Abstract
The Mycobacterium fortuitum group (MFG) consists of about 15 species of fast-growing nontuberculous mycobacteria (NTM). These globally distributed microorganisms can cause diseases in humans and animals, especially fish. The increase in the number of species belonging to MFG and the diagnostic techniques panel do not allow to clarify their real clinical significance. In this study, biomolecular techniques were adopted for species determination of 130 isolates derived from fish initially identified through biochemical tests as NTM belonging to MFG. Specifically, gene sequencing and phylogenetic analysis were used based on a fragment of the gene encoding the 65 KDa heat shock protein (hsp65). The analyzes made it possible to confirm that all the isolates belong to MFG, allowing to identify the strains at species level. Phylogenetic analysis substantially confirmed what was obtained by gene sequencing, except for six strains; this is probably due to the sequences present in NCBI database. Although the methodology used cannot represent a univocal identification system, this study has allowed us to evaluate its effectiveness as regards the species of MFG. Future studies will be necessary to apply these methods with other gene fragments and to clarify the real pathogenic significance of the individual species of this group of microorganisms.
1. Introduction
The Mycobacterium fortuitum group (MFG) includes several rapidly growing nontuberculous mycobacteria (NTM) with very similar cultural, biochemical, and molecular features. The group takes its name from the most representative species, Mycobacterium fortuitum, described for the first time in 1938 [1] and included in group IV (rapid growers mycobacteria) of the Runyon’s classification [2]. With the improvement of diagnostic techniques, this group has undergone several additions and taxonomic changes as happened for most species of the genus Mycobacterium. This is evident also in the founding species of MFG, divided into two subspecies, M. fortuitum subsp. fortuitum and M. fortuitum subsp. acetamidolyticum [3]. Based on biochemical and cultural characteristics, several authors had proposed a division of M. fortuitum isolates into three biovars [4], subsequently separated into two distinct species (M. fortuitum and M. peregrinum) and a third biovariant complex with two unnamed taxa [5]. However, Schinsky and collaborators [6] demonstrated that the aforementioned “third biovariant complex” was composed of different species, subsequently named M. boenickei, M. brisbanense, M. houstonense, and M. neworleansense. Based on Bergey’s Manual and papers of the International Journal of Systematic and Evolutionary Microbiology regarding the official nomenclature, the MFG currently includes 13 NTM species: M. alvei, M. boenickei, M. brisbanense, M. conceptionense, M. farcinogenes, M. fortuitum (subs. fortuitum/acetamidolyticum), M. houstonense, M. neworleansense, M. peregrinum, M. porcinum, M. senegalense, M. septicum, and M. setense [7,8,9,10,11,12,13]. Moreover, other species closely related to MFG are reported, although not officially included in the group: specifically, these mycobacteria are M. arceuilense, M. lutetiense and M. montmartrense [14].
The importance of MGT studying lies in that several of its members are implicated in human and animal diseases. Being ubiquitous bacteria, the infection sources are represented by both natural (water and soil) [15,16] and anthropic environments (water distribution systems, swimming pools) [17,18,19,20]. Concerning human medicine, mycobacteria belonging to MFG are mainly the cause of dermal infections in immunocompromised patients [21]. Mycobacterium fortuitum is recognized as an etiological agent of pulmonary [22], bone [23], skin and soft tissue diseases following surgery [24] and catheter-associated infections [25]. Albeit sporadically, cases of infections in human due to members of the MFG are also reported in literature for M. alvei [26], M. brisbanense [27], M. conceptionense [28], M. houstonense [29], M. peregrinum [30,31], M. porcinum [32], M. senegalense [33,34], and M. septicum [35].
As mentioned, other animal species are also susceptible to NTM infection belonging to the MFG. M. fortuitum has been found in several terrestrial animal species, including bovine with mastitis [36,37], wild boars [38], dogs [39], and cats [40]. Mycobacterium senegalense and M. farcinogenes are well known in Africa as bovine pathogens, in which they are causative agents of a cutaneous diseases called “farcy” [7,41]. These same mycobacteria caused abscesses and death in guinea pigs following experimental infection [42]. Similar to what has been described for humans, other members of MFG have also been reported sporadically for wild and domestic animals [8,43,44,45,46].
However, based on scientific literature, the highest reports number of MFG members infections are in fishes. Mycobacterium fortuitum, like M. marinum and M. chelonae, is recognized as the main causative agent of fish mycobacteriosis [47]. Usually, these pathologies begin with nonspecific clinical signs (emaciation, abnormal behavior, scale loss, ulcerative skin lesion, skeletal deformities), then can become chronic with the formation of whitish military nodules in parenchymatous organs (liver, spleen, kidney) [48]. Mycobacteriosis caused by this group of pathogens affects both fresh and saltwater fishes, especially with regard to ornamental species [49,50,51]. Despite the large number of studies, MFG members isolated from fish are not always identified at the species level. In fact, it is possible to find works in which is not possible to discriminate between closely related species or the isolates are indicated as mycobacteria belonging to the MFG [52,53]. This is particularly evident as regards the isolates recognized with the sole use of biochemical tests, which do not allow to discriminate between the various species of MFG (Table 1).

Table 1.
Cultural and biochemical characteristics of the MFG members. The information about the main cultural and biochemical tests used comes from Bergey’s Manual of Systematics Bacteriology, 2nd ed. [54], with additions on the species subsequently described [14].
Therefore, it is likely that over the years there has been an underestimation of the less known and more recent Mycobacterium species of belonging to MFG. In addition, causing a loss of information, this lack of identification does not allow to attribute an accurate clinical significance to the different species of MFG. Thus, the aim of this study is to identify at species level the mycobacteria isolated from fish previously recognized by biochemical tests as M. fortuitum or as members belonging to the MFG.
2. Materials and Methods
2.1. Strains Collection and Preliminary Identification
To perform this study, strains of NTM isolated from fish identified by biochemical tests as MFG members were selected during the years 2015–2019. The strains were isolated from parenchymatous organs (spleen, liver, kidney) of fish with external clinical signs attributable to mycobacteriosis. Before inoculation on Löwenstein–Jensen (Microbiol, Uta-Cagliari, Italy) and Stonebrink medium (Microbiol, Uta-Cagliari, Italy), the organs were homogenized, washed with a 1.5% aqueous solution of cetylpyridinium chloride monohydrate (AppliChem, Darmstadt, Germany) for 30 min and centrifuged for 20 min to obtain a pellet. For the first isolation, two tubes for each medium were incubated at 28 ± 1 °C and at 37 ± 1 °C for two months, respectively. All tubes were constantly checked for evaluating microorganisms growth. At the end of the incubation, colonies grown were then subjected to Ziehl-Neelsen (ZN) staining and biochemical tests as described by Kent and Kubica [55] with updates of Magee and Ward [54]. The tests carried out are those listed previously in Table 1. Following preliminary identification, the strains were stored in cryobank at −80 °C pending subsequent analyses.
2.2. DNA Extraction and Amplification
The stored strains were reactivated using Middlebrook 7H9 broth (Microbiol, Uta-Cagliari, Italy) incubated at 28 ± 1 °C. Then, the broth was inoculated by a 10 μL sterile disposable loop in Löwenstein–Jensen medium to allow a better visualization of the colonies; this medium was incubated again at 28 ± 1 °C. Colonies were placed in a sterile eppendorf containing 200 μL of nuclease-free water (Sigma-Aldrich, St. Louis, MO, USA) and the bacterial DNA was extracted by ExtractMe Genomic DNA kit (Blirt S.A., Gdańsk Poland) following the manufacturer’s guidelines. The extracted nucleic acid was immediately tested or stored at −20 °C before amplification.
For species identification, the protocol proposed by Telenti et al. [56] for the amplification of a 441 bp fragment of the 65 kDa heat shock protein (hsp65) gene was chosen. Compared to the original protocol, the PCRs were conducted in a volume of 50 μL using 25 μL of PrimeDirect™ Probe RT-qPCR Mix, with UNG (Takara Bio Inc., Shiga, Japan), primers Tb11 (5′-ACCAACGATGGTGTGTCCAT) and Tb12 (5′-CTTGTCGAACCGC-ATACCCT) at the concentration of 10 μM, 5 μL of template and nuclease-free water (Sigma-Aldrich, St. Louis, MO, USA) to bring to volume. A reference strain of M. fortuitum (M. fortuitum subsp. fortuitum da Costa Cruz, ATCC® 6841™) was used as PCR positive control and water as negative control. PCR were performed on a 2720 Thermal Cycler (Applied Biosystems, Waltham, MA, USA) using the following thermal protocol: 50 °C for 2 min for Uracil-DNA Glycosylase (UDG) activation, 96 °C for 2 min for the initial denaturation, 40 amplification cycles (denaturation: 95 °C × 30′; annealing: 59 °C × 30′; extension: 72 °C × 30′) and 72 °C for 5 min for the final elongation.
PCR products were visualized using 2% agarose gel (Merck, Darmstadt, Germany), prepared using tris acetate-EDTA buffer 1× (Merck Millipore, Darmstadt, Germany) and GelRed® Nucleic Acid Gel Stain (Biotium, Fremont, CA, USA). Fragment size was assessed using AmpliSize molecular ruler 50–2000 bp ladder (Bio-Rad, Segrate, Italy).
2.3. Sanger Sequencing and Species Determination
PCR products were purified by columns using Extractme DNA Gel-Out kit (Blirt S.A., Gdańsk, Poland). Then, the purified amplicons were subjected to Cycle Sequencing using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer instruction concerning mix preparation and thermal protocol. Cycle sequencing products were purified using DyeEx 2.0 Spin Kit (Quiagen, Hilden, Germany). A total of 5 μL of DNA were loaded into a 96-well plate with 10 μL of formamide (Merck Millipore, Darmstadt, Germany) and analyzed by Abi Prism 310 Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA). A consensus sequence was generated using ClustalW [57], which was then compared with the data on NCBI (National Center for Biotechnology Information) using Nucleotide Blast (Blastn) [58] for species determination. Following identification, the obtained sequences were deposited on the GenBank database.
2.4. Hsp65 Phylogenetic Tree
As a further analysis for species determination, a phylogenetic tree was constructed using the data obtained by sequencing the hsp65 gene fragment. Alignment and phylogenetic analysis were performed with a 421 bp fragment using MEGAX software [59]. The tree was built using sequences of reference strains of the various members of MFG, plus M. chelonae strain ATCC 35,749 with the function of outgroup. The statistical method used was maximum likelihood analysis with the general time reversible (GTR) nucleotide substitution model; 1000 bootstrap replications were performed.
3. Results
3.1. Strains Collection after Preliminary Identification
Following identification by biochemical tests, 130 strains of NTM belonging to MFG were isolated from diseased fish. All isolates were fast-growing (growth in less than seven days), non-chromogenic (no pigment production), alcohol-acid-resistant (ZN+) bacilli. The isolates were identified from 25 different fish species, mostly freshwater (110/130, p = 84.6%). Furthermore, the analyzed mycobacteria were mainly recovered from aquarium fish (109/130, p = 83.8%); the remaining microorganisms came from farmed fish species (21/130, p = 16.2%). All the strains collected were analyzed with biomolecular methods for the species determination.
3.2. Sanger Sequencing and Species Determination
The DNA extracted from all the isolates was amplified by the method described in the previous section (see Material and Methods, Section 2.2). Agarose gel electrophoresis allowed to verify the correct size of all the amplicons. Then, all 130 stored strains were subjected to Sanger sequencing. The comparison of the sequences obtained with those deposited in the NCBI database allowed to identify all the mycobacteria as species belonging to the MFG, with an identity percentage between 98.34% and 100%. Specifically, the isolates belonged to 9 different species, divided as follows: 63 M. peregrinum (p = 48.5%), 38 M. fortuitum (p = 29.2%), 12 M. senegalense (p = 9.2%), four M. arceuilense (p = 3.1%), four M. brisbanense (p = 3.1%), four M. conceptionense (p = 3.1%), two M. alvei (p = 1.5%), two M. setense (p = 1.5%), and one M. septicum (p = 0.8%). Table 2 summarizes the results of the species identification, in relation to the fish species analyzed. Details of isolates, including GenBank Accession Number of the deposited sequences, are available in Supplementary Materials (Table S1).

Table 2.
Results of the identification by hsp65 gene sequencing of the isolated strains.
3.3. Hsp65 Phylogenetic Tree
As a further analysis to determine the species of the isolates, a phylogenetic tree was construct with the obtained sequences compared with those of selected reference strains. The phylogenetic analysis substantially confirmed what previously obtained, except for 6 isolates (indicated by “♦” in the tree). The aforementioned strains are respectively MYC M5-4 (isolated from Aulonocara sp., identified as M. fortuitum, ID% 99.05), MYC 39 (isolated from Colisa lalia, identified as M. conceptionense, ID% 98.34), MYC 80 (isolated from Acipenser ruthenus, identified as M. peregrinum, ID% 99.76), MYC 81 (isolated from Misgurnus sp., identified as M. peregrinum, ID% 99.76), MYC 119 (isolated from Hypostomus plecostomus, identified as M. peregrinum, ID% 99.76), and MYC 153 (isolated from Carassius auratus, identified as M. peregrinum, ID% 99.76). Furthermore, with regard to the correctly-identified strains, the tree allowed to distinguish the M. fortuitum isolates in the two subspecies (Figure 1).
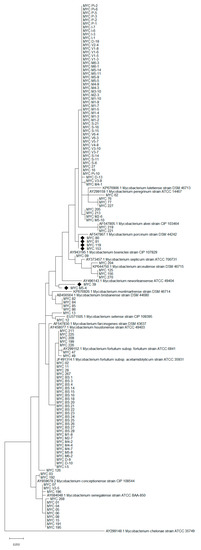
Figure 1.
Phylogenetic tree obtained with a 421 bp fragment of the hsp65 gene. The strains indicated with the symbol “♦” gave conflicting results between gene sequencing and phylogenetic analysis.
4. Discussion
MFG members are globally distributed rapidly growing non-chromogenic mycobacteria. Since the first identification of the founding member of the group, M. fortuitum, the number of related species recognized has gradually increased in relation to the new identification techniques adopted. Given the wide distribution of these microorganisms, humans and animals can easily come into contact with them. In some cases, certain species of MFG are known to cause disease; among them, there are fish mycobacteriosis. Although M. fortuitum is one of the major etiological agents of these pathologies, the other MFG species are not to be excluded. In the present study, we tried to clarify the possible presence and role of the other species of MFG causing fish mycobacteriosis, in addition to M. fortuitum, using biomolecular techniques as an identification tool.
In the first instance, 130 NTM strains preliminarily recognized with biochemical methods as MFG members were analyzed. These strains were isolated from fish that showed external and/or internal (granulomas) clinical signs. Analyzing the positive fish species and holding conditions, we have a fairly accurate representation of what is reported in the literature on fish mycobacteriosis. In fact, most of the species (109/130, p = 83.8%) come from aquaria, whose conditions (limited space, water recirculation, possible overcrowding) are predisposing factors for the onset mycobacteriosis in fish [49,60]. The remaining samples (21/130, p = 16.2%) came from farmed fish: Additoinally, in this case there are predisposing factors for the onset of infectious diseases, including high density in tanks and intensive farming conditions [61]. As further confirmation of the wide distribution of MFG species [15,62,63], the isolates come from both freshwater (110/130, p = 84.6%) and saltwater (20/130, p = 15.4%) fish.
The preliminary identification of these isolates was carried out with biochemical tests. Among them, some are easy to perform and interpret (e.g., growth rate, pigment production, growth at different temperatures), given the presence of exhaustive data in the bibliography. For other tests, however, updated and necessary data are not always available in order to correctly given the result to determining the species of the isolate (see Table 1). In these cases, the data may lack or there may be a variable behavior between isolates of the same species (e.g., Tween-80 hydrolysis for M. fortuitum). It should also be considered that, although rapidly growing, the biochemical tests conducted on members of MFG are time consuming compared to the same ones performed on other more easily cultivable bacterial genera.
In relation to this and to the increase in the number of recognized species [64,65], biomolecular methods are a quick, effective and cheaper way to NTM species determination. Our study appears to confirm this statement: In fact, all the isolates were identified at species level by hsp65 gene sequencing with a percentage between 98.34 and 100%. Based on the study of McNabb and collaborators [66], the identification of species through sequence analysis of a portion of hsp65 gene can be considered valid when the ID percentage is greater than 97%. Based on this, all identifications obtained were considered valid. The most isolated species was M. peregrinum (p = 48.5%). This species is already known as an etiological agent of fish mycobacteriosis, even if it is attributed a secondary role compared to other species (e.g., M. marinum, M. chelonae., M. fortuitum) [47]. Other authors have previously reported high prevalence of this species of mycobacterium [49]: It must therefore be considered whether these are sporadic cases linked to occasional mortality or whether there is an underestimation. To follow, the most identified species was M. fortuitum (p = 29.2%). Being among the species most involved in fish mycobacteriosis onset [47,48], our study confirms when already known from previous studies. As mentioned, the use of biomolecular techniques has led to the recognition of new species of MFG [14,64,67]. The application of these techniques in the diagnosis of fish mycobacteriosis has allowed to highlight nine different MFG species in our study. Previous studies using analytical methodologies comparable to those used in our study were able to identify these same species [52,53]. As several MFG species have also been found in humans [11,12,13,68], correct identification at species level is essential, without limiting to the use of methods such as biochemical tests. In fact, the improvement of diagnostic techniques is fundamental for these poor studied microorganisms to implement our knowledge, especially for their potential zoonotic implication.
As a further test, a phylogenetic analysis was performed to confirm the sequencing results. Compare to gene sequencing, this analysis provides an easily interpretable and immediate result, even on a graphic level. Despite the high identification rates in sequencing, 6 isolates (p = 4.6%) did not confirm the results obtained following phylogenetic analysis. For example, MYC 80, MYC 81, MYC 119 and MYC 153 were all identified as M. peregrinum with an ID% of 99.76 (similarity with strain MK341520.1) following Blastn analysis; instead, the phylogenetic tree shows a greater resemblance to M. porcinum DSM 44242. These cases pose a known problem for those who work with gene sequences databases, that is the quality controls of the deposited sequences [69]. Finally, compared to Blastn identification, the phylogenetic tree allowed the division between the strains of M. fortuitum in the two related subspecies, further implementing the information regarding the identification of these isolates.
5. Conclusions
NTM belonging to the MFG represent both a problem for fish and for public health, being potential zoonotic agents [47,52]. Being a group made up of several species, correct identification is the first step for accurate diagnosis and therapy, as well as to increase knowledge on relatively recent identified species (e.g., M. arceuilense, M. lutetiense, M. montmartrense). These little known and poorly diagnosed species pose several problems, including the use of adequate diagnostic techniques for the identification and assignment of a precise clinical significance. Our study confirmed that the use of a portion of the hsp65 gene is a valid diagnostic tool for MFG species identification, through several hypervariable regions present in its sequence (Table 3).

Table 3.
Mutations in the hsp65 gene sequences of the portion considered in the study found in the NCBI Nucleotide database. Reference strains indicated as type strain in Bergey’s manual [54] were considered. Nucleotide positions refer to those obtained by aligning type strains sequences with the complete genome of M. fortuitum subsp. fortuitum strain ATCC 6841 (Sequence ID: CP014258.1). The dot (·) indicates identical base compared to M. fortuitum subsp. fortuitum strain ATCC 6841 taken as reference species.
Despite the advantages, our study also highlighted several limitations in the methods used. First, the sequences present in the database must be of good quality for a correct identification of the species. Although qualitatively acceptable, some species have only single sequences for the hsp65 gene. Therefore, although the hsp65 gene provides more accurate identifications than other genes (e.g., rRNA16S) for MFG members, the most suitable approach in case of sequencing is the multigenic one [70]. Therefore, in cases of doubtful identification, the use of more fragments of the genome of these microorganisms is recommended, as a single gene able to discriminate all the species of the genus Mycobacterium is not yet known. In addition to sequencing, phylogenetic analysis provides a further diagnostic tool. Although there are advantages, even with respect to gene sequencing, phylogenetically similar species are a limitation for this method. In this perspective, a taxonomic revision of the MFG from a clinical point of view could bring advantages in the diagnostic process [71,72].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9040797/s1, Table S1. Details about the strains analyzed in the study.
Author Contributions
Methodology: D.M., M.T., P.P., G.E., V.M., A.D., and M.P.; investigation: D.M., M.T., P.P., G.E., V.M., and M.P.; resources: A.D. and M.P.; data curation: D.M., M.T., P.P., G.E., V.M., and M.P.; writing—original draft preparation: D.M.; writing—review and editing: M.T., P.P., G.E., V.M., A.D., and M.P.; visualization: D.M., P.P., and M.P.; supervision: A.D. and M.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Da Costa Cruz, J.C. Mycobacterium fortuitum um novo bacillo acidoresistance pathogenico para o homen. Acta Med. Rio Janerio 1938, 1, 297–301. [Google Scholar]
- Runyon, E.H. Anonymous mycobacteria in pulmonary disease. Med. Clin. N. Am. 1959, 43, 273–290. [Google Scholar] [CrossRef]
- Tsukamura, M.; Yano, I.; Imaeda, T. Mycobacterium fortuitum subspecies acetamidolyticum, a new subspecies of Mycobacterium fortuitum. Microbiol. Immunol. 1986, 30, 97–110. [Google Scholar] [CrossRef]
- Bönicke, R. The occurrence of atypical mycobacteria in the environment of man and animal. Bull. Int. Union Tuberc. 1966, 37, 361–368. [Google Scholar]
- Wallace, R.J.; Brown, B.A., Jr.; Silcox, V.A.; Tsukamura, M.; Nash, D.R.; Steele, L.C.; Steingrube, V.A.; Smith, J.; Sumter, G.; Zhang, Y.S.; et al. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 1991, 163, 598–603. [Google Scholar] [CrossRef]
- Schinsky, M.F.; Morey, R.E.; Steigerwalt, A.G.; Douglas, M.P.; Wilson, R.W.; Floyd, M.M.; Butler, W.R.; Daneshvar, M.I.; Brown-Elliott, B.A.; Wallace, R.J., Jr.; et al. Taxonomic variation in the Mycobacterium fortuitum third biovariant complex: Description of Mycobacterium boenickei sp. nov., Mycobacterium houstonense sp. nov., Mycobacterium neworleansense sp. nov. and Mycobacterium brisbanense sp. nov. and recognition of Mycobacterium porcinum from human clinical isolates. Int. J. Syst. Evol. Microbiol. 2004, 54, 1653–1667. [Google Scholar] [CrossRef]
- Chamoiseau, G. Etiology of farcy in African bovines: Nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 1979, 29, 407–410. [Google Scholar] [CrossRef][Green Version]
- Tsukamura, M.; Nemoto, H.; Yugi, H. Mycobacterium porcinum sp. nov., a porcine pathogen. Int. J. Syst. Bacteriol. 1983, 33, 162–165. [Google Scholar] [CrossRef]
- Ausina, V.; Luquin, M.; Barcelo, M.G.; Laneelle, M.A.; Levy-Frebault, V.; Belda, F.; Prats, G. Mycobacterium alvei sp. nov. Int. J. Syst. Bacteriol. 1992, 42, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Kusunoki, S.; Ezaki, T. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status, Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 1992, 42, 240–245. [Google Scholar] [CrossRef]
- Schinsky, M.F.; McNeil, M.M.; Whitney, A.M.; Steigerwalt, A.G.; Lasker, B.A.; Floyd, M.M.; Hogg, G.G.; Brenner, D.J.; Brown, J.M. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. Evol. Microbiol. 2000, 50, 575–581. [Google Scholar] [CrossRef]
- Adékambi, T.; Stein, A.; Carvajal, J.; Raoult, D.; Drancourt, M. Description of Mycobacterium conceptionense sp. nov., a Mycobacterium fortuitum group organism isolated from a posttraumatic osteitis inflammation. J. Clin. Microbiol. 2006, 44, 1268–1273. [Google Scholar] [CrossRef]
- Lamy, B.; Marchandin, H.; Hamitouche, K.; Laurent, F. Mycobacterium setense sp. nov., a Mycobacterium fortuitum-group organism isolated from a patient with soft tissue infection and osteitis. Int. J. Syst. Evol. Microbiol. 2008, 58, 486–490. [Google Scholar] [CrossRef]
- Konjek, J.; Souded, S.; Guerardel, Y.; Trivelli, X.; Bernut, A.; Kremer, L.; Welte, B.; Joyeux, M.; Dubrou, S.; Euzeby, J.P.; et al. Mycobacterium lutetiense sp. nov., Mycobacterium montmartrense sp. nov. and Mycobacterium arcueilense sp. nov., members of a novel group of non-pigmented rapidly growing mycobacteria recovered from a water distribution system. Int. J. Syst. Evol. Microbiol. 2016, 66, 3694–3702. [Google Scholar] [CrossRef]
- Bartram, J.; Cotruvo, J.A.; Dufour, A.; Rees, G.; Pedley, S.; Water, S.; World Health Organization. Pathogenic Mycobacteria in Water: A Guide to Public Health Consequences, Monitoring and Management; IWA Publishing: London, UK, 2004. [Google Scholar]
- De Groote, M.A.; Pace, N.R.; Fulton, K.; Falkinham, J.O., III. Relationship between Mycobacterium isolates from patients with pulmonary mycobacterial infection and potting soils. Appl. Environ. Microbiol. 2006, 72, 7602–7606. [Google Scholar] [CrossRef]
- Emde, K.M.; Chomyc, S.A.; Finch, G.R. Initial investigation on the occurrence of Mycobacterium species in swimming pools. J. Environ. Health 1992, 54, 34–37. [Google Scholar]
- Van Ingen, J.; Boeree, M.J.; Dekhuijzen, P.N.R.; van Soolingen, D. Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin. Microbiol. Infect. 2009, 15, 888–893. [Google Scholar] [CrossRef]
- Halstrom, S.; Price, P.; Thomson, R. Review: Environmental mycobacteria as a cause of human infection. Int. J. Mycobacteriol. 2015, 4, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Millar, B.C.; Moore, J.E. Hospital ice, ice machines, and water as sources of nontuberculous mycobacteria: Description of qualitative risk assessment models to determine host-Nontuberculous mycobacteria interplay. Int. J. Mycobacteriol. 2020, 9, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C.; Somoskovi, A. Human pathogenic mycobacteria. In Reference Module in Biomedical Sciences; Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Matsumoto, T.; Otsuka, K.; Tomii, K. Mycobacterium fortuitum thoracic empyema: A case report and review of the literature. J. Infect. Chemother. 2015, 21, 747–750. [Google Scholar] [CrossRef] [PubMed]
- Staneck, J.L.; Frame, P.T.; Altemeier, W.A.; Miller, E.H. Infection of bone by Mycobacterium fortuitum masquerading as Nocardia asteroides. Am. J. Clin. Pathol. 1981, 76, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, J.; Mougari, F.; Picot, S.; Boukerrou, M.; Barau, G.; Ali Ahmed, S.A.; Raskine, L.; Camuset, G.; Michault, A.; Simac, C.; et al. A case of postoperative breast infection by Mycobacterium fortuitum associated with the hospital water supply. Am. J. Infect. Control 2015, 43, 406–408. [Google Scholar] [CrossRef]
- Rathor, N.; Khillan, V.; Panda, D. Catheter associated mycobacteremia: Opening new fronts in infection control. Indian J. Crit. Care Med. 2015, 19, 350–352. [Google Scholar] [CrossRef]
- Lee, C.H.; You, H.L.; Wang, J.W.; Tang, Y.F.; Liu, J.W. Prosthetic joint infection caused by Mycobacterium alvei in an elderly patient. J. Clin. Microbiol. 2011, 49, 3096–3098. [Google Scholar] [CrossRef] [PubMed]
- Gynthersen, R.; Qvist, T.; Andersen, Å.B.; Katzenstein, T.L. Mycobacterium brisbanense lung infection facilitated by steroid induced adrenal insufficiency. APMIS 2019, 127, 106–108. [Google Scholar] [CrossRef]
- Michienzi, S.M.; Burgos, R.M.; Novak, R.M. Mycobacterium conceptionense pneumonitis in patient with HIV/AIDS1. Emerg. Infect. Dis. 2019, 25, 1986–1988. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, Z.; Chen, Z.; Sun, Z. Surgical wound infection following open humeral fracture caused by Mycobacterium houstonense: A case report. BMC Infect. Dis. 2019, 19, 333. [Google Scholar] [CrossRef]
- Wallace, R.J., Jr.; Swenson, J.M.; Silcox, V.A.; Good, R.C.; Tschen, J.A.; Stone, M.S. Spectrum of disease due to rapidly growing mycobacteria. Rev. Infect. Dis. 1983, 5, 657–679. [Google Scholar] [CrossRef]
- Griffith, D.E.; Girard, W.M.; Wallace, R.J., Jr. Clinical features of pulmonary disease caused by rapidly growing mycobacteria: Analysis of 154 patients. Am. Rev. Respir. Dis. 1993, 147, 1271–1278. [Google Scholar] [CrossRef]
- Patil, R.; Patil, T.; Schenfeld, L.; Massoud, S. Mycobacterium porcinum peritonitis in a patient on continuous ambulatory peritoneal dialysis. J. Gen. Intern. Med. 2011, 26, 346–348. [Google Scholar] [CrossRef]
- Klomjit, N.; Chewcharat, A.; D’Uscio, M.; Kattah, A.G. Mycobacterium septicum associated peritonitis: A case report. Perit. Dial. Int. 2020, 40, 600–602. [Google Scholar] [CrossRef]
- Carretero, O.; Reyes, C.; San-Juan, R.; Chaves, F.; López-Roa, P. Mycobacterium senegalense infection after implant-based breast reconstruction, Spain. Emerg. Infect. Dis. 2020, 26, 611–613. [Google Scholar] [CrossRef]
- Hogg, G.G.; Schinsky, M.F.; McNeil, M.M.; Lasker, B.A.; Silcox, V.A.; Brown, J.M. Central line sepsis in a child due to a previously unidentified mycobacterium. J. Clin. Microbiol. 1994, 37, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.J. Mycobacterium fortuitum as a cause of bovine mastitis: Tuberculin sensitivity following experimental infections. J. Am. Vet. Med. Assoc. 1965, 147, 1600–1607. [Google Scholar] [PubMed]
- Koehne, G.; Maddux, R.; Britt, J. Rapidly growing mycobacteria associated with bovine mastitis. Am. J. Vet. Res. 1981, 42, 1238–1239. [Google Scholar]
- Kalensky, P. Isolation of mycobacteria from wild boar. Veterinárství 1992, 42, 346–347. [Google Scholar]
- Leissinger, M.K.; Garber, J.B.; Fowlkes, N.; Grooters, A.M.; Royal, A.B.; Gaunt, S.D. Mycobacterium fortuitum lipoid pneumonia in a dog. Vet. Pathol. 2015, 52, 356–359. [Google Scholar] [CrossRef]
- Couto, S.S.; Artacho, C.A. Mycobacterium fortuitum pneumonia in a cat and the role of lipid in the pathogenesis of atypical mycobacterial infections. Vet. Pathol. 2007, 44, 543–546. [Google Scholar] [CrossRef]
- Ridell, M.; Goodfellow, M.; Abdulle, M.A. Identification of actinomycetes isolated from cases of bovine farcy in the Sudan. Zent. Bakteriol. Mikrobiol. Hyg. A 1985, 259, 43–50. [Google Scholar] [CrossRef]
- Bercovier, H.; Vincent, V. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. Rev. Sci. Tech. 2001, 20, 265–290. [Google Scholar] [CrossRef]
- Youssef, S.; Archambault, M.; Parker, W.; Yager, J. Pyogranulomatous panniculitis in a cat associated with infection by the Mycobacterium fortuitum/peregrinum group. Can. Vet. J. 2002, 43, 285–287. [Google Scholar]
- Malik, R.; Shaw, S.E.; Griffin, C.; Stanley, B.; Burrows, A.K.; Bryden, S.L.; Titmarsh, J.; Stutsel, M.J.; Carter, S.A.; Warner, A.; et al. Infections of the subcutis and skin of dogs caused by rapidly growing mycobacteria. J. Small Anim. Pract. 2004, 45, 485–494. [Google Scholar] [CrossRef]
- Asaava, L.L.A.; Githui, W.A.; Mwangi, M.; Mwangi, E.; Juma, E.; Moraa, R.; Halakhe, A.; Gicheru, M.M. Isolation, identification and associated risk factors of non-tuberculous mycobacteria infection in humans and dromedary camels in Samburu County, Kenya. Zoonoses Public Health 2020, 67, 713–731. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Udenberg, T.J. Mycobacterium porcinum causing panniculitis in the cat. Can. Vet. J. 2020, 61, 39–43. [Google Scholar] [PubMed]
- Delghandi, M.R.; El-Matbouli, M.; Menanteau-Ledouble, S. Mycobacteriosis and infections with non-tuberculous mycobacteria in aquatic organisms: A review. Microorganisms 2020, 8, 1368. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, D.T.; Rhodes, M.W. Mycobacteriosis in fishes: A review. Vet. J. 2009, 180, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, R.G.; Florio, D.; Fioravanti, M.L.; Rossi, M.; Prearo, M. Occurrence of Mycobacterium spp. in ornamental fish in Italy. J. Fish Dis. 2008, 31, 433–441. [Google Scholar] [CrossRef]
- Akbari, S.; Mosavari, N.; Tadayon, K.; Rahmati-Holasoo, H. Isolation of Mycobacterium fortuitum from fish tanks in Alborz, Iran. Iran. J. Microbiol. 2014, 6, 234–239. [Google Scholar]
- Puk, K.; Banach, T.; Wawrzyniak, A.; Adaszek, Ł.; Ziętek, J.; Winiarczyk, S.; Guz, L. Detection of Mycobacterium marinum, M. peregrinum, M. fortuitum and M. abscessus in aquarium fish. J. Fish Dis. 2018, 41, 153–156. [Google Scholar] [CrossRef]
- Kušar, D.; Zajc, U.; Jenčič, V.; Ocepek, M.; Higgins, J.; Žolnir-Dovč, M.; Pate, M. Mycobacteria in aquarium fish: Results of a 3-year survey indicate caution required in handling pet-shop fish. J. Fish Dis. 2017, 40, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Gcebe, N.; Michel, A.L.; Hlokwe, T.M. Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa. BMC Microbiol. 2018, 18, 32. [Google Scholar] [CrossRef]
- Magee, J.G.; Ward, A.C. Family III: Mycobacteriaceae chester 1897. In Bergey’s Manual of Systematics Bacteriology, 2nd ed.; Kämpfer, P., Busse, H.J., Trujillo, M.E., Suzuki, K., Ludwig, W., Whitman, W.B., Eds.; Springer: New York, NY, USA, 2012; Volume 5, pp. 312–375. [Google Scholar]
- Kent, P.T.; Kubica, G.P. Public Health Mycobacteriology. A Guide for the Level III Laboratory; US Department of Health and Human Services, Centers for Disease Control: Atlanta, GA, USA, 1985. [Google Scholar]
- Telenti, A.; Marchesi, F.; Balz, M.; Bally, F.; Böttger, E.C.; Bodmer, T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 1993, 31, 175–178. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega, accurate alignment of very large numbers of sequences. In Multiple Sequence Alignment Methods Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2013; Volume 1079, pp. 105–116. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Beran, V.; Matlova, L.; Dvorska, L.; Svastova, P.; Pavlik, I. Distribution of mycobacteria in clinically healthy ornamental fish and their aquarium environment. J. Fish. Dis. 2006, 29, 383–393. [Google Scholar] [CrossRef]
- Aranaz, A.; Gibello, A.; Alvarez, J.; Mata, A.I.; Rodríguez, A.; Fallola, C.; Fernández-Garayzábal, J.F.; Domínguez, L. Mycobacterium peregrinum infection in farmed European tench (Tinca tinca L.). Vet. Microbiol. 2008, 131, 393–399. [Google Scholar] [CrossRef]
- Mediel, M.J.; Rodriguez, V.; Codina, G.; Martin-Casabona, N. Isolation of mycobacteria from frozen fish destined for human consumption. Appl. Environ. Microbiol. 2000, 66, 3637–3638. [Google Scholar] [CrossRef] [PubMed]
- Lorencova, A.; Klanicova, B.; Makovcova, J.; Slana, I.; Vojkovska, H.; Babak, V.; Pavlik, I.; Slany, M. Nontuberculous mycobacteria in freshwater fish and fish products intended for human consumption. Foodborne Pathog. Dis. 2013, 10, 573–576. [Google Scholar] [CrossRef]
- Tortoli, E. Impact of genotypic studies on mycobacterial taxonomy: The new mycobacteria of the 1990s. Clin. Microbiol. Rev. 2003, 16, 319–354. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. The new mycobacteria: An update. FEMS Immunol. Microbiol. 2006, 48, 159–178. [Google Scholar] [CrossRef]
- McNabb, A.; Eisler, D.; Adie, K.; Amos, M.; Rodrigues, M.; Stephens, G.; Black, W.A.; Isaac-Renton, J. Assessment of partial sequencing of the 65-kilodalton heat shock protein gene (hsp65) for routine identification of Mycobacterium species isolated from clinical sources. J. Clin. Microbiol. 2004, 42, 3000–3011. [Google Scholar] [CrossRef] [PubMed]
- Tortoli, E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev. 2014, 27, 727–752. [Google Scholar] [CrossRef] [PubMed]
- Talavlikar, R.; Carson, J.; Meatherill, B.; Desai, S.; Sharma, M.; Shandro, C.; Tyrrell, G.J.; Kuhn, S. Mycobacterium senegalense tissue infection in a child after fish tank exposure. Can. J. Infect. Dis. Med. Microbiol. 2011, 22, 101–103. [Google Scholar] [CrossRef]
- Turenne, C.Y.; Tschetter, L.; Wolfe, J.; Kabani, A. Necessity of quality-controlled 16S rRNA gene sequence databases: Identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 2001, 39, 3637–3648. [Google Scholar] [CrossRef]
- Gomila, M.; Ramirez, A.; Lalucat, J. Diversity of environmental Mycobacterium isolates from hemodialysis water as shown by a multigene sequencing approach. Appl. Environ. Microbiol. 2007, 73, 3787–3797. [Google Scholar] [CrossRef]
- Tortoli, E.; Fedrizzi, T.; Meehan, C.J.; Trovato, A.; Grottola, A.; Giacobazzi, E.; Serpini, G.F.; Tagliazucchi, S.; Fabio, A.; Bettua, C.; et al. The new phylogeny of the genus Mycobacterium: The old and the news. Infect. Genet. Evol. 2017, 56, 19–25. [Google Scholar] [CrossRef]
- Paul, G.R.; Leber, A.; Nemastil, C.J.; Novak, K.J.; Brady, M.; Stack-Simone, S.; Greninger, A.L.; Antonara, S. Identification of Mycobacterium porcinum in patients with cystic Fibrosis: Pathogen or contaminant? J. Cyst. Fibros. 2020, 19, 580–586. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).