmid- and chromosome-derived sequences for single species. Microb. Genom. 2018, 4, e000224. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, X.; Xie, Y.; Bi, D.; Sun, J.; Li, J.; Tai, C.; Deng, Z.; Ou, H.-Y. ICEberg 2.0: An updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019, 47, D660–D665. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Ekseth, O.K.; Kuiper, M.; Mironov, V. orthAgogue: An agile tool for the rapid prediction of orthology relations. Bioinformatics 2014, 30, 734–736. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Mohamed, K.; Fan, Y.; Agama Study Group; Achtman, M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef]
- Clermont, O.; Gordon, D.; Denamur, E. Guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology 2015, 161, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a Web Tool for Subtyping of Extraintestinal Pathogenic Escherichia coli Based on the fumC and fimH Alleles. J. Clin. Microbiol. 2018, 56, e00063-18. [Google Scholar] [CrossRef]
- Dahbi, G.; Mora, A.; Mamani, R.; López, C.; Alonso, M.P.; Marzoa, J.; Blanco, M.; Herrera, A.; Viso, S.; García-Garrote, F.; et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: Comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int. J. Med. Microbiol. 2014, 304, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Kaesbohrer, A.; Bakran-Lebl, K.; Irrgang, A.; Fischer, J.; Kämpf, P.; Schiffmann, A.; Werckenthin, C.; Busch, M.; Kreienbrock, L.; Hille, K. Diversity in prevalence and characteristics of ESBL/pAmpC producing E. coli in food in Germany. Vet. Microbiol. 2019, 233, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Feng, J.; Pu, J.; Xu, X.; Lu, S.; Yang, J.; Wang, Y.; Jin, D.; Du, X.; Meng, X.; et al. Genomic and molecular characterisation of Escherichia marmotae from wild rodents in Qinghai-Tibet plateau as a potential pathogen. Sci. Rep. 2019, 9, 10619. [Google Scholar] [CrossRef] [PubMed]
- Gornatti Churria, C.D.; Arias, N.; Origlia, J.; Netri, C.; Marcantoni, H.; Píscopo, M.; Loyola, H.; Petruccelli, M. Citrobacter freundii infection in two captive Australian king parrots (Alisterus scapularis). J. Zoo Aquarium Res. 2014, 2, 52–53. [Google Scholar]
- Marques, C.; Belas, A.; Aboim, C.; Cavaco-Silva, P.; Trigueiro, G.; Gama, L.T.; Pomba, C. Evidence of Sharing of Klebsiella pneumoniae Strains between Healthy Companion Animals and Cohabiting Humans. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Conrad, C.C.; Stanford, K.; Narvaez-Bravo, C.; Neumann, N.F.; Munns, K.; Tymensen, L.; Jokinen, C.; McAllister, T.A. Zoonotic Fecal Pathogens and Antimicrobial Resistance in Canadian Petting Zoos. Microorganisms 2018, 6, 70. [Google Scholar] [CrossRef]
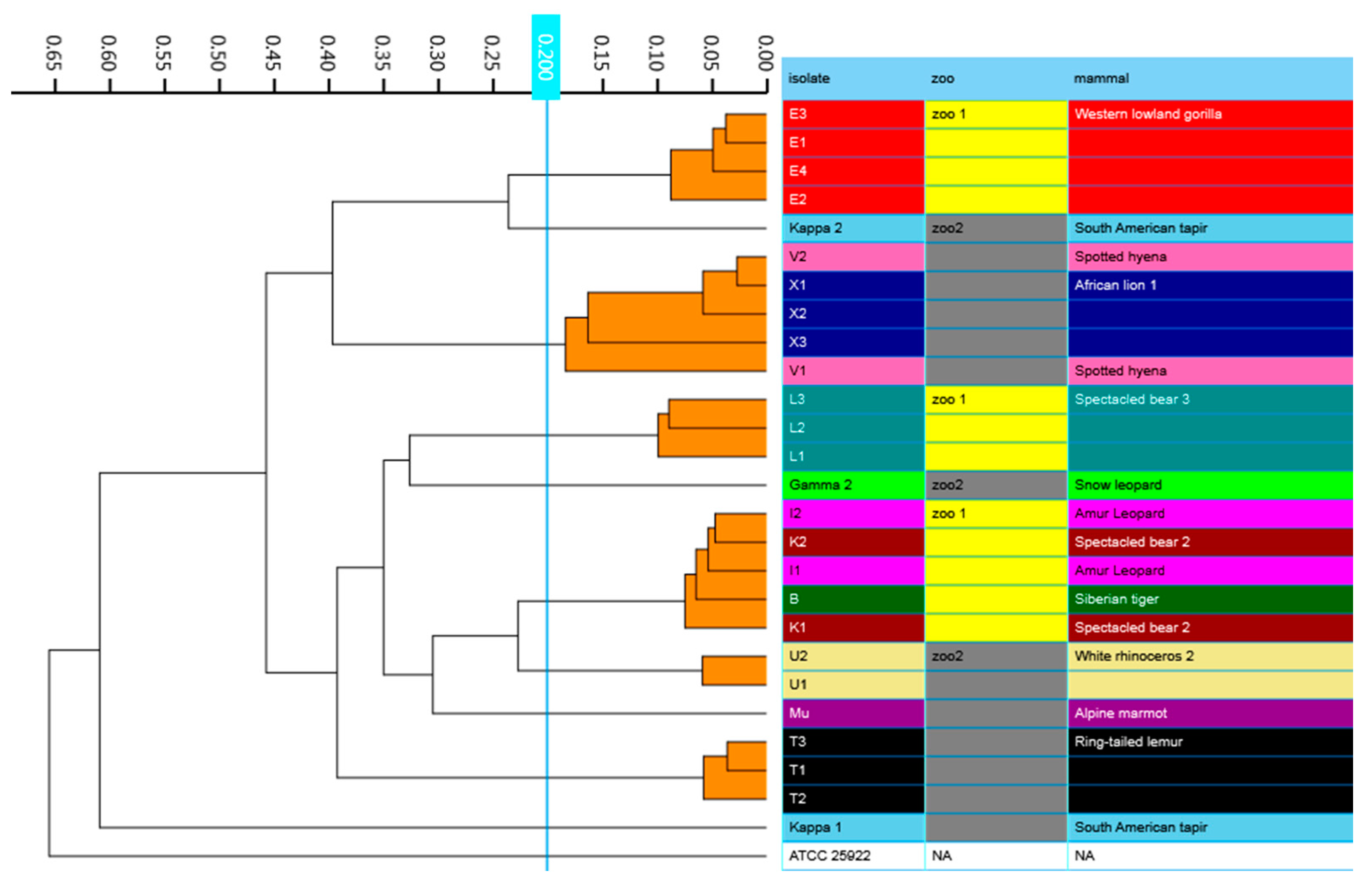
| Zoo | Zoo Mammal | Isolate | Identification | bla-Genes |
|---|---|---|---|---|
| 1 | Amur tiger | B | E. coli | CTX-M-1 |
| Western lowland gorilla | E1 | E. coli | TEM-1 | |
| E2 | E. coli | TEM-1 | ||
| E3 | E. coli | TEM-1 | ||
| E4 | E. coli | TEM-1 | ||
| Amur leopard | I1 | E. coli | CTX-M-1 | |
| I2 | E. coli | CTX-M-1 | ||
| Spectacled bear_B | K1 | E. coli | CTX-M-1 | |
| K2 | E. coli | CTX-M-1 | ||
| Spectacled bear_C | L1 | E. coli | CTX-M-1 | |
| L2 | E. coli | CTX-M-1 | ||
| L3 | E. coli | CTX-M-1 | ||
| 2 | Ring-tailed lemur | T1 | E. coli | TEM-1 |
| T2 | E. coli | TEM-1 | ||
| T3 | E. coli | TEM-1 | ||
| White rhinoceros_B | U1 | E. coli | CTX-M-1 | |
| U2 | E. coli | CTX-M-1 | ||
| Spotted hyena | V1 | E. coli | CTX-M-1 | |
| V2 | E. coli | CTX-M-1 | ||
| African lion_A | X1 | E. coli | CTX-M-1 | |
| X2 | E. coli | CTX-M-1 | ||
| X3 | E. coli | CTX-M-1 | ||
| Snow leopard | γ2 | E. coli | TEM-1; CTX-M-1 | |
| South American tapir | κ1 | E. coli | CTX-M-1 | |
| κ2 | E. coli | CTX-M-1 | ||
| Alpine marmot | μ | E. marmotae | CTX-M-1 | |
| African lion_B | δ | Klebsiella pneumoniae | CTX-M-15; SHV-32 | |
| Tasmanian devil | θ | Citrobacter freundii | CMY-124 | |
| 1 | Dromedaries (n = 3) | M | Pseudomonas sp. | / |
| 2 | Asian elephant_A | N | Pseudomonas sp. | / |
| Asian elephant_B | Z1 | Pseudomonas sp. | / | |
| Z2 | Pseudomonas sp. | / | ||
| Sumatran orangutan | β | Pseudomonas sp. | / | |
| Hippopotamus_A | η | Pseudomonas sp. | / | |
| 1 | Eastern lowland gorillas (n = 2) | F | Achromobacter spanius | / |
| Isolate | AMP | AMC | CFO | CFLEX | CFQUI | CFTIO | IMI | GEN | ENROF | TRIM | SxT | NEOMY | DOX | TET | STR | FFC | AMI | NI | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | B | R | S | S | R | R | R | S | S | S | S | R | S | R | R | I | S | S | S |
| E1 | R | R | R | R | S | R | S | S | S | R | R | I | R | R | R | S | S | S | |
| E2 | R | R | R | R | S | R | S | S | S | R | R | S | R | R | R | S | S | S | |
| E3 | R | R | R | R | S | R | S | S | I | R | R | S | R | R | R | S | S | S | |
| E4 | R | R | R | R | S | R | S | S | I | R | R | S | R | R | R | S | I | S | |
| I1 | R | S | S | R | R | R | S | S | S | S | R | I | R | R | I | S | S | S | |
| I2 | R | S | S | R | R | R | S | S | R | S | S | I | R | R | R | S | I | S | |
| K1 | R | S | S | R | R | R | S | S | S | S | R | S | R | R | I | S | S | S | |
| K2 | R | S | S | R | R | R | S | S | S | S | R | I | R | R | I | S | S | S | |
| L1 | R | S | S | R | R | R | S | S | S | S | R | S | R | R | I | S | S | S | |
| L2 | R | S | S | R | R | R | S | S | S | S | R | S | R | R | I | S | S | S | |
| L3 | R | S | S | R | R | R | S | S | S | S | R | S | R | R | I | S | S | S | |
| T1 | R | S | S | R | R | R | S | R | R | R | R | R | R | R | R | I | S | S | |
| T2 | R | S | S | R | R | R | S | R | R | R | R | R | R | R | R | S | S | S | |
| T3 | R | S | S | R | R | R | S | R | R | R | R | R | R | R | R | S | S | S | |
| U1 | R | S | S | R | R | R | S | S | S | S | S | I | S | S | R | S | S | S | |
| U2 | R | S | S | R | R | R | S | S | S | S | S | S | S | S | R | S | S | S | |
| V1 | R | S | S | R | R | R | S | S | S | S | R | S | S | S | I | S | S | S | |
| V2 | R | S | S | R | R | R | S | S | S | S | R | S | S | S | I | S | S | S | |
| X1 | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | S | S | S | |
| X2 | R | S | S | R | R | R | S | S | S | S | R | I | S | S | I | S | S | S | |
| X3 | R | S | S | R | R | R | S | S | S | S | R | S | S | S | S | S | S | S | |
| γ2 | R | S | S | R | R | R | S | S | S | S | R | S | S | R | S | S | S | S | |
| κ1 | R | S | S | R | R | R | S | S | S | R | S | I | S | S | S | S | S | S | |
| κ2 | R | S | S | R | R | R | S | S | S | S | S | S | S | S | S | S | S | S | |
| E. marmotae | μ | R | S | S | R | R | R | S | S | S | R | R | I | S | S | S | S | S | S |
| E. coli ATCC 25922 | S | S | S | S | S | S | S | S | S | S | S | S | S | S | I | S | S | S | |
| Klebsiella pneumoniae | δ | R * | S | S | R | R | R | S | S | R | S | S | I | S | S | S | S | S | S |
| Citrobacter freundii | θ | R * | R * | R | R * | S | R | S | S | S | S | S | I | R | S | R | S | S | S |
| Pseudomonas sp. | M | R * | R * | R * | R * | S | R * | S | S | S | R * | R * | R * | R * | R * | R * | R * | S | R * |
| N | R * | R * | R * | R * | S | R * | S | S | S | R * | R * | R * | R * | R * | R * | R * | S | R * | |
| β | R * | R * | R * | R * | S | R * | S | S | R | R * | R * | R * | R * | R * | R * | R * | S | R * | |
| Z1 | R * | R * | R * | R * | S | R * | S | S | I | R * | R * | R * | R * | R * | R * | R * | S | R * | |
| Z2 | R * | R * | R * | R * | S | R * | S | S | I | R * | R * | R * | R * | R * | R * | R * | S | R * | |
| η | R * | R * | R * | R * | I | R * | S | S | R | R * | R * | R * | R * | R * | R * | R * | S | R * | |
| Achromobacter spanius | F | R * | S | R * | R * | S | R * | S | S | S | R | R | S | S | S | R * | S | S | S |
| Isolate | Potential Important AMR Genes | Phenotypic Resistance | Clustered AMR Genes † | Linked Transposases or IME ‡ | Predicted Contig Origin § |
|---|---|---|---|---|---|
| B | β-lactams: ampC1, CTX-M-1 | Yes | CTX-M-1, tet(C) and sul 2 | IS1294, ISEcp1, IS186B, and IS5075 | Plasmid |
| Tetracyclines: tet(A), tet(C) | Yes | ||||
| Sulphonamides: sul2 | Yes | ||||
| E1-4 | β-lactams: ampC1, TEM-1, DHA-1 | Yes | TEM-1, tet(B), tetR, sul2, APH(3″)-Ib and APH(6)-ld | Tn2 and IS5 | Plasmid |
| Tetracyclines: tet(B), tetR | Yes | ||||
| Sulphonamides: sul1, sul2 | Yes | ||||
| Trimethoprim: dfrA8, dfrA17 | Yes | ||||
| Aminoglycosides: APH(6)-Id, APH(3″)-Ib | Yes | QnrB4, sul1, mphA, and dfrA17 | IS186B, IS1R, and Tn2 | Plasmid | |
| Macrolides: mphA | / | ||||
| Fluoroquinolones: QnrB4 | No | ||||
| I1-2 | β-lactams: ampC1, CTX-M-1 | Yes | CTX-M-1, tet(C) and sul 2 | ISEcp1, IS1294, and IS186B | Plasmid |
| Tetracyclines: tet(C) | Yes | ||||
| Sulphonamides: sul2 | Yes | ||||
| Aminoglycosides: efflux pumps | Yes | ||||
| Fluorquinolones: efflux pumps | Yes | ||||
| K1-2 | β-lactams: ampC1, ampC, CTX-M-1 | Yes | CTX-M-1, tet(A), tet(D) and sul2 | ISEcp1, IS1294, and IS186B | Plasmid |
| Tetracyclines: tet(A), tet(D) | Yes | ||||
| Sulphonamides: sul2 | Yes | ||||
| L1-3 | β-lactams: ampC1, CTX-M-1 | Yes | CTX-M-1 | ISEcp1; putative ICE with T4SS | Chromosome |
| Tetracyclines: tet(A) | Yes | ||||
| Sulphonamides: sul2 | Yes | tet(A) and sul2 | IS1294 and IS5075 | Plasmid | |
| T1-3 | β-lactams: ampC1, CTX-M-3, TEM-1 | Yes | CTX-M-3, mphA, and AAC(3)-IIc | Tn2; Putative IME | Chromosome |
| Tetracyclines: tet(B), tetR | Yes | ||||
| Sulphonamides: sul2 | Yes | ||||
| Trimethoprim: dfrA17 | Yes | ||||
| Aminoglycosides: APH(3′)-Ia, APH(6)-Id, APH(3″)-Ib, aadA5, AAC(3)-IIc | Yes | TEM-1, tet(B), dfrA17, sul2, APH(3′)-Ia, APH(6)-Id, APH(3″)-Ib and aadA5 | Tn2, ISEc8, IS3411, IS1R, and IS2 | Plasmid | |
| Macrolides: mphA | / | ||||
| Fluoroquinolones: gyrA, parC | Yes | ||||
| U1-2 | β-lactams: ampC1, ampC, CTX-M-1 | Yes | CTX-M-1 | ISEcp1 and IS1294 | Plasmid |
| Aminoglycosides: efflux pumps | Yes | ||||
| Fluoroquinolones: gyrA | No | ||||
| V1-2 | β-lactams: ampC1, CTX-M-1 | Yes | CTX-M-1, sul2 | IS5075, IS1294, and ISEcp1 | Plasmid |
| Sulphonamides: sul2 | Yes | ||||
| X1-3 | β-lactams: ampC, CTX-M-1 | Yes | CTX-M-1, sul2 | IS5075, IS1294, and ISEcp1 | Plasmid |
| Sulphonamides: sul2 | Yes | ||||
| γ2 | β-lactams: ampC1, ampC, CTX-M-61, TEM-1 | Yes | CTX-M-61, TEM-1, sul2 | Tn2, ISEcp1, IS2, IS1294, and IS5075 | Plasmid |
| Tetracyclines: efflux pumps | Yes | ||||
| Sulphonamides: sul2 | Yes | ||||
| κ1 | β-lactams: ampC1, CTX-M-1 | Yes | CTX-M-1 | ISEcp1 and IS1294 | Plasmid |
| Trimethoprim: dfrA5 | Yes | ||||
| Κ2 | β-lactams: ampC1, ampC, CTX-M-1 | Yes | CXT-M-1 | ISEcp1, IS1294 | Plasmid |
| μ | β-lactams: CTX-M-1 | Yes | CTX-M-1 | ISEcp1 and IS1294 | Plasmid |
| Sulphonamides: sul2 | Yes | ||||
| Trimethoprim: dfrA17 | Yes | sul2, dfrA17, aadA5 | IS5075 and TnEc3 | Plasmid | |
| Aminoglycosides: aadA5 | No | ||||
| ATCC 25922 | / | / | / | / | / |
| Isolate | Pasteur ST | Warwick ST | Phylogroup | Serotype | CHTyper | Virotype |
|---|---|---|---|---|---|---|
| B | 294 | 162 * | B1 | O8:H28 | fumC65 fimH32 | B |
| E1 | 2 | 10929 * | C | O16:H48 | fumC11 fimH475 | / |
| E2 | 2 | 10929 * | C | O16:H48 | fumC11 fimH475 | / |
| E3 | 2 | 10929 * | C | O16:H48 | fumC11 fimH475 | / |
| E4 | 2 | 10929 * | C | O16:H48 | fumC11 fimH475 | / |
| I1 | 294 | 162 * | B1 | O8:H28 | fumC65 fimH32 | B |
| I2 | 294 | 162 * | B1 | O8:H28 | fumC65 fimH32 | B |
| K1 | 294 | 162 * | B1 | O8:H28 | fumC65 fimH32 | B |
| K2 | 294 | 162 * | B1 | O8:H28 | fumC65 fimH32 | B |
| L1 | 529 | Unknown ST; Nearest match: 180, 675 | B1 | O22:H16 | fumC23 fimH32 | B |
| L2 | 529 | Unknown ST; Nearest match: 180, 675 | B1 | O22:H16 | fumC23 fimH32 | B |
| L3 | 529 | Unknown ST; Nearest match: 180, 675 | B1 | O22:H16 | fumC23 fimH32 | B |
| T1 | 355 | 162 * | B1 | O55:H10 | fumC65 fimH32 | B |
| T2 | 355 | 162 * | B1 | O55:H10 | fumC65 fimH32 | B |
| T3 | 355 | 162 * | B1 | O55:H10 | fumC65 fimH32 | B |
| U1 | Unknown ST; Nearest match: 325 | 1844 | B1 | O8:H49 | fumC29 fimH38 | B |
| U2 | Unknown ST; Nearest match: 325 | 1844 | B1 | O8:H49 | fumC29 fimH38 | B |
| V1 | 42 | 57 * | E | O140:H25 | fumC31 fimH27 | B |
| V2 | 42 | 57 * | E | O140:H25 | fumC31 fimH27 | B |
| X1 | 42 | 57 * | E | O140:H25 | fumC31 fimH27 | B |
| X2 | 42 | 57 * | E | O140:H25 | fumC31 fimH27 | B |
| X3 | 42 | 57 * | E | O140:H25 | fumC31 fimH27 | B |
| γ2 | 843 | 1564 * | A | 0-:H21 | fumC252 fimH / | B |
| κ1 | 24 | Unknown ST; Nearest match: 58, 223 | Unknown | O8:H25 | fumC4 fimH32 | B |
| κ2 | 165 | 10 * | C | O16:H12 | fumC11 fimH24 | / |
| μ | Unknown ST; Nearest match: 606 | 8370 * | F | O13:H56 | fumC48 fimH150/160 | / |
| ATCC 25922 | 52 | Unknown ST; Nearest match: 73, 5999 | B2 | O6:H1 | fumC24 fimH30 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Witte, C.; Vereecke, N.; Theuns, S.; De Ruyck, C.; Vercammen, F.; Bouts, T.; Boyen, F.; Nauwynck, H.; Haesebrouck, F. Presence of Broad-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Zoo Mammals. Microorganisms 2021, 9, 834. https://doi.org/10.3390/microorganisms9040834
De Witte C, Vereecke N, Theuns S, De Ruyck C, Vercammen F, Bouts T, Boyen F, Nauwynck H, Haesebrouck F. Presence of Broad-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Zoo Mammals. Microorganisms. 2021; 9(4):834. https://doi.org/10.3390/microorganisms9040834
Chicago/Turabian StyleDe Witte, Chloë, Nick Vereecke, Sebastiaan Theuns, Claudia De Ruyck, Francis Vercammen, Tim Bouts, Filip Boyen, Hans Nauwynck, and Freddy Haesebrouck. 2021. "Presence of Broad-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Zoo Mammals" Microorganisms 9, no. 4: 834. https://doi.org/10.3390/microorganisms9040834
APA StyleDe Witte, C., Vereecke, N., Theuns, S., De Ruyck, C., Vercammen, F., Bouts, T., Boyen, F., Nauwynck, H., & Haesebrouck, F. (2021). Presence of Broad-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Zoo Mammals. Microorganisms, 9(4), 834. https://doi.org/10.3390/microorganisms9040834








