Abstract
Mycobacterium fortuitum group (MFG) members are able to cause clinical mycobacteriosis in fish and other animals including humans. M. alvei, M. arceuilense, M. brisbanense, M. conceptionense, M. fortuitum, M. peregrinum, M. porcinum, M. senegalense, M. septicum, and M. setense were isolated from fish with mycobacteriosis. In other animals only three MFG species have been isolated: M. arceuilense from camels’ milk, M. farcinogenes from cutaneous infections often described as “farcy”, and M. fortuitum from different domestic and wild mammals’ species. Out of 17, only 3 MFG species (M. arceuilense, M. lutetiense and M. montmartrense) have never been reported in humans. A total of eight MFG members (M. alvei, M. brisbanense, M. conceptionense, M. fortuitum subsp. acetamidolyticum, M. houstonense, M. peregrinum, M. porcinum, and M. septicum) have been isolated from both pulmonary and extrathoracic locations. In extrathoracic tissues five MFG species (M. boenickei, M. farcinogenes, M. neworleansense, M. senegalense, and M. setense) have been diagnosed and only one MFG member (M. fortuitum subsp. acetamidolyticum) has been isolated from pulmonary infection.
We would like to comment on a recent article published in your journal by Mugetti et al. [1]. This article is well written and discusses the diagnostics of all 17 members of the Mycobacterium fortuitum Group (MFG) currently known. The aim of the study was the improvement of species diagnostics with respect to pathogenicity of all MFG members in 130 freshwater and saltwater fish with pathognomonic pathological lesions in parenchymatous organs and the skin. The field is divided by the fact that while many publications include most MFG members as fish pathogens, another significant number of publications describe these MFG members only as saprophytic mycobacteria present in the water environment (biofilm, sediment, water plants, non-vertebrates and other matrices) or on healthy animals including fish. To this end we have summarised published results concerning the prevalence of MFG members in the environment and their clinical relevance to humans and animals (including fish). Specifically, we question whether these MFG members are not only pathogenic/virulent to fish, but also to other animals and humans from an epidemiological and epizootiological point of view.
1. Clinical Relevance in Fish
Mycobacterium fortuitum was first described in 1938 [2] and since that time, during last two decades in particular, the remaining MFG members have been identified (Table 1). A total of 11 MFG members have been isolated from fish. Prior to a study by Mugetti et al. [1], only M. fortuitum, M. peregrinum, M. porcinum, M. septicum and M. setense were described as fish pathogens. In their study, Mugetti et al. [1] described for the first time five MFG members (M. alvei, M. arceuilense, M. brisbanense, M. conceptionense and M. senegalense) as species isolated from fish with mycobacteriosis (Table 1). These five MFG species were isolated from 10 genera of freshwater fish originally living in tropical and temperate zones. M. alvei and M. brisbanense were detected in one fish genus each. Three MFG species were isolated from fish of three (M. arceuilense), four (M. conceptionense) and six (M. senegalense) genera (Table 2). This finding could indicate that these MFG members could be considered as emerging fish pathogens.

Table 1.
Mycobacterium fortuitum Group members’ characteristics (year of description, pathogenicity/virulence for humans and animals, environmental prevalence and detection in fish).

Table 2.
Characteristics of examined infected freshwater and saltwater fish with mycobacteriosis in the study carried out by Mugetti et al. [1].
Three MFG species (M. conceptionense, M. houstonense and M. senegalense) were previously isolated only rarely from fish directed for consumption, without known clinical relevance for the fish [7]. In the study of Mugetti et al. the pathogenicity for fish was documented in two of these MFG species: M. conceptionense and M. senegalense [1]. Two MFG species (M. fortuitum and M. peregrinum) in this study were diagnosed in 10 and 13 fish genera, respectively (Table 2); however, the relevance of this observation is questionable. This high prevalence in fish with mycobacteriosis was more frequently diagnosed in significantly earlier studies, as shown in Table 1, when diagnostics were not as sophisticated as they are now [1] and it is possible that misidentification of these species has occurred in some of these cases.
2. Clinical Relevance in Other Animals
Surprisingly in other animals (specifically mammal, not including humans), only three MFG species have been isolated: M. arceuilense from camels’ milk, M. farcinogenes from cutaneous infections often described as “farcy” and M. fortuitum from different domestic and wild mammal species (Table 1). These relatively rare cases are in contrast with MFG species detection in humans, described below.
3. Clinical Relevance in Humans
Out of 17, only 3 MFG species (M. arceuilense, M. lutetiense and M. montmartrense) have not ever been reported in humans. A total of eight MFG members (M. alvei, M. brisbanense, M. conceptionense, M. fortuitum subsp. acetamidolyticum, M. houstonense, M. peregrinum, M. porcinum, and M. septicum) have been isolated from both pulmonary and extrathoracic tissues. In extrathoracic tissues, five MFG species (M. boenickei, M. farcinogenes, M. neworleansense, M. senegalense, and M. setense) have been diagnosed and only one MFG member (M. fortuitum subsp. acetamidolyticum) has been isolated in pulmonary infection. Three MFG species (M. arceuilense, M. lutetiense, and M. montmartrense) have not ever been reported in humans (Table 1).
The fact that most MFG species have been isolated in humans, and more rarely in animals and the environment, is likely due to a lack of research. The source of mycobacteriosis caused by MFG member infections reported in humans is often suspected to be from the immediate environment, such as living spaces, hospitals, gardens, swimming pools, surface waters, etc. [31]; however, few research groups actively demonstrate that the environment is the source. Only a study by Konjek et al. [4], focused on the water distribution system in which the presence of M. lutetiense and M. montmartrense was described and research done by Davarpanah et al. [30] studying the presence of MFG members in hospital soil and dust in Iran are exceptions to this. This work by Mugetti et al. [1] should encourage further investigation into environmental sources of MFG infections. Further research should also be conducted to evaluate the clinical relevance of remaining MFG species previously not reported in humans.
4. Environmental Prevalence
The ecology of mycobacteria, including all known MFG species (up until 2009) was described by Kazda et al. [31]. At that time 14 of 17 MFG members had been described previously (Table 1). Over the subsequent decade we have continued the study of mycobacterial ecology concerning MFG members. We have described 12 MFG members isolated from the environment and non-vertebrates, including from peat [32,33], soil and earthworms [34], aquariums and fishponds [35], prawns used for fish feed and aquarium plants such as Vesicularia dubyana and Cryptocoryne wendtii [36], alluvial wooden material in karstic caves [37] and in other matrices (unpublished data). Only five MFG members (M. boenickei, M. farcinogenes, M. fortuitum subsp. acetamidolyticum, M. houstonense, and M. neworleansense) have not ever been reported in the environment (Table 1). From an epidemiological and epizootiological point of view, we suspect that the environmental presence of MFG members plays an important role in the spreading and host exposure of MFG members.
5. Conclusions
In the publication of Mugetti et al., the current taxonomy of MFG members is shown and an improvement to the methods for identifying clinically relevant and environmental isolates is demonstrated [1]. As well as fish, MFG members are able to cause clinical mycobacteriosis in other animals including humans. Therefore, the detection of new MFG species in fish could indicates new risks for humans, in particular for fish handlers, aquarists and their family members (Figure 1). MFG species have often been diagnosed in human skin infections in a condition previously known as “swimming pool granuloma”. A decrease in the prevalence of this condition is thought to be due to improvements in swimming pool management. Consequently, this condition is mostly seen in infected people associated with aquarium fish, and currently skin mycobacteriosis is most often described as “fish tank granuloma” [31]. Results presented by Mugetti et al. open new perspectives in the rapid identification of MFG members not only in fish, but also in other hosts and the environment [1]. We envisage that this added information concerning the clinical relevance of MFG members and their environmental prevalence will significantly extend the knowledge of this field.
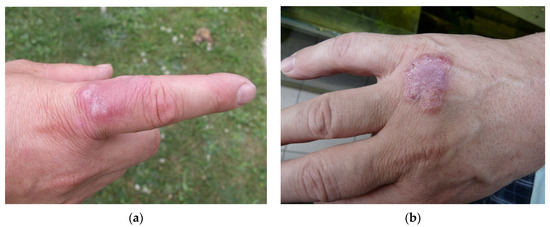
Figure 1.
Fish tank granuloma diagnosed in professional aquarist on two places on his right hand: (a) infection on 17 June 2003; (b) infection on 2 July 2008 (photo I. Pavlik).
Author Contributions
Conceptualization: I.P. and R.T.W.; clinical relevance in humans: V.U. and I.P.; clinical relevance in animals: I.P.; environmental prevalence: R.T.W. and I.P.; writing—review and editing: R.T.W. and I.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by the Czech Health Research Council No. NU20-09-00114.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of Mendel University in Brno, Czech Republic (Ethics Committee Resolution No. 9, 6 November 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Availability of data and materials correspondence be addressed to the corresponding author.
Conflicts of Interest
The author declares no conflict of interest.
References
- Mugetti, D.; Tomasoni, M.; Pastorino, P.; Esposito, G.; Menconi, V.; Dondo, A.; Prearo, M. Gene sequencing and phylogenetic analysis: Powerful tools for an improved diagnosis of fish mycobacteriosis caused by Mycobacterium fortuitum group members. Microorganisms 2021, 9, 797. [Google Scholar] [CrossRef] [PubMed]
- LPSN: List of Prokaryotic Names with Standing in Nomenclature. Available online: https://lpsn.dsmz.de/ (accessed on 6 October 2021).
- Varghese, B.; Al-Hajoj, S. A global update on rare non-tuberculous mycobacteria in humans: Epidemiology and emergence. Int. J. Tuberc. Lung Dis. 2020, 24, 214–223. [Google Scholar] [CrossRef]
- Konjek, J.; Souded, S.; Guerardel, Y.; Trivelli, X.; Bernut, A.; Kremer, L.; Welte, B.; Joyeux, M.; Dubrou, S.; Euzeby, J.P.; et al. Mycobacterium lutetiense sp. nov., Mycobacterium montmartrense sp. nov. and Mycobacterium arcueilense sp. nov., members of a novel group of non-pigmented rapidly growing mycobacteria recovered from a water distribution system. Int. J. Syst. Evol. Microbiol. 2016, 66, 3694–3702. [Google Scholar] [CrossRef]
- Asaava, L.L.A.; Githui, W.A.; Mwangi, M.; Mwangi, E.; Juma, E.; Moraa, R.; Halakhe, A.; Gicheru, M.M. Isolation, identification and associated risk factors of non-tuberculous mycobacteria infection in humans and dromedary camels in Samburu County, Kenya. Zoonoses Public Health 2020, 67, 713–731. [Google Scholar] [CrossRef]
- Wee, W.Y.; Tan, T.K.; Jakubovics, N.S.; Choo, S.W. Whole-Genome sequencing and comparative analysis of Mycobacterium brisbanense reveals a possible soil origin and capability in fertiliser synthesis. PLoS ONE 2016, 11, e0152682. [Google Scholar] [CrossRef]
- Lorencova, A.; Klanicova, B.; Makovcova, J.; Slana, I.; Vojkovska, H.; Babak, V.; Pavlik, I.; Slany, M. Nontuberculous mycobacteria in freshwater fish and fish products intended for human consumption. Foodborne Pathog. Dis. 2013, 10, 573–576. [Google Scholar] [CrossRef]
- Oda, G.; Winters, M.A.; Pacheco, S.M.; Sikka, M.K.; Bleasdale, S.C.; Dunn, B.; Boswell, E.; Winters, B.; Bumsted, A.; Frisch, J.; et al. Clusters of nontuberculous mycobacteria linked to water sources at three Veterans Affairs Medical Centers. Infect. Control Hosp. Epidemiol. 2020, 41, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Tan, W.; Deng, Y.; Gao, D.; Li, L.; Lu, Z.; Graviss, E.A.; Ma, X. Diagnostic delay of pulmonary nontuberculous mycobacterial infection in China. Multidiscip. Respir. Med. 2014, 9, 48. [Google Scholar] [CrossRef]
- Chamoiseau, G. Etiology of farcy in African bovines: Nomenclature of the causal organisms Mycobacterium farcinogenes Chamoiseau and Mycobacterium senegalense (Chamoiseau) comb. nov. Int. J. Syst. Bacteriol. 1979, 29, 407–410. [Google Scholar] [CrossRef][Green Version]
- Ridell, M.; Goodfellow, M.; Abdulle, M.A. Identification of Actinomycetes isolated from cases of bovine farcy in the Sudan. Zent. Bakteriol. Mikrobiol. Hyg. A 1985, 259, 43–50. [Google Scholar] [CrossRef]
- Bercovier, H.; Vincent, V. Mycobacterial infections in domestic and wild animals due to Mycobacterium marinum, M. fortuitum, M. chelonae, M. porcinum, M. farcinogenes, M. smegmatis, M. scrofulaceum, M. xenopi, M. kansasii, M. simiae and M. genavense. Rev. Sci. Tech. 2001, 20, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Tsukamura, M.; Yano, I.; Imaeda, T. Mycobacterium fortuitum subspecies acetamidolyticum, a new subspecies of Mycobacterium fortuitum. Microbiol. Immunol. 1986, 30, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Staneck, J.L.; Frame, P.T.; Altemeier, W.A.; Miller, E.H. Infection of bone by Mycobacterium fortuitum masquerading as Nocardia asteroides. Am. J. Clin. Pathol. 1981, 76, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Jaubert, J.; Mougari, F.; Picot, S.; Boukerrou, M.; Barau, G.; Ali Ahmed, S.A.; Raskine, L.; Camuset, G.; Michault, A.; Simac, C.; et al. A case of postoperative breast infection by Mycobacterium fortuitum associated with the hospital water supply. Am. J. Infect. Control 2015, 43, 406–408. [Google Scholar] [CrossRef]
- Matsumoto, T.; Otsuka, K.; Tomii, K. Mycobacterium fortuitum thoracic empyema: A case report and review of the literature. J. Infect. Chemother. 2015, 21, 747–750. [Google Scholar] [CrossRef]
- Rathor, N.; Khillan, V.; Panda, D. Catheter associated mycobacteremia: Opening new fronts in infection control. Indian J. Crit. Care Med. 2015, 19, 350–352. [Google Scholar]
- Peterson, K.J. Mycobacterium fortuitum as a cause of bovine mastitis: Tuberculin sensitivity following experimental infections. J. Am. Vet. Med. Assoc. 1965, 147, 1600–1607. [Google Scholar]
- Koehne, G.; Maddux, R.; Britt, J. Rapidly growing mycobacteria associated with bovine mastitis. Am. J. Vet. Res. 1981, 42, 1238–1239. [Google Scholar]
- Machackova, M.; Matlova, L.; Lamka, J.; Smolik, J.; Melicharek, I.; Hanzlikova, M.; Docekal, J.; Cvetnic, Z.; Nagy, G.; Lipiec, M.; et al. Wild boar (Sus scrofa) as a possible vector of mycobacterial infections: Review of literature and critical analysis of data from Central Europe between 1983 to 2001. Vet. Med.-Czech. 2003, 48, 51–65. [Google Scholar] [CrossRef]
- Couto, S.S.; Artacho, C.A. Mycobacterium fortuitum pneumonia in a cat and the role of lipid in the pathogenesis of atypical mycobacterial infections. Vet. Pathol. 2007, 44, 543–546. [Google Scholar] [CrossRef]
- Leissinger, M.K.; Garber, J.B.; Fowlkes, N.; Grooters, A.M.; Royal, A.B.; Gaunt, S.D. Mycobacterium fortuitum lipoid pneumonia in a dog. Vet. Pathol. 2015, 52, 356–359. [Google Scholar] [CrossRef]
- Delghandi, M.R.; El-Matbouli, M.; Menanteau-Ledouble, S. Mycobacteriosis and infections with non-tuberculous mycobacteria in aquatic organisms: A review. Microorganisms 2020, 8, 1368. [Google Scholar] [CrossRef]
- Pavlik, I.; Falkinham, J.; Kazda, J. Environments providing favourable conditions for the multiplication and transmission of mycobacteria. Chapter 5; In The Ecology of Mycobacteria: Impact on Animal’s and Human’s Health, 1st ed.; Kazda, J., Pavlik, I., Falkinham, J., Hruska, K., Eds.; Springer: Dordrecht, The Netherlands; Heidelberg, Germany; London, UK; New York, NY, USA, 2009; pp. 89–198. [Google Scholar]
- Schinsky, M.F.; Morey, R.E.; Steigerwalt, A.G.; Douglas, M.P.; Wilson, R.W.; Floyd, M.M.; Butler, W.R.; Daneshvar, M.I.; Brown-Elliott, B.A.; Wallace, R.J., Jr.; et al. Taxonomic variation in the Mycobacterium fortuitum third biovariant complex: Description of Mycobacterium boenickei sp. nov., Mycobacterium houstonense sp. nov., Mycobacterium neworleansense sp. nov. and Mycobacterium brisbanense sp. nov. and recognition of Mycobacterium porcinum from human clinical isolates. Int. J. Syst. Evol. Microbiol. 2004, 54, 1653–1667. [Google Scholar] [PubMed]
- Kim, S.Y.; Moon, S.M.; Jhun, B.W.; Kwon, O.J.; Huh, H.J.; Lee, N.Y.; Lee, S.H.; Shin, S.J.; Kasperbauer, S.H.; Huitt, G.A.; et al. Species distribution and macrolide susceptibility of Mycobacterium fortuitum complex clinical isolates. Antimicrob. Agents Chemother. 2019, 63, e02331-18. [Google Scholar] [CrossRef] [PubMed]
- Lazo-Vasquez, A.F.; Gonzales-Zamora, J.A. Mycobacterium peregrinum pacemaker pocket infection: A case report and review of the literature. Case Rep. Infect. Dis. 2020, 2020, 8831026. [Google Scholar] [CrossRef]
- Gcebe, N.; Michel, A.L.; Hlokwe, T.M. Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa. BMC Microbiol. 2018, 18, 32. [Google Scholar] [CrossRef]
- Kusar, D.; Zajc, U.; Jencic, V.; Ocepek, M.; Higgins, J.; Zolnir-Dovc, M.; Pate, M. Mycobacteria in aquarium fish: Results of a 3-year survey indicate caution required in handling pet-shop fish. J. Fish Dis. 2017, 40, 773–784. [Google Scholar] [CrossRef]
- Davarpanah, M.; Azadi, D.; Shojaei, H. Prevalence and molecular characterization of non-tuberculous mycobacteria in hospital soil and dust of a developing country, Iran. Microbiology (Reading) 2019, 165, 1306–1314. [Google Scholar] [CrossRef] [PubMed]
- Kazda, J.; Pavlik, I.; Falkinham, J.; Hruska, K. The Ecology of Mycobacteria: Impact on Animal’s and Human’s Health, 1st ed.; Springer: Dordrecht, The Netherlands; Heidelberg, Germany; London, UK; New York, NY, USA, 2009; 520p. [Google Scholar]
- Matlova, L.; Kaevska, M.; Moravkova, M.; Beran, V.; Shitaye, J.E.; Pavlik, I. Mycobacteria in peat used as a supplement for pigs: Failure of different decontamination methods to eliminate the risk. Vet. Med.-Czech. 2012, 57, 212–217. [Google Scholar] [CrossRef]
- Skoric, M.; Mrlik, V.; Svobodova, J.; Beran, V.; Slany, M.; Fictum, P.; Pokorny, J.; Pavlik, I. Infection in a Komodo dragon (Varanus komodoensis) caused by Mycobacterium intracellulare: A case report. Vet. Med.-Czech. 2012, 57, 163–168. [Google Scholar] [CrossRef]
- Moravkova, M.; Lamka, J.; Kriz, P.; Pavlik, I. The presence of Mycobacterium avium subsp. avium in common pheasants (Phasianus colchicus) living in captivity and in other birds, vertebrates, non-vertebrates and the environment. Vet. Med.-Czech. 2011, 56, 333–343. [Google Scholar]
- Slany, M.; Makovcova, J.; Jezek, P.; Bodnarova, M.; Pavlik, I. Relative prevalence of Mycobacterium marinum in fish collected from aquaria and natural freshwaters in central Europe. J. Fish Dis. 2014, 37, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Slany, M.; Jezek, P.; Fiserova, V.; Bodnarova, M.; Stork, J.; Havelkova, M.; Kalat, F.; Pavlik, I. Mycobacterium marinum infections in humans and tracing of its possible environmental sources. Can. J. Microbiol. 2012, 58, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Modra, H.; Bartos, M.; Hribova, P.; Ulmann, V.; Hubelova, D.; Konecny, O.; Gersl, M.; Kudelka, J.; Voros, D.; Pavlik, I. Detection of mycobacteria in the environment of the Moravian Karst (Bull Rock Cave and the relevant water catchment area): The impact of water sediment, earthworm castings and bat guano. Vet. Med.-Czech. 2017, 62, 153–168. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).